Abstract
Purpose
The development of second primary tumors (SPT) or recurrence alters prognosis for curatively-treated head and neck squamous cell carcinoma (HNSCC) patients. 13-cis-retnoic acid (13-cRA) has been tested as a chemoprevention agent in clinical trials with mixed results. Therefore, we investigated if genetic variants in the PI3K/PTEN/AKT/MTOR pathway could serve as biomarkers to identify which patients are at high risk of an SPT/recurrence while also predicting response to 13-cRA chemoprevention.
Experimental Design
A total of 137 pathway SNPs were genotyped in 440 patients from the Retinoid Head and Neck Second Primary Trial and assessed for SPT/recurrence risk and response to 13-cRA. Risk models were created based on epidemiology, clinical, and genetic data.
Results
Twenty-two genetic loci were associated with increased SPT/recurrence risk with six also being associated with a significant benefit following chemoprevention. Combined analysis of these high-risk/high-benefit loci identified a significant (P = 1.54×10−4) dose-response relationship for SPT/recurrence risk, with patients carrying 4–5 high-risk genotypes having a 3.76-fold (95%CI:1.87–7.57) increase in risk in the placebo group (n=215). Patients carrying 4–5 high-risk loci showed the most benefit from 13-cRA chemoprevention with a 73% reduction in SPT/recurrence (95%CI:0.13–0.58) compared to those with the same number of high-risk genotypes who were randomized to receive placebo. Incorporation of these loci into a risk model significantly improved the discriminatory ability over models with epidemiology, clinical, and previously identified genetic variables.
Conclusions
These results demonstrate that loci within this important pathway could identify individuals with a high-risk/high-benefit profile and are a step towards personalized chemoprevention for HNSCC patients.
INTRODUCTION
Head and neck squamous cell carcinoma (HNSCC) is estimated to be diagnosed in over 48,000 individuals in the United States alone in 2010 (1). World-wide, over 600,000 diagnoses are made each year (2, 3). Treatment of early stage (I and II) HNSCC with surgery with or without radiotherapy is successful in 60–80% of these patients leading to a favorable long-term prognosis (4). The major challenge in these patients is the development of second primary tumors (SPT) and recurrences of the primary tumor, which develop in up to 20% of the patients within 5 years of curative treatment (5, 6). Because of this high risk in HNSCC patients, there has been a focus on retinoid-based chemoprevention to reduce or eliminate these events.
In a clinical trial of 13-cis-retinoic acid (13-cRA, also known as isotretinoin) for the prevention of SPT in 103 HNSCC patients who were disease-free at time of enrollment, a significant reduction in SPTs were observed following treatment with 13-cRA (7, 8). This finding led to a larger phase III clinical trial utilizing low dose 13-cRA in 1,190 early stage HNSCC patients. The results of this study showed no significant difference between incidence of SPT/recurrence in the placebo and chemoprevention arms (9). In follow-up analysis, our group identified that common genotypes within RXRA (retinoid X receptor), JAK2 (Janus kinase 2), and CDC25C (cell division cycle 25 homolog C phosphatase) that could identify HNSCC patients at high risk of SPT/recurrence while also predicting a favorable response to 13-cRA (10), indicating that biomarkers could be used to select patients who would be in the most need of and respond best to 13-cRA chemoprevention.
Although the most significant predictors identified were RXRA, JAK2, and CDC25C, several genetic loci within TSC1 (tuberous sclerosis 1) also showed a similar pattern. TSC1 plays a major role in regulating the PI3K/PTEN/AKT/MTOR pathway through the activation/suppression of MTOR activity (11, 12) and loss of TSC1 function results in uncontrolled cell growth and proliferation (13). This pathway has also been shown to be the key modulator of cell death and survival and is often dysregulated in cancer, including in cancers of the head and neck (14, 15), resulting in tumorigenesis, cell invasion, and drug resistance (16). 13-cRA and other retinoids exert their effects through the various RAR and RXR retinoid receptors. Interestingly, RAR receptors have been shown to be downregulated in head and neck cancers(17). One mechanism for this repression is through AKT-driven phosphorylation(18,19). Futhermore, retinol and retinoids have been shown to directly activate the PI3K/PTEN/AKT/MTOR pathway in several different cell types(20, 21).
Because of the importance of this pathway in cancer development and progression, we hypothesized that a pathway-based approach to analyze the effect of genetic variation within this pathway may be highly predictive of SPT/recurrence risk in HNSCC patients and also response to 13-cRA chemoprevention. A total of 137 genetic loci from 20 genes functioning in the PI3K/PTEN/AKT/MTOR pathway were included in this analysis of SPT/recurrence risk and 13-cRA response in 440 patients participating in the Retinoid Head and Neck Second Primary Trial.
RESULTS
Patient population
The patient population for this study has been well described previously (5, 10, 22). Briefly, of the 440 HNSCC patients included in the analysis, 215 were randomized to the placebo arm and 225 were selected to receive 13-cRA chemoprevention. There were no significant differences between patient characteristics of the treatment arms for age, gender, ethnicity, smoking status, years smoked, number of cigarettes/day, pack-year, alcohol consumption, tumor site, stage, and previous treatment regimens.
