My son enjoys visiting my lab on weekends because the university internet connection is much faster than our home modem connection. He also likes to read DNA sequences to find that single base change that often spells the cause of human genetic disease. And, when he finds a change, he asks me how it occurred. We discuss DNA damage, repair, and replication, but I can never exactly answer that “how” question for him. It takes me back to the 1970s! I vividly remember the end of a backpacking trip in the Great Smoky Mountains National Park, enjoying our first non-freeze-dried meal in 10 days and reading the newspaper article about the discovery of a mouse germ cell “supermutagen.” It was N-ethyl-N-nitrosourea (ENU) inducing specific-locus mutations in mouse spermatogonia cells. Russell et al. (1) concluded that “ENU can serve as a model compound in exploring the effect of such factors as dose-response, dose fractionation, sex and cell stage on the mutagenic action of a chemical.” Subsequent studies showed that ENU is also a powerful rodent carcinogen, inducing mutations in oncogenes (2, 3). But, the question still hangs in the air: Why is ENU such a potent mutagen? It damages DNA, but what happens to yield that gene mutation? Twenty years of research have brought us to the brink of understanding the molecular “how” for ENU and many other mutagens. And now we have a new aid for this quest. In a paper in this issue of PNAS, Bielas and Heddle (4) use ENU as the mutagenic agent in an elegant study of in vitro mutation induction in transgenic mouse cells that carry the bacterial lacI gene. What I find so novel is that the method allows them to determine when the DNA damage is converted or fixed into a heritable mutation, and how much damage is repaired.
The method developed by Bielas and Heddle (4) is based on the phenotype of mutations in the lacI gene. Genomic DNA is isolated from the mouse cells and the lacI gene “packaged” into lambda bacteriophage for phenotypic analysis in Escherichia coli. When the lacI gene is mutated, the phage forms blue plaques. However, as shown in their figure 1, if the mutation was fixed while the transgene was in the mouse cell (cellular mutation), the plaque is homogeneous (>90%) mutant. If the DNA damage is converted into a mutation during the subsequent phage replication in the bacteria (bacterial mutation), the plaque is mosaic, with a 50:50 distribution of mutant and wild-type phage. On its own, this observation provides a method to follow fixation of “potentially mutagenic adducts” on the DNA strand into double-stranded mutations in the mouse cells. But, it gets better. Using this approach with quiescent cells arrested for DNA synthesis and cell division allows Bielas and Heddle to study the role of DNA replication in mutation fixation, as well as the kinetics of adduct repair. The frequency of bacterial mutations defines the mutagenic adduct level, and the frequency of cellular mutations defines the extent of mutation fixation by the mouse cells. This method allows measurement of both repair and fixation in the absence of DNA replication. DNA replication effects then are studied by inducing the quiescent cells to proliferate. The results shown in their figures 5–7 clearly demonstrate that, although proliferating cells quickly convert bacterial mutations to cellular mutations, quiescent cells do not, i.e., cell proliferation (DNA replication) is necessary for mutation fixation. Remarkably, the frequency of bacterial mutations (which defines the level of potentially mutagenic adducts) remains stable over time in the quiescent cells and these adducts do not appear to be repaired. In addition, after inducing the quiescent cells to proliferate, only a fraction of these adducts are converted to cellular mutations. Cell proliferation results in both the repair of a portion of the potentially mutagenic adducts and the fixation into mutations of the remainder of the adducts. These could, of course, be different adducts.
