Abstract
Protein function is often regulated by post-translational modifications (PTMs) and recent advances in mass-spectrometry have resulted in an exponential increase in PTM identification. However, the functional significance of the vast majority of these modifications remains unknown. To address this problem, we compiled nearly 200,000 phosphorylation, acetylation and ubiquitination sites from 11 eukaryotic species, including 2,500 novel ubiquitylation sites for S. cerevisiae. We developed methods to prioritize the functional relevance of these PTMs by predicting those that likely participate in cross-regulatory events, regulate domain activity or mediate protein-protein interactions. PTM conservation within domain families identifies regulatory ‘hot-spots’ that overlap with functionally important regions, a concept we experimentally validated on the HSP70 domain family. Finally, our analysis of the evolution of PTM regulation highlights potential routes for neutral drift in regulatory interactions and suggests that only a fraction of modification sites are likely to have a significant biological role.
Introduction
The activity and localization of proteins inside the cell can be regulated by reversible post-translational modifications (PTMs), including protein phosphorylation, acetylation and ubiquitylation. How these modifications regulate protein function and how this regulation diverges during evolution is crucial for understanding signaling systems. Recent advances in mass-spectrometry (MS) have increased the ability to identify PTMs with thousands of sites now routinely discovered per study (Choudhary and Mann, 2010). However, the functional characterization of these modifications is now rate limiting, a fact further complicated by the recent findings that they can be highly divergent across species (Beltrao et al., 2009; Holt et al., 2009; Landry et al., 2009; Tan et al., 2009). Despite the poor conservation within single proteins, the overall number of phosphosites per protein within different functional modules (i.e. protein complex or pathways) is conserved (Beltrao et al., 2009). This phenomenon could be explained by compensatory turn-over of phosphorylation sites, similar to documented cases of compensatory turn-over of transcription factor binding sites in promoter regions (Ludwig et al., 2000). The similarities in the evolutionary properties of transcriptional and post-translational regulatory networks (Moses and Landry, 2010) lend credence to the idea that phenotypic diversity is primarily driven by changes in regulatory networks (Carroll, 2005).
Although phosphosites observed in high-throughput studies are, on average, poorly conserved, sites with a known function are more significantly constrained (Ba and Moses, 2010; Landry et al., 2009). These trends have led some to speculate that there is a substantial fraction of phosphorylation sites that are non-functional (Landry et al., 2009; Lienhard, 2008). Conservation of modification sites or regulatory interactions can be used to prioritize experimental validation (Tan et al., 2009) but do not provide a putative functional consequence for the modification. Therefore, developing approaches to dissect the functional importance of PTMs is currently the most significant bottleneck in proteomic studies of post-translational regulation.
In this study, we experimentally determined 2,500 ubiquitylation sites for S. cerevisiae and compiled a list of nearly 200,000 modification sites across 11 eukaryotic species in order to develop predictors of PTM functional relevance. These data, as well as structural information, were used to identify modifications that might regulate protein-protein interactions, mediate domain activity or be part of cross-regulatory events between different PTMs. We show that sites with predicted function are more likely to be conserved and that conservation of PTMs within domain families identifies important regulatory regions (termed here regulatory “hot-spots”). We validate these approaches by experimentally characterizing novel regulatory hot-spots within the HSP70 chaperone domain family and characterizing a phosphosite within Skp1 (part of the Skp1/Cullin/F-box E3 ligase) as likely regulating the interaction between Skp1 and the Met30 F-box protein. In summary, the resource developed in this study, which is accessible online (http://ptmfunc.com), can provide mechanistic functional annotations to PTMs and generate specific predictions for experimental validation. This analysis also allows for a better understanding of the evolution of post-translational networks and suggests that only a fraction of PTMs is likely to have a regulatory role.
Results
A resource of eukaryotic PTMs for functional and evolutionary studies
In order to study the evolutionary properties and functional role of protein post-translational regulation, we compiled previously published in-vivo, mass-spectrometry derived, PTMs (Table S1). We compiled a total of 153,478 phosphorylation sites for 11 eukaryotic species, retaining only sites that have high site localization probability (Methods). The phosphorylation dataset covers a broad evolutionary time scale with information for 3 fungi (S. cerevisiae, S. pombe and C. albicans), 2 plant species (A. thaliana and O. sativa), 3 mammals (H. sapiens, M. musculus and R. norvegicus) as well as X. laevis, D. melanogaster and C. elegans. We also compiled 13,133 lysine acetylation sites (covering H. sapiens, M. musculus and D. melanogaster) and 22,000 human ubiquitylation sites (Emanuele et al., 2011; Kim et al., 2011; Wagner et al., 2011). In addition, we used a mass spectrometry (MS) approach to experimentally determine 2,500 ubiquitylation sites in S. cerevisiae to facilitate comparative studies. Using a set of 12 different S. cerevisiae phosphoproteomics experiments, we estimate that the curated datasets should have less than 4% of false positive sites (Table S2).
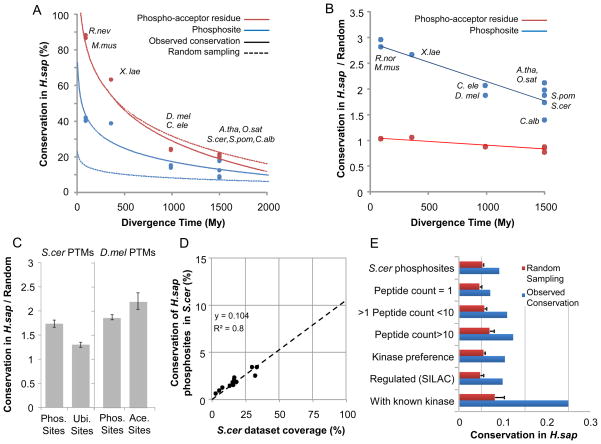
Previous studies have used sequence conservation to study the evolution of phosphosites (Ba and Moses, 2010; Holt et al., 2009; Landry et al., 2009). In this work, we used the compiled data to directly compare the phosphoproteomes across these 11 species and to evaluate the impact of data quality on the evolutionary observations. We selected one of the species with the highest coverage, the human dataset, as reference and compared the data from all other species to it (Figure 1A). We aligned 1-to-1 orthologs of each species to H. sapiens proteins, and for each phosphosite, determined the conservation in the human protein of both the phospho-acceptor residue (i.e. sequence conservation) and the phosphorylation site (i.e. phosphosite conservation). In order to account for potential errors in MS phosphosite positional assignments, we considered a phosphosite conserved if the corresponding human ortholog was also phosphorylated within a window of +/−2 alignment positions. Both residue and phosphosite conservation were found to be proportional to the divergence age away from H. sapiens (Figure 1A). Phosphosite conservation ranged from ~8–18% for the distantly related plants and fungi to ~40% for the closely related mouse and rat. We then asked if the observed value was higher than expected by randomly re-assigning the same number of phosphosites within each orthologous protein. As previously described by Landry and colleagues, we observed that the sequence conservation of the phospho-acceptor residue was not higher than expected by chance (Figure 1B) (Landry et al., 2009). However, the conservation of the phosphorylation sites was approximately 2 to 3 times higher than random (Figure 1B). This difference suggests that the conservation of the phosphorylation state is a better indicator of functional importance than sequence conservation of the phospho-acceptor residue. For all the following analysis, we used the conservation of the PTM state as the comparative metric.