Genetic variation identifying high risk HNSCC patients
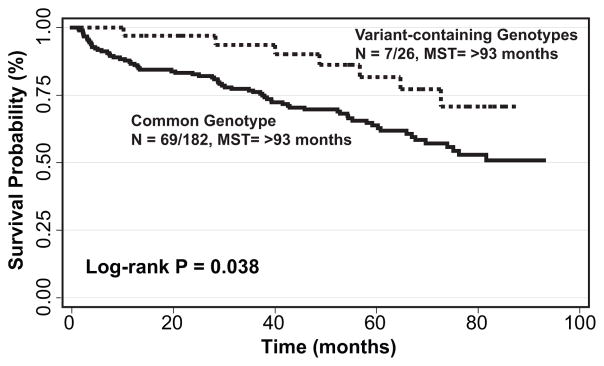
To identify genetic variation that could serve as markers to identify patients at high risk for developing SPT or recurrence, we analyzed the effect of the 1387 pathway SNPs in the 215 patients randomized to the placebo arm. A total of 22 SNPs were significantly associated with risk of development of SPT/recurrence. Of particular interest were the fifteen genetic loci that were able to identify the majority of the population – those with the common genotype – that had an increased risk of SPT/recurrence ranging from 3.57- to 1.56-fold (Table 1). Interestingly, of these, 10 were located within TSC1. The 182 (84.7% of the population) HNSCC patients with the common genotype for the most significant genetic locus, TSC1:rs7040593, had a greater than 3-fold increased risk of developing a SPT or recurrence (HR:3.03, 95%CI:1.37–6.67). Furthermore, these patients had a significant SPT/recurrence-free survival disadvantage compared to those with at least one variant allele (P = 0.038, Figure 1).
Table 1.
Effect of PI3K/PTEN/AKT/mTOR Genetic Variation
| SNP | Gene | SNP Location | in Placebo Group | Effect of Genotype in Placebo Group | Effect of 13-cRA by Genotype | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Common Genotype | Variant-containing Genotype | ||||||||||||
| MAF (%) | Genotype Counts (cc/cv/vv) | Risk Genotype | Model | * HR(95% CI) | P-value | MST (risk genotype) | HR(95% CI) | P- value | HR(95% CI) | P-value | |||
| rs9521509 | IRS2 | intron | 0.44 | 76/90/49 | common | dominant | 1.82(1.14–2.86) | 0.012 | 74.0 | 0.64(0.37–1.12) | 0.12 | ||
| rs9515119 | IRS2 | intron | 0.31 | 105/86/24 | common | dominant | 1.67(1.04–2.70) | 0.035 | 75.1 | 0.70(0.45–1.10) | 0.12 | ||
| rs7999797 | IRS2 | intron | 0.41 | 79/94/42 | common | dominant | 1.59(1.00–2.56) | 0.050 | 81.6 | 0.77(0.46–1.28) | 0.31 | ||
| rs10274 | RPS6KB2 | 3-UTR | 0.37 | 85/98/30 | common | dominant | 1.75(1.10–2.78) | 0.020 | 64.7 | 0.62(0.36–1.07) | 0.088 | ||
| rs7158047 | RPS6KL1 | 5-UTR | 0.40 | 77/102/36 | common | recessive | 2.17(1.01–4.55) | 0.048 | >93 | 0.91(0.52–1.59) | 0.73 | ||
| rs7040593 | TSC1 | 5-FR | 0.08 | 182/33/0 | common | dominant | 3.03(1.37–6.67) | 0.0066 | >93 | 0.73(0.51–1.04) | 0.081 | ||
| rs3827665 | TSC1 | intron | 0.06 | 188/27/0 | common | dominant | 3.57(1.41–9.09) | 0.0075 | >93 | 0.73(0.51–1.05) | 0.088 | ||
| rs739442 | TSC1 | 3-UTR | 0.44 | 70/100/45 | common | recessive | 1.56(1.12–2.17) | 0.0089 | 74.0 | 0.49(0.28–0.87) | 0.014 | ||
| rs2519760 | TSC1 | 3-FR | 0.41 | 71/113/31 | common | recessive | 1.61(1.12–2.33) | 0.0095 | 75.1 | 0.69(0.40–1.19) | 0.18 | ||
| rs2809243 | TSC1 | 3-UTR | 0.37 | 86/98/29 | common | recessive | 1.64(1.12–2.38) | 0.010 | 75.1 | 0.70(0.43–1.15) | 0.16 | ||
| rs4962225 | TSC1 | 5-FR | 0.20 | 137/71/7 | common | dominant | 1.92(1.15–3.23) | 0.013 | 75.1 | 0.57(0.37–0.88) | 0.011 | ||
| rs7035940 | TSC1 | 5-FR | 0.20 | 137/71/7 | common | dominant | 1.92(1.15–3.23) | 0.013 | 75.1 | 0.57(0.37–0.88) | 0.011 | ||
| rs10491534 | TSC1 | 3-UTR | 0.10 | 172/41/2 | common | dominant | 2.00(1.08–3.70) | 0.028 | >93 | 0.71(0.49–1.03) | 0.074 | ||
| rs2073869 | TSC1 | 3-FR | 0.13 | 159/54/2 | common | dominant | 1.89(1.08–3.33) | 0.028 | >93 | 0.70(0.47–1.03) | 0.072 | ||
| rs7874234 | TSC1 | intron | 0.20 | 135/73/7 | common | dominant | 1.72(1.03–2.86) | 0.036 | 76.3 | 0.59(0.38–0.92) | 0.020 | ||
|
| |||||||||||||
| rs892119 | AKT2 | intron | 0.16 | 151/58/6 | variant | additive | 3.06(1.04–8.99) | 0.042 | 76.3 | 0.78(0.43–1.43) | 0.43 | ||
| rs4972842 | PDK1 | intron | 0.18 | 146/60/9 | variant | additive | 5.16(2.21–12.03) | 0.00015 | >93 | 0.74(0.42–1.33) | 0.31 | ||
| rs4972839 | PDK1 | 5-FR | 0.21 | 132/71/10 | variant | recessive | 1.56(1.08–2.26) | 0.017 | 67.7 | 0.77(0.47–1.28) | 0.31 | ||
| rs4129341 | PIK3CD | 5-FR | 0.18 | 145/61/7 | variant | dominant | 1.68(1.04–2.71) | 0.035 | 81.6 | 0.48(0.26–0.87) | 0.016 | ||
| rs1234221 | PTEN | 5-FR | 0.22 | 130/76/9 | variant | recessive | 1.55(1.05–2.30) | 0.029 | 75.1 | 0.56(0.34–0.94) | 0.029 | ||
| rs869116 | TSC1 | 5-FR | 0.50 | 54/108/53 | variant | recessive | 1.52(1.08–2.13) | 0.016 | 81.6 | 0.76(0.52–1.10) | 0.14 | ||
| rs4367688 | TSC1 | intron | 0.50 | 55/107/53 | variant | recessive | 1.45(1.04–2.02) | 0.030 | 81.6 | 0.71(0.48–1.04) | 0.076 | ||
adjusted for age, gender, ethnicity, smoking status, tumor site, and tumor stage
underlined SNPs are in linkage disequilibrium, bold values are significant at P<0.05
Figure 1.