The importance of this technique goes beyond the present results with ENU. It offers a new approach to understanding the mutagenic mechanism of the many chemicals that are ubiquitous in our environment. We know a great deal about the DNA damage caused by alkylating agents such as ENU and related chemicals. Their reaction with DNA results in methyl, ethyl, or larger molecular weight adducts on the DNA. This DNA alkylation can cause both cancer and inherited mutations. DNA repair systems have evolved to recognize and “repair” these damages. ENU reaction with DNA yields many adducts, the major ones being N7-ethylguanine (N7-G), O6-ethyl G (O6-G), O2-ethyl thymine (O2-T), N3-ethyl adenine (N3-A), O2-ethyl cytosine (O2-C), and O4-ethyl T (O4-T). Other alkylating agents are similar in their adduct profile, differing primarily in relative amounts of the various adducts (5, 6). Repair pathways include direct removal of the alkyl adducts from O6-G, and perhaps O4-T, through alkyl DNA alkyltransferase (AGT; designated MGMT in humans). Base excision repair (BER) and nucleotide excision repair (NER) also recognize adducts in DNA, probably because of the distortion of the base-pairing stability. As Bielas and Heddle (4) point out, it has become almost dogma that DNA repair either restores the adducted base to its normal structure (e.g., through the action of AGT) or removes the adducted base and inserts a correct base (e.g., through the action of BER or NER). A connection between this DNA repair and RNA transcription also has been found and is termed transcription-coupled repair (7). It is assumed that mutations occur only if DNA replication occurs before the damage has been repaired, i.e., restored to normal. Bielas and Heddle's results with quiescent cells induced to proliferate (their figure 7) can be viewed as a measure of the effect of persistent, unrepaired adducts. (The apparent repair of a fraction of these adducts after proliferation is induced must be related to DNA replication because the lacI gene is not transcribed in the mouse cells.) Now the identity of both the persistent adducts and the specific mutations can be determined.
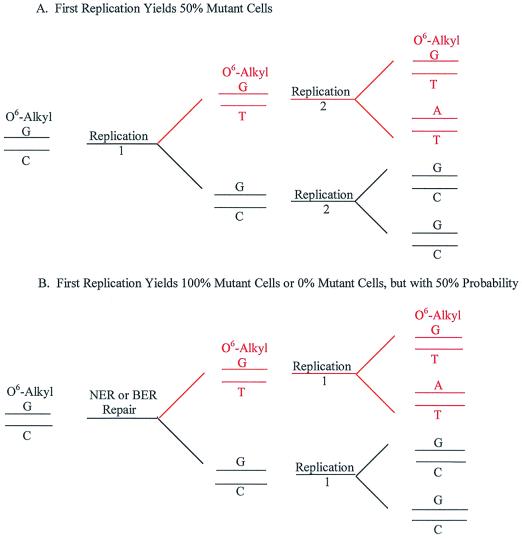
In AGT-deficient (AGT−) cells, mutation induction correlates primarily with O6-alkyl G and these mutations are G:C→A:T transition mutations resulting from the mispairing of O6-alkyl G with T during DNA replication (5, 6). Because most early studies of mutation induction in vitro used AGT− cells and because O6-alkyl G is the predominant promutagenic adduct, the G:C→A:T transition was thought to be the primary effect of alkylating agents. More recent studies with in vivo mutagenicity assays in rodents have found that the predominant mutations are A:T→T:A transversions. Representative studies are summarized in Table 1. These in vivo studies measure mutations in actively proliferating T lymphocytes. In vivo, 44% of the ENU induced mutations are A:T→T:A transversions, thought to be caused by the miscoding properties of O2-alkyl T. In toto, 81% of the mutations occur at A:T base pairs, and only 19% at G:C base pairs (Table 1). In in vitro studies with AGT+ cells, a similar spectrum of 40% A:T→T:A transversions and 69% of mutations at A:T base pairs is found (Table 1). However, in in vitro studies with AGT− cells (which show higher mutation induction per unit dose than is found with AGT+ cells), the spectrum shifts to 46% G:C→A:T transitions, because of the absence of repair of O6-alkyl G adducts. In addition, in the absence of AGT, there is evidence that O6-alkyl G adducts induce mutations in the absence of DNA replication. In studies with division-arrested Chinese hamster ovary (CHO) AGT− cells, HPRT mutations induced by the ethylating agent ethyl methane sulfonate were fixed and expressed in