Figure 1. Evolutionary properties of eukaryotic post-translational modification sites.
A) We analyzed the conservation of 10 phosphoproteomes against that of H.sapiens using protein alignments of 1-to-1 orthologs. For each species, we compared the conservation of the phosphoacceptor residues (i.e. sequence conservation) with the conservation of phosphorylation site based on the MS experimental evidence. A random expectation or null model for each case was determined based on the random shuffling of phosphosite positions within each human protein. B) Ratio of observed conservation over random expectation. C) Ratio of conservation over random expectation ratio for different PTMs. D) Predicted coverage and conservation in H.sapiens for 12 different previously reported phosphoproteomics experiments for S.cerevisiae. E) To access the impact of data quality on the conservation values we made use of different criteria to define subsets of S.cerevisiae phosphosites. For each subset we calculated the observed conservation in H.sapiens as well as the expected value based on random sampling.
In order to evaluate the generality of these evolutionary observations across different PTMs, we studied the conservation over random expectation of ubiquitylation and acetylation sites (Figure 1C). We compared the conservation of S. cerevisiae phosphorylation and ubiquitylation sites in H. sapiens over a random expectation calculated based on random sampling of similar number of modification acceptor residues within the same proteins. Similarly, we compared the conservation of D. melanogaster phosphorylation and acetylation sites in human orthologs. The three modifications show a low level of evolutionary constraint, ranging from 1.3 to 2.2 times higher conservation than expected based on an equivalent random sample of PTM acceptor residues (Figure 1C). Protein acetylation shows a higher value of conservation over random when compared to phosphorylation, consistent with previous work (Weinert et al., 2011), whereas ubiquitylation appears to have a lower evolutionary constraint when compared to phosphorylation (Figure 1C).
Given that these datasets gathered so far are likely to be incomplete, the conservation values presented here are under-estimated. We used 12 different S. cerevisiae phosphoproteomics experiments to evaluate the error in the conservation estimates by plotting the values of conservation versus coverage for each S. cerevisiae phosphoproteomic dataset (Figure 1D). Extrapolating from the regression analysis we estimate that, when corrected for coverage, approximately 10% of H. sapiens phosphosites would be conserved in S. cerevisiae. We also calculated the corrected conservation value for each dataset independently and estimated the corrected median conservation value for H. sapiens phosphosites in S. cerevisiae as ~13% (Figure 1D). This value is higher but comparable to the observed 8% conservation measured with the complete dataset. This suggests that, at least for the extensively studied phosphoproteomes of S. cerevisiae and H. sapiens, additional data is unlikely to dramatically change the conservation estimates.
Besides coverage (i.e. false negatives), data quality (i.e. false positives) and low phosphosite abundance are also important factors when estimating phosphoproteome conservation. We compared the conservation of S. cerevisiae phosphosites in H. sapiens across different data quality criteria, including the number of spectral counts, match to known kinase recognition motifs and information on dynamically regulated phosphosites (Figure 1E). The conservation of different classes of phosphorylation sites (Figure 1E, blue bars) was compared to an equivalent random sample (Figure 1E, red bars). To determine statistical significance of the results, the ratios of conserved over expected values for the different phosphosite groups were compared using a Mann-Whitney ranked test. We assumed that spectra and/or peptide count for each phosphosite is correlated with data quality and/or phosphosite abundance and observed that phosphosites supported by multiple spectra/peptides (Figure 1E, peptide count >1) are more likely to be conserved than those observed only once (Figure 1E, peptide count = 1, p-value<10−8). Additionally, sites that represent well-matched kinase-recognition motifs (Figure 1E, “Kinase preference”) or are known to be regulated (Figure 1E, “Regulated”), as measured in quantitative mass-spectrometry studies (Holt et al., 2009; Huber et al., 2009; Soufi et al., 2009), are moderately more conserved than average sites and more highly conserved than expected by chance (Figure 1E. “S.cer phosphosites”, p-value<10−9). Finally, sites that are known to be functionally important (Ba and Moses, 2010) or have described in-vivo kinase regulators (www.phosphogrid.org) (Stark et al., 2010) are more than 3 times more conserved than average sites (Figure 1E, “With known kinases” vs. “S.cer phosphosites”, p-value<10−16). These results imply that higher phosphosite functionality, quality and/or abundance are correlated with conservation and support previous observations made with sequence analysis (Landry et al., 2009). It is likely that low abundance and/or nonfunctional phosphosites, with low conservation, dominate the overall measured divergence. These results further underscore the need to devise methods to assign functional roles to PTM sites.
Phosphorylation coupled to other classes of modifications
On average, approximately 75% of known phosphorylation sites, 40% of acetylation sites and 45% of ubiquitylation sites occur outside known PFAM globular domains (Table S1). It has become increasingly apparent that phosphosites within unstructured regions not only recruit phospho-binding domains but also often regulate other PTMs or localization signals (Hunter, 2007). This complex interplay between PTMs has been previously described in histone tails where they have been proposed to form a code to be read by different effectors and control gene chromatin states (Strahl and Allis, 2000). More recently, examples of cross-regulation between adjacent PTMs have been observed in several other proteins suggesting this to be a universal mode of protein regulation (Hunter, 2007). Examples include the promotion of sumoylation by a priming phosphorylation in several transcription factors (Yang and Gregoire, 2006), the cross-inhibition between adjacent phosphorylation and methylation sites in DNMT1 (Esteve et al., 2011) and the positive role of lysine acetylation on the phosphorylation of Cdc6 (Paolinelli et al., 2009).