Effect of TSC1:rs7040593 genotypes on event-free survival in placebo group. N: number of events/total number of individuals; MST: median event-free survival time in months.
Seven additional genetic loci were also significantly associated with SPT/recurrence risk. However for these SNPs, in contrast to the common genotype conferring risk, the variant-containing genotypes were the risk genotypes representing a minority of the population at increased risk of SPT/recurrence (Table 1). The largest increase in risk for this type of genetic loci was rs4972842 located within an intron of the gene encoding pyruvate dehydrogenase kinase 1 (PDK1) and carried by 69 of the 215 (33.7%) HNSCC patients. Patients with variant genotypes had a 5.16-fold increase in risk (95%CI:2.21–12.03). Other significant increases in risk were observed for genetic loci in AKT2, PIK3CD, PTEN, and TSC1.
Effect of 13-cRA chemoprevention on patients at high risk
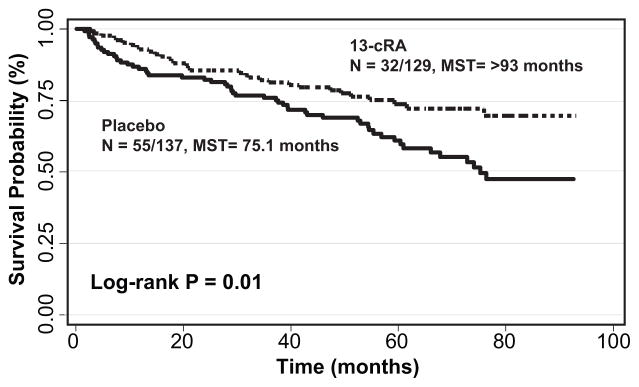
We next determined the effect of 13-cRA treatment compared to placebo for those carrying the high-risk genotypes (Table 1). Of these 22 genetic loci, six were also associated with a significant benefit for patients who received 13-cRA chemoprevention. Two TSC1 loci, rs4962225 and rs7035940, are in high linkage disequilibrium and resulted in a 43% reduction in SPT/recurrence following 13-cRA treatment in those with the common genotype (HR:0.57, 95%CI:0.37–0.88) compared to patients with the common genotype who did not receive chemoprevention. This significant decrease in SPT/recurrence incidence resulted in a greater than 17.9 month increase in event-free survival time from 75.1 months to over 93 months (Figure 2). Similar significant reductions in SPT/recurrence risk were associated with the common genotypes for TSC1:rs739442 and TSC1:rs7874234 (HR:0.49, 95%CI:0.27–0.87 and HR:0.59, 95%CI:0.38–0.92, respectively) following 13-cRA chemoprevention. These findings are similar to the reduction in risk when assessing the effect of the high-risk genotypes in the 13-cRA treatment arm, with the exception of TSC1:rs739442 which was not significantly associated with SPT/recurrence risk (Supplemental Table).
Figure 2.
Effect of 13-cRA treatment in patients with the common genotype for TSC1:rs4962225. N: number of events/total number of individuals; MST: median event-free survival time in months.
Carriers of the variant-containing genotypes for PIK3CD:rs4129341 and PTEN:rs1234221 had a significantly higher risk of developing SPT/recurrence compared to those with the common genotype (Table 1). These same variant genotypes were also associated with significant benefit following 13-cRA intervention. PIK3CD:rs4129341 was associated with a 52% reduction in SPT/recurrence (HR:0.48, 95%CI:0.26–0.87) and PTEN:rs1234221 associated with a 44% reduction (HR:0.56, 95%CI:0.34–0.94) in patients who received 13-cRA compared to those who received placebo.
The findings for these genetic loci with regard to response to 13-cRA were robust and remained significant (P < 0.05) for a majority of the 500 bootstrap resampling iterations for TSC1:rs739442 (80.8%), TSC1:rs4962225/rs7035940 (98.2%), TSC1:rs7874234 (87.0%), PIK3CD:rs4129341(87.0%), and PTEN:rs1234221 (67.4%).