the absence of DNA replication (15). (Mutations induced in proliferating CHO cells by alkylating agents correlate with O6-G adducts and are primarily G:C→A:T transition mutations.) A novel in situ study of alkylation induced mutations at the glucose-6-phosphate dehydrogenase (G6PD) gene in CHO cells that allowed analysis of the mutant phenotype in single cell colonies showed both mosaic colonies, i.e., 50% mutant and 50% wild type, and pure colonies, i.e., 100% mutant (16, 17). (The mosaic colonies are parallels to Bielas and Heddle's bacterial mutations, and the pure colonies parallel their cellular mutations.) Both these CHO cell results suggest that there is a “mutational mechanism which fixes the mutation in both strands of the DNA before the next replication cycle” (17). In hindsight, it would have been informative to study these G6PD mutations with quiescent cells. Fig. 1 presents a mechanism for this mutation fixation in the absence of DNA replication. Replication of the O6-alkyl G:C base pair yields a wild-type G:C base pair and a mutated O6-alkyl G:T base pair (Fig. 1A, mutation in red). Subsequent replication results in a “fixed,” mutant A:T base pair (G:C→A:T transition) and a mosaic 50:50 mutant colony. In the absence of AGT repair, repair of the O6-alkyl G:C “mismatch” either removes the O6-alkyl G and “repairs” with a G, or removes the C and “misrepairs” with a T (Fig. 1B, mutation in red). Subsequent replication yields either a pure wild-type colony after repair or a pure mutant colony after misrepair. Both processes yield the same frequency of mutations, but through different mechanisms. This mechanism of mutation induction places the importance of AGT activity in a new light. In its absence (AGT−), mutations can result in the absence of DNA replication. Perhaps in quiescent cells in vivo, this misrepair is a general mechanism of mutation induction resulting from O6-alkyl G and other adducts, which create apparent base pair mismatches, which are not repaired by specific repair enzymes such as AGT. Perhaps transcription-coupled repair has more precise discrimination between the alkylated base and the normal base, and preferentially removes the adducted base.
Table 1.
Single base substitution mutation spectrum resulting from ENU treatment
| Species | Gene | Repair | Single base
substitution, %a
|
Ref. | |||||
|---|---|---|---|---|---|---|---|---|---|
| G∶C→A∶T TSb (O6–G)c | G∶C→T∶A TVb | G∶C→C∶G TVb | A∶T→G∶C TSb (O4–T)c | A∶T→T∶A TVb (O2–T)c | A∶T→C∶G TVb | ||||
| In Vivo | |||||||||
| Mouse | lacI | AGT+ | 17 | 11 | 2 | 29 | 29 | 11 | 8 |
| Mouse | HPRT | AGT+ | 4 | 2 | 0 | 28 | 55 | 12 | 9 |
| Rat | HPRT | AGT+ | 10 | 4 | 0 | 25 | 48 | 13 | 10 |
| Ratd | HPRT | AGT+ | 16 | 9 | 0 | 13 | 44 | 19 | 11 |
| Mean | 12 | 7 | 0 | 24 | 44 | 13 | |||
| Mean | 19 (G∶C) | 81 (A∶T) | |||||||
| In Vitro | |||||||||
| Ratd | HPRT | AGT+ | 27 | 5 | 0 | 14 | 45 | 9 | 12 |
| Humane | HPRT | AGT+ | 21 | 0 | 6 | 12 | 52 | 9 | 13 |
| Humanf | HPRT | AGT+ | 19 | 15 | 0 | 31 | 23 | 12 | 14 |
| Mean | 22 | 7 | 2 | 19 | 40 | 10 | |||
| Mean | 31 (G∶C) | 69 (A∶T) | |||||||
| Humane | HPRT | AGT− | 42 | 3 | 0 | 11 | 34 | 11 | 13 |
| Humanf | HPRT | AGT− | 50 | 1 | 7 | 7 | 21 | 11 | 14 |
| Mean | 46 | 2 | 4 | 9 | 28 | 11 | |||
| Mean | 52 (G∶C) | 48 (A∶T) | |||||||
G∶C→A∶T mutations occurring in CpG dinucleotides have not been included because they arise through a different mechanism involving the deamination of 5-methyl C.
TS = transition mutation and TV = transversion mutation.
In vitro treatment with ENU yields the following levels of specific DNA adducts (relative to N7-G = 1.00): O6–G = 0.93, O2–T = 0.33, N3–A = 0.23, O2–C = 0.14, and O4–T = 0.06. The O6–G adducts are removed through AGT. The O2–T adducts appear to persist.
Same cells in vivo and in vitro.