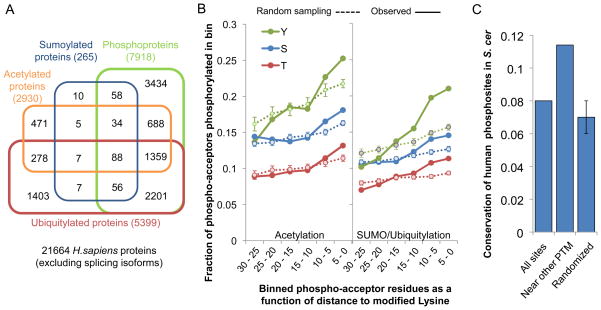
We hypothesized that it is possible to assign a functional role to PTM sites by searching for the co-occurrence of different modifications within the same protein. For the human proteome, using the information on lysine acetylation, ubiquitylation and sumoylation, we observed a significant overlap between proteins containing these lysine modifications and the phosphoproteome (Figure 2A). While 36% of all proteins are phosphoproteins, over 69% of proteins containing any of these lysine modifications is also phosphorylated (Figure 2A). This enrichment is highly significant (p-value < 1×10−70, with a Fisher’s exact test) and not merely due to MS detection bias for abundant proteins (Table S3). We next asked if these PTMs tend to cluster within the protein sequence (Figure 2B). Given the small number of currently characterized sumoylation sites, we grouped these together with ubiquitylation for the analysis. We binned phospho-acceptor residues (i.e. serine, threonine and tyrosine) according to their smallest distance to a modified lysine residue. In each distance bin, we then calculated the fraction of acceptor residues that is phosphorylated and compared this observed value with a random expectation by randomly re-assigning the same number of phosphorylation sites within each protein. We observed that, on average, phospho-acceptors near modified lysines are preferentially phosphorylated when compared to more distant residues or an equivalent random sample of sites. These results show that the different PTMs tend to cluster within protein sequences. This result is not merely due to preferential accumulation of PTMs in unstructured regions (Figure S1) and was also observed using phosphorylation and lysine acetylation data for mouse and phosphorylation and ubiquitylation data for S. cerevisiae (Figure S1). If phosphorylation sites near other PTMs are more likely to be functionally relevant, then we assumed that these should also show higher conservation. We tested this by comparing the conservation, in S. cerevisiae, of all human phosphorylation sites with those within 15 amino acids of another PTM. The conservation of phosphorylation sites near modified lysines were higher than for average phosphosites and higher than an equivalent random sample (p-value<10−7) (Figure 2C).
Figure 2. Association of protein phosphorylation with lysine post-translational modifications.
A) A Venn diagram representing the overlap of the different lysine modified proteins with the human phosphoproteome. While 33% of the human proteins are phosphorylated, 71% of the acetylated proteins, 69% of the ubiquitylated proteins and 89% of the sumoylated proteins are also phosphorylated. B) The fraction of the phosphoacceptor residues was plotted as function of the distance to modified lysine residues. Observed values were compared with expected values based on random sampling. C) Human phosphorylation sites that are near an acetylated lysine residue (<15 amino-acid distance) were more likely conserved in S. cerevisiae than average sites or an equivalent random sample. See also Figure S1 and Table S3.
Regulation of protein-protein interactions
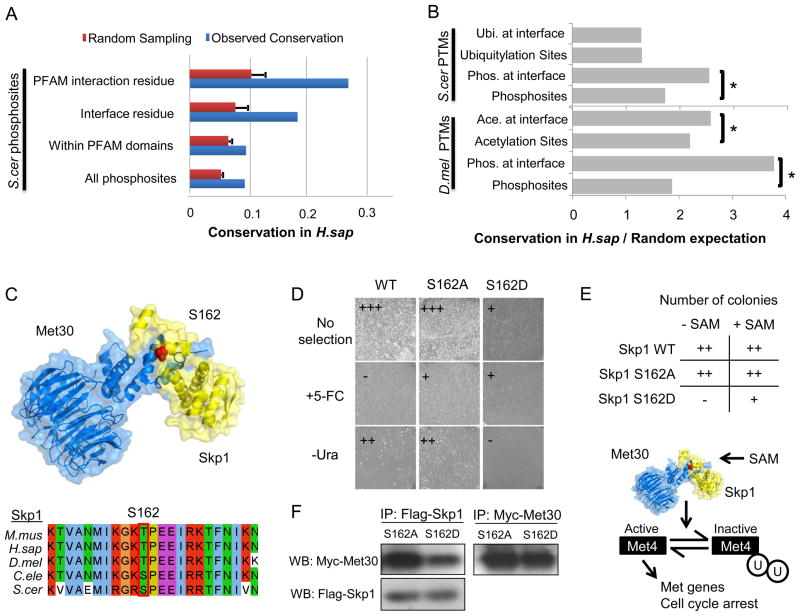
For the PTM sites that occur within structured regions, we can make use of the growing structural knowledge deposited in the PDB (www.pdb.org) to assign putative functional roles for the protein modifications, especially those found within protein interfaces, since they may be involved in the regulation of protein-protein interactions. For human or S. cerevisiae protein-pairs that are known to physically interact, we used x-ray structures, homology models or docking solutions to define the most likely interface regions (Methods). Using these models, we identified 870 phosphorylation, 632 ubiquitylation and 263 acetylation sites at putative interface residues that can potentially regulate protein-protein interactions (available at ptmfunc.com). To expand the number of putative interface residues, we made use of the 3DID database of domain-domain interactions (Stein et al., 2011). For each domain family, as annotated in PFAM (pfam.sanger.ac.uk), 3DID contains annotations of what residues have been shown to participate in physical interactions in x-ray structures. We have used these annotations to assign interaction residues for PFAM domains in the 11 proteomes (Methods) and identified 3968 phosphorylation, 1802 ubiquitylation and 1691 acetylation sites that potentially regulate protein-protein interactions (ptmfunc.com). Using either of these definitions, we observed that S. cerevisiae phosphosites at interface residues are approximately 2 to 3 times more likely to be conserved in H. sapiens than average phosphosites (Figure 3A, “Interface residue” or “PFAM interaction residue” vs. “All phosphosites”, p-value<10−14). It is known that globular domain regions are easier to align than the unstructured regions where most phosphosites occur. However, the higher conservation of phosphosites at interface residues is not merely due to alignment issues since phosphosites that occur within PFAM domains are not more conserved than average sites (Figure 3A). We confirmed these evolutionary trends using interface models for human protein-protein interactions (Figure S2A)
Figure 3. Regulation of interface residues by PTMs.
A) The conservation of all S.cerevisiae phosphosites in H.sapiens was compared with the conservation of phosphosites within PFAM domain interface residues (from the 3DID database) and at putative interface residues (from interaction models). B) We compared the conservation over random expectation of different PTMs at PFAM interface residues (from the 3DID database). The ratios of conservation over random expectation were compared using a Mann-Whitney rank test. (*) p-value <0.05. C) The Skp1:Met30 interaction model. The S162 position of the S. cerevisiae Skp1 was found to be phosphorylated in S. cerevisiae, H. sapiens, M. musculus and C. elegans. D) A protein complementation assay of cytosine deaminase activity reports on the Skp1:Met30 interaction. The phosphomimetic mutation reversed the growth pattern on selective media reporting on interaction strength by growing on +5-FC plates and without any observable growth on –Ura plates suggesting that the in vivo affinity of the Skp1:Met30 complex is reduced by phosphorylation. F) Skp1 S162D mutation shows a significant decrease in bound Met30 when compared to the S162A mutant by co-immunoprecipitation. E) Skp1 wild-type and mutants (S162A and S162D) were plated in the presence or absence of SAM. Overexpression of the S162D mutant resulted in poor growth, a phenotype that was relieved in the presence of SAM. See also Figure S2.