Combined analysis of PI3K/PTEN/AKT/MTOR pathway genetic variation
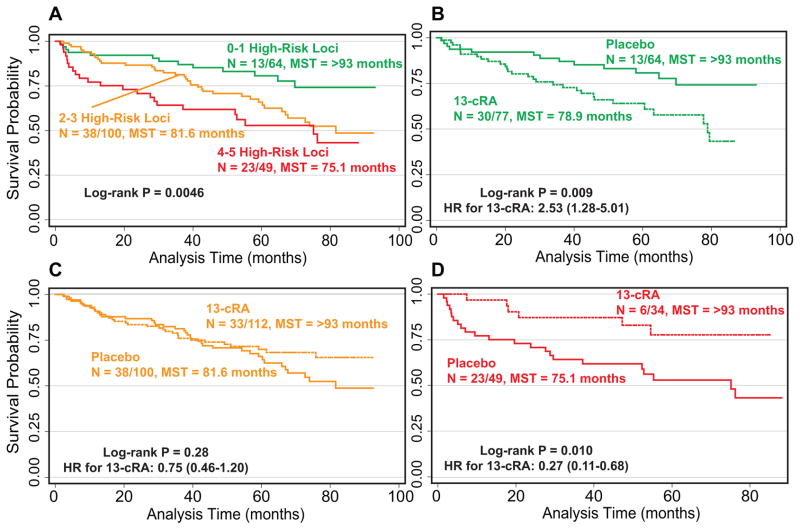
We performed a combined analysis of the six genetic loci (TSC1:rs739442, TSC1:rs4962225, TSC1:rs7035940, TSC1:rs7874234, PIK3CD:rs4129341, and PTEN:rs1234221) that identified HNSCC patients at high-risk of SPT/recurrence while also predicting response to 13-cRA (high-risk/high-benefit loci) to determine if the cumulative effect of these genotypes within the pathway could better identify those patients at high risk of SPT/recurrence (Table 2). Because the two TSC1 SNPs rs4962225 and rs7035940 are in high linkage disequilibrium, we only included one of these loci in the analysis. A significant (P = 1.54×10−4) dose-response relationship was evident for SPT/recurrence risk in the placebo group. Patients with 2 or 3 high-risk genotypes (46.9% of the population) were at a 2.20-fold increased risk of SPT/recurrence (95%CI:1.16–4.15) compared to those with 0 or 1 high-risk genotypes. This increase in SPT/recurrence risk jumpedreached to 3.76-fold in patients carrying 4 or 5 high-risk genotypes (95%CI:1.87–7.57). These patients comprised 23% of the population had a significantly shorter median time to event of 75.1 months as well (P = 0.0046) (Figure 3A)
Table 2.
Effect of number of high-risk genotypes on SPT/recurrence risk in the placebo group
| Number of high-risk genotypes | No SPT/Recurrence, N(%) | SPT/Recurrence, N(%) | * HR(95% CI) | P-value |
|---|---|---|---|---|
| 0–1 | 51(79.7) | 13(20.3) | 1.00(reference) | |
| 2–3 | 62(62.0) | 38(38.0) | 2.20(1.16–4.15) | 0.015 |
| 4–5 | 26(53.1) | 23(46.9) | 3.76(1.87–7.57) | 2.1×10−4 |
| P for trend | 1.54×10−4 |
adjusted for age, gender, ethnicity, smoking status, tumor site, and tumor stage
Figure 3.
Cumulative effect of high-risk/high-benefit genotypes (TSC1:rs739442, TSC1:rs4962225, TSC1:rs7035940, TSC1:rs7874234, PIK3CD:rs4129341, and PTEN:rs1234221) on A) risk of SPT/recurrence in the placebo group, and response to 13-cRA for individuals with B) 0–1, C) 2–3, or D) 4 or 5 high-risk/high-benefit genotypes. N: number of events/total number of individuals; MST: median event-free survival time in months. Curves shown in dashed lines represent patients receiving 13-cRA.
When we assessed the effect of 13-cRA compared to placebo in patients with different number of high-risk loci, patients at lowest risk of developing a SPT/recurrence (0–1 high-risk loci) received no benefit from 13-cRA. In fact, these individuals were at a significant 2.53-fold increased risk (95%CI:1.28–5.01) of SPT/recurrence when receiving 13-cRA compared to the placebo arm patients (Figure 3B). Carriers of 2–3 high-risk loci did not appear to benefit from 13-cRA treatment with no difference in event-free survival durations (P = 0.28) (Figure 3C). This group of patients would carry an equal number of low-risk/low-benefit genetic loci, essentially cancelling out the effect of the high-risk/high-benefit loci. However, 13-cRA had a dramatic benefit for HNSCC patients with the highest number of high-risk genotypes. Individuals with 4 or 5 high-risk genotypes had a highly significant (P = 0.0056) 73% reduction in SPT/recurrence (95%CI:0.11–0.68) when receiving 13-cRA chemoprevention compared to those with the same number of high-risk genotypes who were randomized to receive placebo, resulting in a significant event-free survival difference between the two groups (P = 0.010) (Figure 3D).