Same cells AGT+ and AGT−.
Same cells AGT+ and AGT−.
Figure 1.
Fate of O6-alkyl G adducts in AGT-deficient cells (G:C→A:T transition mutations). In AGT-proficient cells, the O6-alkyl adduct is removed from G through direct transfer to an acceptor protein. In AGT-deficient cells, there are two possible fates for this and similar adducts. If the cellular DNA is replicated, the O6-alkyl G is misreplicated as an A and a T is incorporated in the new DNA strand. Subsequent replication “fixes” the G:C→A:T mutation (A, mutation in red). This would yield a colony of 50% mutant cells. In the absence of DNA replication, nucleotide excision repair (NER) or base excision repair (BER) processes might recognize the “mismatched” base pair. Correct repair would remove the O6-alkyl G and insert the correct G. Misrepair would remove the C and insert a T opposite O6-alkyl G. The now fixed G:C→A:T mutation would be propagated during subsequent DNA replication (B, mutation in red). The total mutant yield through misrepair would be the same, assuming a 50% probability of either repair or misrepair. However, the mutant colony would be “pure,” i.e., 100% mutant cells.
The method developed by Bielas and Heddle provides a way to answer many of these questions. The use of AGT− transgenic mouse cells in the proliferating versus quiescent cell assay will allow quantitation of the direct fixation of O6-alkyl G:C base pairs to A:T transition mutations in the quiescent cells resulting in the cellular mutations not found in the AGT+ cells. This direct fixation also may apply to O4-alkyl T, thought to be the adduct responsible for the A:T→G:C transitions.
The results showing persistent adducts in the quiescent cells that yield bacterial mutations but not cellular mutations plead for a spectrum of these mutations. Is the persistent adduct O2-alkyl T and will the mutations then be A:T→T:A transversions? Will these mutations show the strand bias toward the nontranscribed strand found for these transversions at A:T base pairs induced both in vivo and in vitro in other systems including lacI transgenic mice by ENU (8, 9, 11, 14, 18)? Perhaps not, if this bias is caused by transcription-coupled repair, because the lacI gene is not transcribed. Perhaps so, if this bias is caused by a difference in fidelity of DNA replication between the two strands of the gene (19). The Bielas and Heddle technique offers the opportunity to address these issues with a unique approach.
Lastly, the differentiation between bacterial mutations and cellular mutations could be applied to lacI mutations induced in vivo in lacI transgenic mice. To my knowledge, this has not been done in mutation induction studies, although this approach was used to demonstrate that most spontaneous lacI mutations isolated from mouse small intestine arose in vivo, i.e., were cellular mutations (20). Analyzing induced mutations would differentiate persistent, potentially mutagenic adducts (as bacterial mutations) from in vivo fixed mutations (as cellular mutations).
To come full circle to my son's question, the “how” of genetic change may have several answers. Mutations induced in nondividing cells through DNA repair errors may occur in vivo and be different in spectrum from those induced in dividing cells. Returning to mouse germ cell mutations and ENU, Russell and Russell (21) found that ENU is uniquely mutagenic in arrested, as well as maturing oocytes. This mutagenicity may be caused by persistent, unrepaired adducts that are subsequently converted to mutations through DNA replication or to mutation fixation through misrepair. Perhaps the latter is a first step in the development of some cancers. The paper by Bielas and Heddle presents a new approach to address these important questions.
Acknowledgments
I thank Inge Gobel for patiently processing my handwriting into printed text and Drs. Janice Nicklas, Thomas Skopek, and Vernon Walker for helpful suggestions on the content of this commentary. However, any mis-statements or misleading conclusions are entirely my responsibility. My research is supported by the Lesch-Nyhan Syndrome Children's Research Foundation and by National Institutes of Health Grants RO1 CA68288 and RO1 HD33648.