In order to test the generality of some of these observations across different post-translational modifications, we compared the conservation (over random expectation) of all acetylation, ubiquitylation and phosphorylation sites with those occurring at predicted interface residues (Figure 3B). In line with the observations made for protein phosphorylation, lysine acetylation at interface residues is more likely to be conserved (Figure 3B, p-value<10−5), however, ubiquitylation at interface residues shows a similar level of constraint when compared to average ubiquitylation sites. These results suggest that phosphorylation and acetylation but not ubiquitylation sites at interface residues are more likely to be functionally important than average sites, suggesting that these PTMs are commonly used by the cell to reversible regulate the binding affinity of protein interactions.
The analysis of protein-protein interfaces creates specific predictions for the functional role of PTMs. For example, several alpha subunits of the proteasome are phosphorylated at interface regions (Figure S2 B and C). Serine 13 and tyrosine 5 of Pre8 (the S.cerevisiae alpha 2) are phosphorylated in yeast and human, respectively, and could potentially regulate the interactions with Pre9 (alpha 3). The N-terminus of alpha 5 is also phosphorylated in 7 of the 11 species. This N-terminal region has been shown to be important for proteasome activity (Groll et al., 2000) indicating that these N-terminal phosphorylations might regulate the interactions between alpha subunits or the activity of the proteasome (Figure S2 B and C). Similarly, we predicted that a phosphosite at position S162 in the S.cerevisaie Skp1 could regulate the interaction with Met30 (Figure 3C). Skp1 is a highly conserved protein that is part of the Skp1/Cullin/F-box (SCF) multi-subunit E3 ubiquitin ligase complex (Petroski and Deshaies, 2005). Skp1 interacts with different F-box domain containing proteins that can modulate the ubiquitylation substrate specificity (Petroski and Deshaies, 2005). In S. cerevisiae, Skp1 can interact with the Met30 F-box protein to regulate proteins involved in sulfur metabolism (Jonkers and Rep, 2009) an interaction that is known to be regulated under different stress conditions (Jonkers and Rep, 2009). We postulated that the highly conserved phosphorylation site in Skp1 might regulate the interaction with Met30 and/or other F-box proteins (Figure 3C). We note that given the position of the residue at the end of the helix, it would have to partially unwind to adopt a coil conformation that can access the kinase active site.
To experimentally probe the dependency of the Skp1:Met30 interaction on the phosphorylation status of S162, we used a protein complementation assay (Ear and Michnick, 2009; Michnick et al., 2010) that reports on the strength of the protein-protein interaction in vivo. We fused Skp1 and Met30 to two fragments of the yeast cytosine deaminase and transformed the constructs into a strain that lacks the endogenous enzyme. Skp1 and Met30 interact directly in vivo allowing the two fragments to reconstitute cytosine deaminase activity. Reconstitution of enzyme activity permits growth on media lacking uracil (-Ura) and leads to death on media containing 5-fluorocytosine (+5-FC) (Figure 3D). To test the idea that phosphorylation reversibly regulates the assembly of this interaction, we mutated S162 to alanine (S162A) or the phosphomimetic aspartic acid (S162D). The S162A mutant, similar to wild-type, supported growth on –Ura media, which selects positively for interacting proteins, and grew poorly on +5-FC media, which counter-selects for interacting proteins, indicating that the unphosphorylated state binds Met30 (Figure 3D). In contrast, the S162D mutant grew better on +5-FC than on –Ura media, indicating that the phosphorylated state binds Met30 weaker than the unphosphorylated state (Figure 3D). In order to validate this result, Flag-tagged Skp1-S162A and Skp1-S162D were immunoprecipitated in the presence of Met30-Myc and the Met30:Skp1 interaction was monitored using an α-Myc antibody. We found that the Skp1:Met30 interaction is impaired in the phosphomimetic mutant but not in the alanine mutant (Figure 3F), suggesting that the phosphorylation of S162 acts as a reversible switch for Met30 affinity.
The Skp1:Met30 interaction is required to keep the Met4 transcription factor inactivated via ubiquitylation (Kaiser et al., 2006). The activation of Met4 regulates genes involved in the biosynthesis of sulfur-containing amino acids and glutathione metabolism but it also results in cell cycle arrest (Aghajan et al., 2010). During our interaction studies, we observed that overexpression of Skp1 S162D resulted in poor growth, a phenotype that was not observed with the overexpression of Skp1 WT or S162A mutant (Figure 3E). These results suggest that Skp1 S162D impairs the interaction with Met30 resulting in an activation of Met4 and cell cycle arrest. The Met4 inactivation by Skp1:Met30 is known to be promoted by SAM (Kaiser et al., 2006). Consistent with the hypothesis that Skp1 S162D overexpression results in Met4 activation and cell cycle arrest, growth in the presence of SAM relieves the impaired growth (Figure 3E), presumably by further activating the available pool of Skp1:Met30 and/or relieving independently a cell cycle block. These collective results strongly suggest that the phosphorylation of Skp1 on residue S162 has the potential to reversibly alter the binding affinity of Skp1 with Met30 and regulate the function of Skp1:Met30.
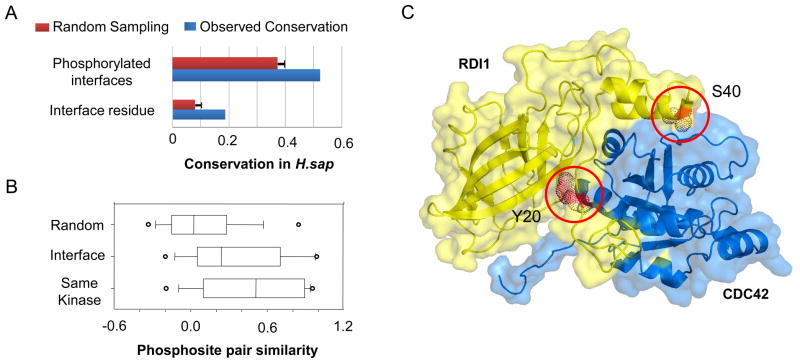
Next, we used the interface models to differentiate between the conservation of phosphorylation sites at interface residues from the conservation of the predicted function (i.e. regulation of the interaction). Intuitively, one can imagine that the phosphorylation of an interface might be conserved despite the divergence of the actual phosphosite position. We observed that over 50% of the interfaces that are phosphorylated in S. cerevisiae are also phosphorylated in H. sapiens, despite only approximately 18% of the interface phosphorylation sites show positional conservation (Figure 4A). However, given that the current phosphoproteomes are likely to be incomplete, we cannot rule out that some of the observed positional divergence is not due to a coverage issue. A similar trend is observed using human interface models (Figure S2A). If the conservation of function with divergence of phosphosite position is mostly the product of a neutral variation, we might expect to observe a conservation of the kinase recognition for the phosphosites at the same interface. To study this issue, we devised a metric of phosphosite similarity based on the models of binding preferences of 63 S. cerevisiae kinases and calculated the similarity of S. cerevisiae interface phosphosites with human phosphosites at the same interface (Methods). We then compared these scores with the similarity scores for random pairs of phosphosites and sites known to be regulated by the same kinases (Figure 4B). The distribution of phosphosite similarity for interface phosphosites is higher than for random pairs (p-value < 2×10−16 with a Kolmogorov-Smirnov test) suggesting that a significant fraction of phosphosites observed at the same interface in different species are phosphorylated by kinases of similar specificity.