Risk Prediction Model for SPT/recurrence Risk
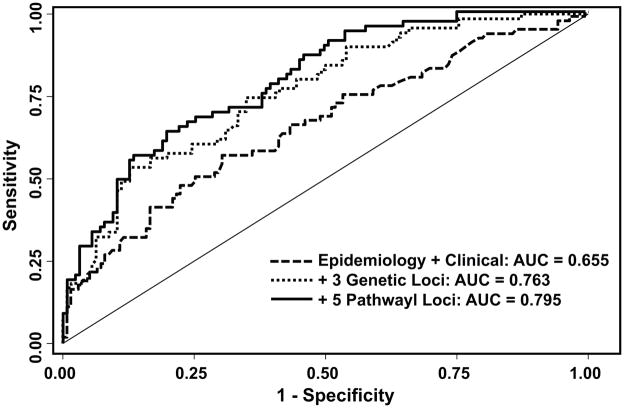
To better assess the ability of the five high-risk/high-benefit genetic loci in identifying HNSCC patients at high risk of SPT/recurrence, we constructed three risk prediction models based on the data from the placebo group. The first model included clinical and epidemiology variables associated with risk and adjusted for in our analysis: age, gender, ethnicity, smoking status, tumor site, and tumor stage (Figure 4). The corresponding AUC was 65.5% and significantly increased to 76.3% (P < 0.001) with the addition of the three previously identified genetic loci (RXRA:rs3118570, JAK2:rs1887427, and CDC25C:rs6596428 (10)). There was an additional significant increase of 3.2% to 79.5% (P = 0.002) with the inclusion of the five high-risk/high-benefit genetic loci. These increases in the AUC with the addition of genetic information remained significant following bootstrap resampling.
Figure 4.
Risk model for SPT/recurrence risk. Receiver operating characteristic curves for three models with significantly different discrimination abilities. AUC: area under the curve.
DISCUSSION
In this study, we demonstrated that genetic variation within the PI3K/PTEN/AKT/MTOR pathway is able to identify HNSCC patients at high risk of SPT/recurrence while also predicting favorable response to 13-cRA chemoprevention. A total of 22 genetic loci were identified as high-risk genotypes with 15 of these significant associations for the common genotype representing a majority of the patient population. A clear beneficial association between receiving 13-cRA chemoprevention compared to placebo was evident for four common genotypes and two variant-containing genotypes. Furthermore, a combinatory effect among these six genetic loci was observed that better stratified the patient population into those with a high-risk/high-benefit genetic background from those with a low-risk/no-benefit profile. A risk model was developed based on these results and showed excellent discriminatory ability. These results suggest that genetic variation within this important cellular signaling pathway could be used as biomarkers to select HNSCC patients to receive 13-cRA chemoprevention.
TSC1 complexes with TSC2 to regulate mTOR activity with inactivation of TSC1/2 resulting in increased mTOR signaling causing unrestrained cell growth and metabolism (13). The highest number of significant associations was for genetic variation with TSC1. Our results suggest that genetic variation within TSC1 plays an important role in modulating risk of SPT/recurrence as well as response to 13-cRA. In particular, the four high-risk/high-benefit genetic loci (rs739442, rs4962225, rs7035940, and rs7874234) were for the common genotype, suggesting that a majority of the patient population would be classified into this group and recommended to receive 13-cRA. These results also suggest that the common genotype for these loci is not sufficient to maintain TSC1 activity during tumor development and recurrence, and thus unable to properly regulate mTOR signaling resulting in increased cell growth and survival signals.
In contrast to common genotypes for the four TSC1 loci, the variant genotypes of both PI3KCD:rs4129341 and PTEN:rs1234221 conferred high-risk/high-benefit. PIK3CD encodes for one of the catalytic domains of the PI3K heterodimer, p110-delta. The PI3K-directed phosphorylation of PIP3 is the key initiation step that transmits signals from receptor tyrosine kinases and other cell surface receptors to downstream molecules such as AKT and mTOR. Overexpression of the wild-type p110-delta is oncogenic in cell culture and overexpression has been observed in several cancer sites, such as AML and glioblastoma. Although not as commonly mutated in solid tumors as p110-alpha (PIK3CA), somatic mutations have been reported in the COSMIC database (www.sanger.ac.uk/genetics/CGP/cosmic/, v57) for head and neck cancer, as well as lung and skin tumors(24). Our results suggest that rs4129341 also behaves similar to oncogenic somatic mutations by increasing the risk of SPT/recurrence in HNSCC. PTEN is a well-known tumor suppressor and often downregulated or inactivated in cancer which results in unrestrained signaling through the pathway. Our results indicate that this may also be the function of the variant rs1234221 allele. Although both of the high-risk/high-benefit PI3KCD:rs4129341 and PTEN:rs1234221 genotypes were the variant-containing genotypes, they were still carried by approximately 30% and 40% of the population, respectively.
Other genes with loci shown to be significant markers for SPT/recurrence risk include IRS2, two isoforms of the S6 kinase (RPS6KB2 and RPS6KL1), AKT2, and PDK1. IRS2 (insulin receptor substrate 2) is an adaptor protein that links receptor tyrosine kinase signaling with downstream pathways, including the PI3K/PTEN/AKT/MTOR pathway (25, 26). Increased activity leads to increased signaling through this pathway and previous studies have shown that IRS2 expression plays a role in metastasis (27). Our results suggest that the IRS2 common genotypes promote tumor progression and thus increase risk of SPT/recurrence in those carrying these genotypes. The S6 serine/threonine kinases are the most downstream effector of signaling through this pathway being activated by AKT and mTOR (28, 29). Increased pathway activity results in ribosomal protein S6 activation that subsequently increases protein translation. This biological mechanism would suggest that the genetic loci significant for increased risk of SPT/recurrence would maintain signaling through this pathway or even enhance it. The three isoforms of the AKT kinase play an important role as a “hub” in transmitting the signals generated through this pathway. Interestingly, our group has previously shown that the variant AKT2:rs892119 was significantly associated with a poor response to chemotherapy, as well as increased risk of recurrence and dying, in a study of esophageal cancer patients (30). The results of this current study are in line with these previous observations where this variant is a marker for a poor outcome. PDK1 is a kinase that activates AKT following PI3K stimulation. The variant rs4972842 was associated with the most significant increase in SPT/recurrence risk, suggesting that this marker results in increased PDK1 activity and enhanced cell survival and proliferation signaling through increased pathway activation.