Footnotes
See companion article on page 11391.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.210383397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.210383397
References
- 1.Russell W L, Kelly E M, Hunsicker P R, Bangham J W, Maddux S C, Phipps E L. Proc Natl Acad Sci USA. 1979;76:5818–5819. doi: 10.1073/pnas.76.11.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peratoni A O, Rice J M, Reed C D, Watatani M, Wenk M L. Proc Natl Acad Sci USA. 1987;84:6317–6321. doi: 10.1073/pnas.84.17.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.You M, Wang Y, Lineen A M, Gunning W T, Stoner G D, Anderson M W. Carcinogenesis. 1992;13:1583–1586. doi: 10.1093/carcin/13.9.1583. [DOI] [PubMed] [Google Scholar]
- 4.Bielas J H, Heddle J A. Proc Natl Acad Sci USA. 2000;97:11391–11396. doi: 10.1073/pnas.190330997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heflich R H, Beranek D T, Kodell R L, Morris S M. Mutat Res. 1982;106:147–161. doi: 10.1016/0027-5107(82)90198-1. [DOI] [PubMed] [Google Scholar]
- 6.Beranek D T, Heflich R H, Kodell R L, Morris S M, Casciano D A. Mutat Res. 1983;110:171–180. doi: 10.1016/0027-5107(83)90026-x. [DOI] [PubMed] [Google Scholar]
- 7.Hanawalt P C. Environ Health Perspect. 1996;104, Suppl. 3:547–551. doi: 10.1289/ehp.96104s3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker V E, Gorelick N J, Andrews J L, Craft T R, deBoer J G, Glickman B W, Skopek T R. Cancer Res. 1996;56:4654–4661. [PubMed] [Google Scholar]
- 9.Skopek T R, Walker V E, Cochrane J E, Craft T R, Cariello N F. Proc Natl Acad Sci USA. 1992;89:7866–7870. doi: 10.1073/pnas.89.17.7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mittelstaedt R A, Smith B A, Heflich R H. Environ Mol Mutagen. 1995;26:261–269. doi: 10.1002/em.2850260402. [DOI] [PubMed] [Google Scholar]
- 11.Jansen J G, Mohn G R, Vrieling H, van Teijlingen C M M, Lohman P H M, van Zeeland A A. Cancer Res. 1994;54:2478–2485. [PubMed] [Google Scholar]
- 12.Jansen J G, van Teijlingen C M, Mohn G R, van Zeeland A A, Vrieling H. Mutagenesis. 1994;9:417–421. doi: 10.1093/mutage/9.5.417. [DOI] [PubMed] [Google Scholar]
- 13.Yang J L, Lee P C, Lin S R, Lin J G. Carcinogenesis. 1994;15:939–945. doi: 10.1093/carcin/15.5.939. [DOI] [PubMed] [Google Scholar]
- 14.Bronstein S M, Cochrane J E, Craft T R, Swenberg J A, Skopek T R. Cancer Res. 1991;51:5188–5197. [PubMed] [Google Scholar]
- 15.O'Neill J P. Mutat Res. 1982;106:113–122. doi: 10.1016/0027-5107(82)90195-6. [DOI] [PubMed] [Google Scholar]
- 16.Stamato T D, MacKenzie L, Pagani J M, Weinstein R. Somatic Cell Genet. 1982;8:643–651. doi: 10.1007/BF01542857. [DOI] [PubMed] [Google Scholar]
- 17.Aronson J F, Stamato T D. Mutat Res. 1987;177:277–281. doi: 10.1016/0027-5107(87)90011-x. [DOI] [PubMed] [Google Scholar]
- 18.Harbach P R, Filipunas A S, Wang Y, Aaron C S. Environ Mol Mutagen. 1992;20:96–105. doi: 10.1002/em.2850200205. [DOI] [PubMed] [Google Scholar]
- 19.Vrieling H, Zhang L-H, van Zeeland A A, Zdzienicka M Z. Mutat Res. 1992;274:147–155. doi: 10.1016/0921-8777(92)90061-7. [DOI] [PubMed] [Google Scholar]
- 20.Paashuis-Lew Y, Zhang X B, Heddle J A. Mutat Res. 1997;373:277–284. doi: 10.1016/s0027-5107(96)00210-2. [DOI] [PubMed] [Google Scholar]
- 21.Russell L B, Russell W L. Mutat Res. 1992;296:107–127. doi: 10.1016/0165-1110(92)90035-8. [DOI] [PubMed] [Google Scholar]