Figure 4. Conservation of interface phosphorylation can be achieved by regulation of different positions.
A) We compared the conservation of all S. cerevisiae interface phosphorylation sites in H. sapiens (Interface residues) with the conservation of the phosphorylation of S. cerevisiae interfaces in H. sapiens without regard to the actual phosphosite position (Phosphorylated interfaces). B) A metric of phosphosite similarity (Methods) was used to compare phosphosite pairs found at the same interfaces in the two different species (S. cerevisiae and H. sapiens) with random phosphosite pairs and pairs known to be regulated by the same kinases. Open circles represent the top and bottom 5th percentiles. C) The model of the S. cerevisiae Cdc42p:Rdi1p interface was annotated with currently known phosphorylation data. The Rdi1p Y20 and S40 positions refer to the S. cerevisiae protein sequence positions. The S40 position is currently known to be phosphorylated in S. cerevisiae and C. albicans but not in human. Conversely the Y20 position is known to be phosphorylated in human but not in fungi.
An example of conserved phosphorylation of an interface at different positions is shown in Figure 4C for the interaction between the S. cerevisiae Rho family GTPase Cdc42p and the Rho inhibitor Rdi1p. Rdi1p is phosphorylated at the S40 position in S. cerevisiae. While the S40 equivalent position is phosphorylated in the C. albicans ortholog it is currently not know to be phosphorylated in human. Instead, the Rdi1p Y20 position is phosphorylated in the human ortholog (Figure 4C) but it is currently not know to be regulated in fungi. Regulation of Rho-inhibitor interactions by phosphorylation has been previously described as an important mechanism for the control the function of Rho proteins (DerMardirossian et al., 2004). Our analysis suggests that the phospho-regulation of the Cdc42:Rdi1 might be highly conserved but achieved by the phosphorylation of different positions in different species.
Post-translational ‘hot-spots’ within domain families
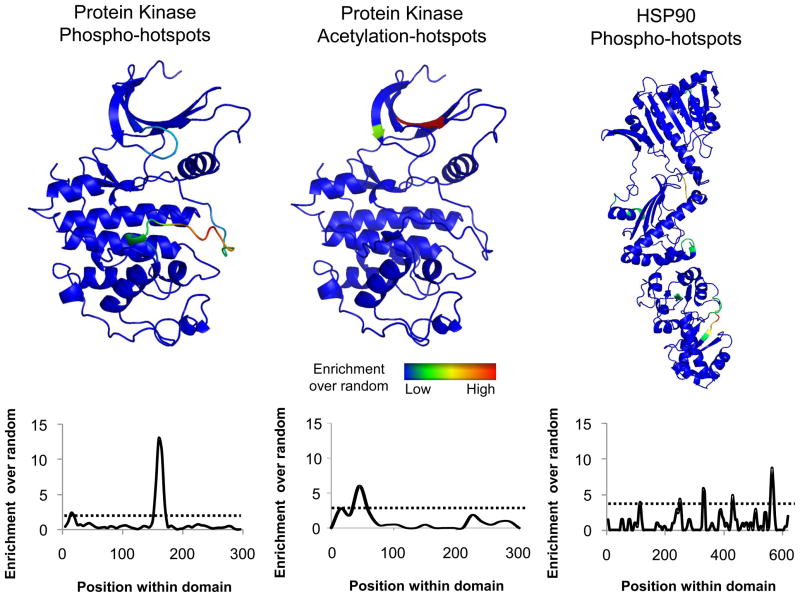
We show above that PTMs with putative functional annotations are more likely to be conserved across species than average sites. We hypothesized that we could use conservation to identify regions within domain families with high regulatory potential. Ten domain families that are extensively phosphorylated across the 11 species with available phosphorylation data were initially selected for this analysis (Table S4). For each domain family, we selected a representative sequence/structure from the PDB (Table S4), then aligned each domain from the 11 species to the representative sequences/structures and mapped to them all phosphorylation sites. Putative regulatory regions were identified by calculating the enrichment of phosphosites over random expectation. Significantly enriched domain regions, or regulatory ‘hot-spots’, were determined based on random sampling with a p-value cut-off of 0.005 or less (Methods). We hypothesize that phosphorylation of residues within these regulatory hot-spots are more likely to regulate domain function. A similar analysis for lysine acetylation was performed for the protein kinase domain.
In Figure 5, we show the sequence and structural mapping for the enrichment of phophorylation or acetylation sites for 2 example domains (protein kinase and HSP90). As expected, the most significantly phosphorylation enriched region for the kinase family is the activation loop region (Nolen et al., 2004). A second hot-spot of phospho-regulation was observed within the ‘glycine-rich’ loop that contributes to ATP binding and has been described to activate or inhibit the activity of kinases, in particular, CDKs (Narayanan and Jacobson, 2009). Interestingly, there is no significant enrichment of acetylation sites within the activation loop of kinases but instead these are preferentially observed within the N-terminal lobe region. This enrichment is primarily due to a catalytic lysine residue that is often observed to be acetylated, a modification that has been previously shown to be important for the regulation of kinase activity (Choudhary et al., 2009).
Figure 5. Phosphorylation enrichment analysis identifies regulatory ‘hot-spots’.
For each domain family under analysis we selected a representative structure from the PDB database (Table S4). Phosphorylation and acetylation data was mapped to the representative sequence/structure using sequence alignments and random sampling was used to calculate the enrichment over random. This value was plotted along the domain sequence position of the representative structure as a moving average and a cut-off of p-value<0.005 was used to identify significant enrichment (dotted line). See also Figure S3.
The HSP90 domain family is a highly conserved dimeric heat-shock protein family that facilitates the folding of client proteins involved in a multitude of biological functions (Taipale et al., 2010). We identified 145 phosphorylation sites within members of the HSP90 domain that were preferentially enriched in the C-terminal region (Figure 5). The strongest enrichment segment corresponds to the residues 600–610 of the yeast HSP90 (HSC82) sequence that projects from the C-terminal region and forms contacts with the equivalent segment of the opposing dimer (Ali et al., 2006). Phosphorylation of this region is therefore likely to regulate HSP90 function. It has been shown that the Ppt1 phosphatase binds to the HSP90 C-terminal region and that the disruption of this interaction results in hyperphosphorylation and misregulation of HSP90 (Wandinger et al., 2006). Consistent with these ideas, Soroka and colleagues validated this prediction by demonstrating that the 600–610 region of the yeast HSP90 is in fact regulated by Ppt1 and that phosphorylation of this region has the potential to regulate HSP90 function (Soroka et al., 2012).