The PI3K/PTEN/AKT/MTOR pathway is complex and plays a role in regulating nearly all essential cellular processes. In this study, we genotyped variants from 20 genes that encode for the major effectors of this pathway. However, there are other proteins that function within this pathway that may also influence risk of SPT/recurrence as well as response to 13-cRA chemoprevention. Some candidate genes include TSC2, PDK2, RPTOR (Raptor), and EIF4EBP1 (4E-BP). A genome-wide approach would be necessary to identify all unknown factors, but would not be possible within the current study design due to the small patient population.
In combined analysis of the six high-risk/high-benefit loci (TSC1:rs739442, TSC1:rs4962225, TSC1:rs7035940, TSC1:rs7874234, PIK3CD:rs4129341, and PTEN:rs1234221), a clear dose-response relationship was evident. Patients with the highest number of high-risk loci from the PI3K/PTEN/AKT/MTOR pathway received the greatest benefit from 13-cRA, providing support that a pathway-based approach to identifying HNSCC patients at high risk who would most benefit from 13-cRA chemoprevention would potentially reduce the incidence of SPT/recurrence in these HNSCC patients. Furthermore, these results suggest that potential combination therapy with a pathway inhibitor in addition 13-cRA could be of benefit in reducing not only risk of SPT/recurrence, but response to 13-cRA as well. Several drugs have shown effectiveness in blocking signaling through this pathway, including mTOR inhibitor rapamyacin (sirolimus) and its analogs, everolimus and temsirolimus, AKT inhibitor MK-2206, and the PI3K inhibitors LY-294002 and wortmannin and their derivatives (31). Unfortunately, the toxicity and side effect profiles for these agents may not be ideal for a chemopreventive agent that will be administered over many years. However, this pathway is of great interest for drug development and targeted agents may be identified in the future that are suitable for chemoprevention. Additional studies will be necessary to test this hypothesis in vitro and to determine whether or not these agents work synergistically with 13-cRA and followed up with clinical trials to determine if indeed they aid in preventing SPT/recurrence.
The utility of these genetic profiles within the clinical setting would be noteworthy since a majority of HNSCC patients would carry high-risk/high-benefit genotypes. Selection of these individuals prior to 13-cRA chemoprevention would be able to avoid exposing patients to unnecessary, and severe, side effects if there is low risk of SPT or recurrence occurring and if the intervention will not provide a benefit to the patient. This is especially true for chemoprevention interventions which are taken by the patient over a long period of time. A strategy of using genetic information to inform whether or not a patient should receive chemoprevention intervention could go a long way towards individualized prevention.
Towards this, we constructed additive risk prediction models and demonstrated that the inclusion of the three previously identified genetic loci (10) and the five identified in this study increased the discriminatory ability of the model to 79.5% from 65.5% for a model based on only clinical and epidemiology. An AUC of >80% is considered to display “excellent” discriminatory ability, demonstrating that this model has great potential for identifying HNSCC patients at high risk of SPT/recurrence. Further model validation and calibration are necessary to determine if this model is appropriate for clinical utility. However, the results are intriguing and may provide a tool to identify patients who would be candidates for 13-cRA chemoprevention.
The Retinoid Head and Neck Primary Trial was the largest randomized, placebo-controlled, double-blind phase III chemoprevention trial of its kind to investigate the effects of 13-cRA on SPT/recurrence in head and neck cancer patients. It has provided the platform to investigate the role of genetic variation in chemoprevention interventions, but it makes it difficult to assess the effects of the identified high-risk/high-benefit genotypes in an independent population. Potentially these variants via could be integrated into a future prospective study. Furthermore, because of the retrospective nature of this secondary analysis, our sample size is limited and there may be slight differences in minor allele frequencies between the two groups (Supplemental Table). However, the effect sizes for each genetic locus are robust (HR > 1.45 in the placebo analysis) suggesting that sample size may not be a major concern.
Long-term prognosis for early stage HNSCC patients is highly dependent on the development of SPT or recurrence. Clinical trials of 13-cRA chemoprevention have given mixed results, but the use of molecular markers to select candidates for this intervention may have a significant clinical impact and help to prevent SPT/recurrences in a majority of HNSCC patients. Here we showed that genetic variation within the important PI3K/PTEN/AKT/MTOR pathway modulates an individual’s risk of SPT/recurrence as well as predicting response to 13-cRA chemoprevention. These results may help in design of future 13-cRA clinical trials as well as determining whether or not a HNSCC patient would be a suitable candidate for 13-cRA chemoprevention.
METHODS
Study population and epidemiologic data
Patients included in this study were stage I and II HNSCC cases enrolled in the Retinoid Head and Neck Second Primary Trial and randomized to receive either daily low dose (30 mg/d) of 13-cRA or placebo for a total of 3 years (5, 9). Patients must have remained cancer-free for at least 16 weeks following surgery and/or radiation treatment to be enrolled in the trial. The primary endpoint was the development of SPT or recurrence within the 4 year follow-up. SPT was defined as a diagnosis of a new cancer with a different histologicaltype, cancer of identical histological type appearing >3 yearsfollowing treatment of the primary tumor, or discovery of a cancer separated from theprimary tumor site by >2 cm of clinically normal epithelium. Recurrence was defined as any tumor of similar histologyoccurring within 2 cm or 3 years of the primary tumor. Written informed consent was obtained from all participants and this study was approved by the University of Texas MD Anderson Cancer Center’s Institutional Review Board.