We believe that this enrichment approach can be used to study the regulatory potential of different domain families and we provide additional examples in Supplementary Information (Figure S3).
Regulation of the HSP70 domain family by protein phosphorylation
The results above strongly suggest that our statistical enrichment analysis can highlight functionally important sites subject to regulation by PTMs. In order to further validate this approach, we studied in more detail the regulation of the heat shock 70kDa (HSP70) domain family. The HSP70 is a highly conserved chaperone that folds client proteins through an ATP-dependent cycle of binding and release (Kampinga and Craig, 2010). HSP70 proteins are constituted of two domains, an N-terminal nucleotide binding domain (NBD) and a C-terminal substrate binding domain (SBD) (Figure 6A). Although the HSP70 family has been extensively studied and is implicated in a myriad of cellular functions (Kampinga and Craig, 2010), its regulation by protein phosphorylation has not been previously explored.
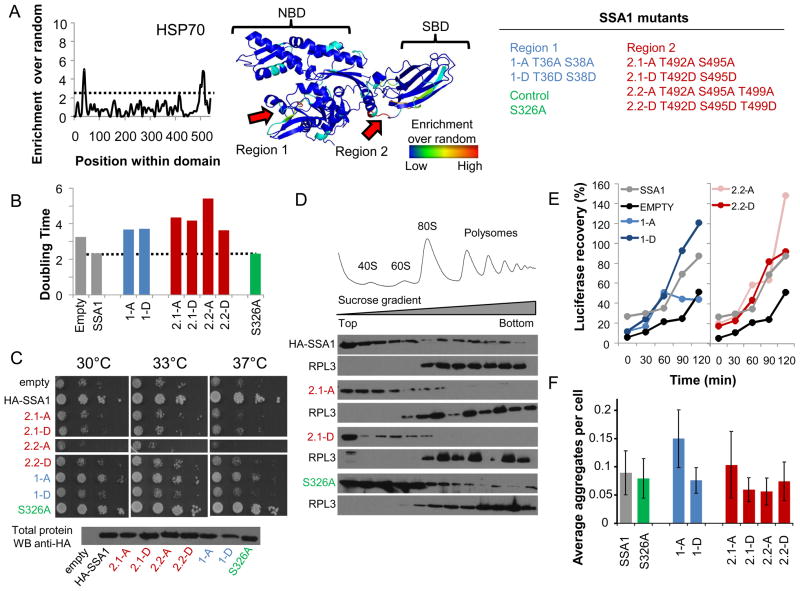
Figure 6. Phosphorylation hot-spots within the HSP70 domain family.
A) A cut-off of p-value<0.005 (dotted line) was used to identify 2 regions that are significantly enriched for phosphosites in the HSP70 family. B) Phosphorylation hot-spot mutants do not complement SSA1 functions as measured by growth in liquid media. C) 10-fold dilution series of Ssa1 mutants on -URA plates and incubated at 30°C, 33°C and 37°C for 2 days. The protein abundance of the different mutants was compared to WT Ssa1 by western. D) The association of the indicated SSA1 mutants with polysomes was examined by immunoblot analysis. Ribosomal profiles (top) were determined by OD254nm and confirmed by immunoblot analysis of the ribosomal proteins Rpl3p. E) Recovery of luciferase activity is expressed as a percentage of activity before heat treatment and is an average of 2 experiments. F) Percentage of cells with multiple and single ubc9-2-GFP aggregates after heat shocked at 37°C for 30 minutes. Errors bars quantify the standard deviations of, at least, 4 technical replicates. See also Figure S4.
We identified 313 phosphosites within HSP70 proteins across the 11 species and our enrichment analysis highlighted 2 significant hot-spots (Figure 6A). Strikingly, both of these mapped to functionally and structurally important regions, one near the nucleotide binding pocket (Region 1) and the second near the entrance to the peptide binding groove (Region 2). The two regions were then used to predict the corresponding regulatory phosphosites in SSA1, an abundant cytosolic HSP70 in the budding yeast. SSA1 has been involved in multiple cellular functions, including binding to polysomes and nascent chains, and assisting the refolding of newly made and stress-denatured polypeptides, as well as prevention of protein aggregation, the post-translational translocation of newly synthesized secreted proteins into the endoplasmid reticulum (ER) and mitochondria, and degradation of misfolded proteins (Albanese et al., 2006; Horton et al., 2001). To test the functional relevance of the predicted sites, SSA1 constructs were designed with alanine or phosphomimetic mutations of residues that were known to be phosphorylated and within these hot-spot regions. Two closely spaced phosphorylated threonines were mutated in Region 1 (T36, T38) and three phosphosites were mutated in Region 2 (T492, S495, T499) (Figure 6A). We also mutated serine 326, a position known to be phosphorylated but outside the hot-spot regions to serve as a control. Since the cytosol of yeast contains four nearly-identical SSA homologues (SSA1–4) the different SSA1 mutants were studied in two yeast strains engineered to lack cytosolic Hsp70 function: (i) a strain lacking SSA2–4 and containing a single copy of SSA1 with a temperature sensitive point mutation renders it inactive above 37°C (ssa1–45) and (ii) a strain lacking both SSA1 and SSA2, but containing functional copies of the less abundant SSA3 and SSA4. Similar results were obtained in both types of cells.
Growth of the wild-type and mutant strains were measured in liquid culture (Figure 6B) or using serial spot dilution assays (Figure 6C). While the control phosphorylation mutant behaved like wild-type, none of the phosphorylation mutants in Regions 1 and 2 were able to fully complement the growth even under non-stress conditions of 30°C, indicating that the regulatory hot-spot phosphorylation sites are important for SSA1 function. In addition, we performed serial spot dilution assays on the single alanine and phosphomimetic mutants of both regions (Figure S4A). With the potential exception of S38D, all single mutants displayed a growth defect under heat shock conditions that is not observed when the WT Ssa1 is expressed or when a control mutation T326A is introduced (Figure S4A). Importantly, the protein abundance of the mutants was comparable to WT Ssa1 (Figure 6C). However, no dramatic differences were observed between the alanine or phosphomimetic mutants suggesting that either the phosphorylation cycle is important for the function of Ssa1 or alternatively, the phosphorylation state of region 1 and region 2 could distinctly affect the function of Ssa1 in the multiple distinct cellular tasks required for cell growth. To obtain further insight we explored the effect of the phosphorylation mutants on a small subset of assays reporting on different Hsp70/SSA functions, namely: association with polysomes (Figure 6D); refolding of firefly luciferase following heat-stress (Figure 6E) and prevention of misfolded protein aggregation (Figure 6F). We examined the association of Ssa1 with polysomes by fractionation extracts on 7%–47% sucrose gradients followed by western blot analysis for the presence of WT or mutant Ssa1 as well as the ribosomal protein Rpl3 (Figure 6D). Both Ssa1 WT and the control mutant associated with polysomes as previously reported (Albanese et al., 2006). However Ssa1 mutated in Region 2 were defective in binding to polysomes. The Ssa1 mutants of Region 1 show similar to wild-type association with polysomes (Figure S4B). In addition we observed that a single phosphomimetic mutation in region 2 (S495D) is sufficient to disrupt the association with polysomes (Figure S4C), a phenotype not observed for the equivalent alanine mutant (S495A) (Figure S4C). These data suggest that the regulatory hot-spot we identified in Region 2 may be involved in binding to nascent chains or in mediating interactions between the translational co-factors Sis1 and Pab1 to the ribosome (Horton et al., 2001).