Genotyping
Peripheral blood samples were collected from all participants and used for extraction of genomic DNA. Samples were stored at −80°C until use. A custom iSelect genotyping array (Illumina, San Diego, CA) was generated to query haplotype-tagging and candidate functional SNPs from genes in 12 cancer-related pathways (22). A total of 9,465 SNPs were genotyped in 450 DNA samples blinded to patient outcome status and intervention arm following the standard Infinium II protocol (Illumina). Following quality control measures, data for 8,347 SNPs in 440 participants remained. Twenty genes within the PI3K/PTEN/AKT/mTOR signaling pathway were selected for further analysis: AKT1, AKT2, AKT3, MTOR, IRS1, IRS2, PDK1, PIK3CD, PTEN, RHEB, RPS6KA2, RPS6KA3, RPS6KA4, RPS6KA5, RPS6KA6, RPS6KB1, RPS6KB2, RPS6KC1, RPS6KL1, and TSC1. A total of 137 SNPs were genotyped from these candidate genes.
Statistical analysis
Hazard ratios (HRs) and 95% confidence intervals (95%CIs) for each individual SNP and SPT/recurrence were estimated by fitting the Cox proportional hazards model while adjusting for age, gender, ethnicity, smoking status, tumor site, and tumor stage. We tested the proportional-hazards assumptions for the three analytic approaches (placebo stratified by genotype, common genotype stratified by treatment, and variant genotype stratified by treatment). The proportionality assumptions for were satisfied for with P > 0.4 for all three when all the covariates were included in the analyses. To compare event-free durations, Kaplan-Meier curves and log-rank tests were constructed. High-risk genotypes were defined as those that significantly increased risk of SPT or recurrence for patients randomized to the placebo arm. Bootstrap resampling was used for internal validation of the five high-risk/high-benefit loci. Each bootstrap sample was drawn from the complete dataset with a corresponding P value for each of the 500 iterations. Combined effects of high-risk genotypes were assessed using a genetic risk score based on the main effect analysis within the placebo group and stratified by treatment group (placebo or 13-cRA). Receiver operating characteristic (ROC) curves and corresponding area under the curves (AUC) and P values were constructed for risk models including combinations of epidemiology, clinical, and genetic factors. Bootstrap resampling was performed 10,000 times to assess the differences between the AUCs for each of the models. Statistical analyses were performed using STATA software (v10, STATA Corporation, College Station, TX). All P values were two-sided and a P ≤ 0.05 was considered statistically significant.
Supplementary Material
STATEMENT OF TRANSLATIONAL RELEVANCE.
Patients diagnosed with early-stage head and neck cancer are at high risk of developing a second primary tumor or recurrence following curative-intent treatment. Towards this, retinoid-based chemoprevention has been explored as an approach to minimize these events and improve long term prognosis. However, there are no clear biomarkers that can predict which patients are at high risk of developing a second primary tumor or recurrence, and more importantly, also predict favorable responses to 13-cis retinoic acid chemoprevention. This high-risk/high-benefit profile would go a long ways towards identification of suitable candidates for chemoprevention interventions. In this study, we identified genetic predictors within a key signaling pathway, PI3K/PTEN/AKT/MTOR, that identified high-risk/high-benefit head and neck cancer patients. A risk prediction model generated from these data had a discriminatory ability of nearly 80%, suggesting that the application of genetic information to identify patients that would be candidates for chemoprevention
Acknowledgments
FUNDING
This work was supported by the National Institutes of Health CA52051 to W.K.H., CA97007 to W.K.H. and S.M.L., and CA86390 to M.R.S.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Khuri FR, Jain SR. Novel agents and incremental advances in the treatment of head and neck cancer. Semin Oncol. 2004;31:3–10. doi: 10.1053/j.seminoncol.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 3.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–94. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 5.Khuri FR, Kim ES, Lee JJ, Winn RJ, Benner SE, Lippman SM, et al. The impact of smoking status, disease stage, and index tumor site on second primary tumor incidence and tumor recurrence in the head and neck retinoid chemoprevention trial. Cancer Epidemiol Biomarkers Prev. 2001;10:823–9. [PubMed] [Google Scholar]
- 6.Haughey BH, Gates GA, Arfken CL, Harvey J. Meta-analysis of second malignant tumors in head and neck cancer: the case for an endoscopic screening protocol. Ann Otol Rhinol Laryngol. 1992;101:105–12. doi: 10.1177/000348949210100201. [DOI] [PubMed] [Google Scholar]
- 7.Hong WK, Lippman SM, Itri LM, Karp DD, Lee JS, Byers RM, et al. Prevention of second primary tumors with isotretinoin in squamous-cell carcinoma of the head and neck. N Engl J Med. 1990;323:795–801. doi: 10.1056/NEJM199009203231205. [DOI] [PubMed] [Google Scholar]