Hsp70 also assists the refolding of heat-denatured polypeptides, a function that can be monitored by following the recovery of luciferase enzymatic activity following heat-stress (Methods). As expected, the cells containing wild type Ssa1 showed robust recovery of luciferase activity while little recovery was observed in the SSA-defective cells transformed with the vector control (Figure 6E). Most phosphorylation mutants performed similar to wild type in this assay; however, a phosphorylation incompetent variant in Region 1 (i.e. the nucleotide binding site) was significantly impaired in assisting the recovery of stress-denatured luciferase when compared to WT or a phosphomimetic mutant. Since Hsp70 is also implicated in the prevention of aggregation following misfolding, we also tested the effect of the phosphorylation mutant to prevent the aggregation of a temperature-sensitive (ts) mutant of Ubc9 under non-permissive conditions (Figure 6F). Similar to what was observed for the recovery of stress-denatured luciferase, the phosphorylation incompetent Ssa1 mutant (i.e. alanine mutant) in Region 1 was impaired in the ability to prevent aggregate formation (Figure 6F) when compared to WT or the phosphomimetic mutant. The phosphomimetic mutant appears to have a similar to WT luciferase recovery and aggregation prevention capacity.
Taken together, these results indicate that the two conserved phosphorylation hot-spots in Hsp70 are functionally relevant, and distinctly affect various Hsp70-dependent activities. While phosphorylation of Hsp70 had been previously observed, our approach provides the first evidence that such phosphorylation is important for Hsp70 regulation. Given the many distinct functions of Hsp70 during the life cycle of the cell our results open the way to future studies dissecting the precise contribution of regulation of each region to overall Hsp70 function as well as the modulation of its activity under various growth conditions.
Discussion
We have compiled a resource of nearly 200,000 PTMs covering 11 eukaryotic species and developed approaches to annotate PTMs that are more likely to cross-regulate each other or to regulate protein-interfaces or domain activity. To make this resource easily available to others, these data are available through a website (http://ptmfunc.com) that contains known PTMs, spectral counts, information on conditional regulation, conservation and putative functional assignments. Using these methods, we have identified a phosphorylation site within Skp1 that is likely to reversibly alter the binding affinity of Skp1 to Met30. Given the position in the crystal structure, it is possible that the phosphorylation of Skp1 at S162 acts by sterically or electrostatically repulsing Met30. However, given that Skp1 interacts with other F-box proteins, it is also possible that the phosphorylation increases the affinity for another protein and titrates Skp1 away from Met30. Based on the assumption that conserved sites are more likely to be functionally relevant, we have identified regions within domain families that show significant enrichment of PTM sites across the 11 species analyzed here (regulatory hot-spots). We have experimentally characterized two such regions within the HSP70 chaperone domain family showing that they affect Hsp70 function and provide additional examples for future studies. Putative functional annotations for 8776 phosphosites from these 11 species are available through our website.
Besides the functional prioritization, this resource can also be used to study the evolution of post-translational regulation. Past work on the evolution of cellular interaction networks has shown that, while protein complex membership diverges slowly and mostly through subunit duplication (Pereira-Leal et al., 2007; van Dam and Snel, 2008), cellular interactions of broad specificity such as protein interactions mediated by small peptide ‘linear-motifs’ diverge quickly (Beltrao and Serrano, 2007; Neduva and Russell, 2005). Our analysis of 11 partial phosphoproteomes further validates previous observations regarding the fast divergence of kinase-substrate interactions (Landry et al., 2009). In addition, we show that lysine modifications are equally poorly constrained when compared to an equivalent random sample of lysine acceptor residues.
Although regulatory interactions diverge quickly, it is possible for these changes to be neutral in respect to phenotype, much in the same way that mutations within open-reading frames can be neutral to the coding sequence. Examples include the conservation of the mating-type logic, regulation of the origin of replication complex and ribosome transcriptional regulation, despite the divergence of the underlying interactions in some fungal species (Lavoie et al., 2010; Moses et al., 2007; Tsong et al., 2006). The existence of equivalent or ‘neutral networks’ as described by Andreas Wagner, among others, may in fact be important for the exploration of novel phenotypes (Wagner, 2008). We observed that PTMs that are known to be regulated in vivo or are predicted to have a function are more likely to be conserved across species than average sites. One explanation for these results would be that there is a significant fraction of PTMs that serve no functional role but result simply from the constant evolutionary turn-over of regulatory interactions. We also observed several examples of conservation of the phosphorylation of a protein-protein interface despite the divergence of the actual phosphosite position. Although we cannot rule out that this observation is due to incomplete phosphoproteome coverage, it provides specific examples of possible neutral variation of PTM regulation for future studies.
The development of PTM functional classifiers will improve our fundamental understanding of cellular signaling but could also, in the long term, have practical biomedical applications. This is particularly relevant for the study of disease since it has been recently shown that disease causing mutations are significantly associated with PTM sites (Li et al., 2010). This resource can aid in the understanding of how mutations associated with PTMs result in disease.
Experimental Procedures
Post-translational modification sites and computational methods
All of the sites compiled are provided in a searchable website (ptmfunc.com). Protein sequences, protein identifiers and ortholog assignments were obtained from the Inparanoid database (inparanoid.sbc.su.se, version 7). For the comparative analysis we considered only 1-to-1 orthologs with Inparanoid confidence scores greater than 90%. The total number of human to species ortholog pairs used in this studied are listed in Table S5. Protein sequence alignments were done with muscle version 3.6 (Edgar, 2004). Additional information on the computational methods is provided in the Extended Experimental.