- 8.Benner SE, Pajak TF, Lippman SM, Earley C, Hong WK. Prevention of second primary tumors with isotretinoin in patients with squamous cell carcinoma of the head and neck: long-term follow-up. J Natl Cancer Inst. 1994;86:140–1. doi: 10.1093/jnci/86.2.140. [DOI] [PubMed] [Google Scholar]
- 9.Khuri FR, Lee JJ, Lippman SM, Kim ES, Cooper JS, Benner SE, et al. Randomized phase III trial of low-dose isotretinoin for prevention of second primary tumors in stage I and II head and neck cancer patients. J Natl Cancer Inst. 2006;98:441–50. doi: 10.1093/jnci/djj091. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Wu X, Hildebrandt MA, Yang H, Khuri FR, Kim E, et al. Global assessment of genetic variation influenceing response to retinoid chemoprevention in head and neck cancer patients. Cancer Prev Res (Phila Pa) 2011;4:185–93. doi: 10.1158/1940-6207.CAPR-10-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci USA. 2002;99:13571–6. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–90. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, et al. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J Clin Invest. 2003;112:1223–33. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Molinolo AA, Hewitt SM, Amornphimoltham P, Keelawat S, Rangdaeng S, Meneses Garcia A, et al. Dissecting the Akt/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin Cancer Res. 2007;13:4964–73. doi: 10.1158/1078-0432.CCR-07-1041. [DOI] [PubMed] [Google Scholar]
- 15.Pedrero JM, Carracedo DG, Pinto CM, Zapatero AH, Rodrigo JP, Nieto CS, et al. Frequent genetic and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head and neck squamous cell carcinoma. Int J Cancer. 2005;114:242–8. doi: 10.1002/ijc.20711. [DOI] [PubMed] [Google Scholar]
- 16.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–74. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lotan R, Xu XC, Lippman SM, Ro JY, Lee JS, Lee JJ, et al. Suppression of retinoic acid receptor-beta in premalignant oral lesions and its up-regulation by isotretinoin. N Engl J Med. 1995;332:1405–10. doi: 10.1056/NEJM199505253322103. [DOI] [PubMed] [Google Scholar]
- 18.Srinivas H, Xia D, Moore NL, Uray IP, Kim H, Ma L, et al. Akt phosphorylates and suppresses the transactivation of retinoic acid receptor alpha. Biochem J. 2006;395:653–62. doi: 10.1042/BJ20051794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lefebvre B, Brand C, Flajollet S, Lefebvre P. Down-regulation of the tumor suppressor gene retinoic acid receptor beta2 through the phosphoinositide 3-kinase/Akt signaling pathway. Mol Endocrinol. 2006;20:2109–21. doi: 10.1210/me.2005-0321. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Khillan JS. A novel signaling by vitamin A/retinol promotes self renewal of mouse embryonic stem cells by activating PI3K/Akt signaling pathway via insulin-like growth factor-1 receptor. Stem Cells. 2010;28:57–63. doi: 10.1002/stem.251. [DOI] [PubMed] [Google Scholar]
- 21.Ohashi E, Kogai T, Kagechika H, Brent GA. Activation of the PI3 kinase pathway by retinoic acid mediates sodium/iodide symporter induction and iodide transport in MCF-7 breast cancer cells. Cancer Res. 2009;69:3443–50. doi: 10.1158/0008-5472.CAN-08-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu X, Spitz MR, Lee JJ, Lippman SM, Ye Y, Yang H, et al. Novel susceptibility loci for second primary tumors/recurrence in head and neck cancer patients: large-scale evaluation of genetic variants. Cancer Prev Res. 2009;2:617–24. doi: 10.1158/1940-6207.CAPR-09-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci USA. 2005;102:802–7. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamauchi T, Kaburagi Y, Ueki K, Tsuji Y, Stark GR, Kerr IM, et al. Growth hormone and prolactin stimulate tyrosine phosphorylation of insulin receptor substrate-1, -2, and -3, their association with p85 phosphatidylinositol 3-kinase (PI3-kinase), and concomitantly PI3-kinase activation via JAK2 kinase. J Biol Chem. 1998;273:15719–26. doi: 10.1074/jbc.273.25.15719. [DOI] [PubMed] [Google Scholar]
- 26.Shepherd PR. Mechanisms regulating phosphoinositide 3-kinase signalling in insulin-sensitive tissues. Acta Physiol Scand. 2005;183:3–12. doi: 10.1111/j.1365-201X.2004.01382.x. [DOI] [PubMed] [Google Scholar]
- 27.Gibson SL, Ma Z, Shaw LM. Divergent roles for IRS-1 and IRS-2 in breast cancer metastasis. Cell Cycle. 2007;6:31–7. doi: 10.4161/cc.6.6.3987. [DOI] [PubMed] [Google Scholar]
- 28.Chung J, Grammer TC, Lemon KP, Kazlauskas A, Blenis J. PDGF- and insulin-dependent pp70S6k activation mediated by phosphatidylinositol-3-OH kinase. Nature. 1994;370:71–5. doi: 10.1038/370071a0. [DOI] [PubMed] [Google Scholar]
- 29.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J Biol Chem. 1998;273:14484–94. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 30.Hildebrandt MA, Yang H, Hung MC, Izzo JG, Huang M, Lin J, et al. Genetic variations in the PI3K/PTEN/AKT/mTOR pathway are associated with clinical outcomes in esophageal cancer patients treated with chemoradiotherapy. J Clin Oncol. 2009;27:857–71. doi: 10.1200/JCO.2008.17.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cleary JM, Shapiro GI. Development of phosphoinositide-3 kinase pathway inhibitors for advanced cancer. Curr Oncol Rep. 12:87–94. doi: 10.1007/s11912-010-0091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.