Immunoprecipitation, mass spectrometry and ubiquitylation site identification
S. cerevisiae Sub592 (containing a HisTag modified ubiquitin) and Sub62 were grown separately in YPD and harvested during mid-log phase (OD600 ~1.0). Protein extract from Sub592 cells (~40mg) was enriched for ubiquitylated proteins via HisTag. Sub62 proteins and half of the ubiquitin enriched Sub592 protein were digested overnight with trypsin, while the remaining half was digested with ArgC. After enzymatic digestion, all three samples were desalted and enriched for diGly containing peptides using a polyclonal antibody as previously described (Cell Signaling, Technology, Danvers, MA) (Kim et al., 2011) and analyzed in an Orbitrap Velos mass spectrometer. Raw files were searched with Sequest against the target-decoy S. cerevisiae protein sequence database. Peptide spectral matches were filtered to a 1% false-discovery rate at the peptide and protein level and diGly sites were localized using a version of the Ascore algorithm that can accept any post-translational modification (Ascore > 13) (Beausoleil et al., 2006). See Extended Experimental Procedures for more details.
Protein complementation assays and Co-IP
Skp1 and Met30 were fused to fragments F1 and F2, respectively, of a split cytosine deaminase by gap-repair cloning. Point mutants were constructed using PCR with site directed oligonucleotides. Protein complementation was assayed as previously described (Ear and Michnick, 2009; Michnick et al., 2010). For Co-IP experiments, yeast cells expressing endogenous Myc-tagged Met30 were transformed with a plasmid expressing a Flag-tagged SKP1 S162A or S162D under control of the Gal promoter and selected for Leucine auxotrophy. Detailed information is available in Extended Experimental Procedures.
DNA, yeast strains
S.cerevisiae strains used were as follows: the ssa1 temperature sensitive strain (mat alpha leu2 trp1 ura3 ade2 his3 lys2, ssa1–45BKD, ssa2::LEU2, ssa3::TRP1, ssa4::LYS2) and was a gift from Betty Craig. The Δssa1::KanMX4 Δssa2::NAT was generated by direct replacement of the SSA2 coding region with the NatMX4 cassette in the single deletion strain Δssa1::KanMX4 and confirmed by PCR.
Drop test assay
Cells were grown overnight in selective medium and then diluted to OD600nm O.4. Cells were grown for another 3 hours and then diluted to OD600nm of 0.1. This sample was then subjected to 10-fold serial dilutions. 10μl of each dilution was then spotted onto –URA plates and allowed to grow at 30°C, 33°C and 37°C for 2 days.
Lysate preparation and ribosome fractionation
200 ml yeast in exponential growth was treated with 100μg/ml of cycloheximide, harvested, washed with cold water, resuspended and frozen as drops in liquid nitrogen. The cell lysate was loaded on a 12 ml 7%–47% sucrose gradient and centrifuged for 150 minutes at 39000 rpm at 4°C. Fractions were collected using a UA/6 detector. The fractions were TCA precipitated and separated by SDS-PAGE and subjected to immunoblot analysis. Detailed protocol available in Extended Experimental Procedures.
Luciferase in vivo refolding assay
The ssa1–45 ts cells were transformed with firefly luciferase and a plasmid driving the expression of the wild type SSA1 or the phosphomutants. After growth at 30°C, the cells were shifted to 44°C for 1 hour, which causes the heat-induced denaturation of luciferase. Cycloheximide was added to 10mg/ml 15 minutes before the end of the heat shock to prevent further expression of luciferase. Cells were then transferred to 30°C to recover. At different time point during the recovery, aliquots were taken, centrifuged and frozen in liquid nitrogen. The luciferase acitivity was measured and recovery is expressed as a percentage of the activity before heat shock treatments.
Microscopy and aggregation assay
The ssa1–45 ts strain was transformed with the different SSA1 mutants as well as with the Gal-Ubc9-2-GFP construct. Cells were grown overnight at 30°C and then diluted to OD600nm 0.3 and induced with 2% galactose for 6 hours. Cells were then shifted to 37°C for 30 minutes to induce the misfolding of Ubc9-2-GFP. The formation of Ubc9-2-GFP puncta was then examined by fluorescence microscopy.
Supplementary Material
Article Highlights.
We compiled a resource of nearly 200,000 PTMs across 11 eukaryotic species
Computational methods were developed to assign function to PTMs
Conservation studies identify regulatory regions within domain families
Evolutionary analysis suggest that only a fraction of PTMs are functionally important
Acknowledgments
We thank Stephen Michnick and Peter Kaiser for strains and plasmids, Betty Craig for the ssa1–45 ts cells, Stéphanie Escusa and Jonathan Warner for reagents. This work was supported by grants from the National Institutes of Health (AI090935, GM082250, GM084448, GM084279, GM081879 (NJK); GM55040, GM062583, GM081879, PN2 EY016546 (WAL); GM56433 (JF); DP5 OD009180 (JSF); and RR01614 (AB)), the Howard Hughes Medical Institute (WAL) and the Packard Foundation (WAL). PB is supported by the Human Frontier Science Program, JSF is a QB3@UCSF Fellow and NJK is a Searle Scholar and a Keck Young Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, et al. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese V, Yam AY, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. doi: 10.1016/j.cell.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba AN, Moses AM. Evolution of characterized phosphorylation sites in budding yeast. Mol Biol Evol. 2010;27:2027–2037. doi: 10.1093/molbev/msq090. [DOI] [PubMed] [Google Scholar]
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Beltrao P, Serrano L. Specificity and evolvability in eukaryotic protein interaction networks. PLoS Comput Biol. 2007;3:e25. doi: 10.1371/journal.pcbi.0030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrao P, Trinidad J, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame A, Krogan N. Evolution of Phosphoregulation: Comparison of Phosphorylation Patterns across Yeast Species. PLoS Biology. 2009;7:1–12. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- DerMardirossian C, Schnelzer A, Bokoch GM. Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase. Mol Cell. 2004;15:117–127. doi: 10.1016/j.molcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Ear PH, Michnick SW. A general life-death selection strategy for dissecting protein functions. Nat Methods. 2009;6:813–816. doi: 10.1038/nmeth.1389. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve PO, Chang Y, Samaranayake M, Upadhyay AK, Horton JR, Feehery GR, Cheng X, Pradhan S. A methylation and phosphorylation switch between an adjacent lysine and serine determines human DNMT1 stability. Nat Struct Mol Biol. 2011;18:42–48. doi: 10.1038/nsmb.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- Holt L, Tuch B, Villén J, Johnson A, Gygi S, Morgan D. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton LE, James P, Craig EA, Hensold JO. The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J Biol Chem. 2001;276:14426–14433. doi: 10.1074/jbc.M100266200. [DOI] [PubMed] [Google Scholar]
- Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, Aebersold R, Loewith R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23:1929–1943. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Jonkers W, Rep M. Lessons from fungal F-box proteins. Eukaryot Cell. 2009;8:677–695. doi: 10.1128/EC.00386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P, Su NY, Yen JL, Ouni I, Flick K. The yeast ubiquitin ligase SCFMet30: connecting environmental and intracellular conditions to cell division. Cell Div. 2006;1:16. doi: 10.1186/1747-1028-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Levy ED, Michnick SW. Weak functional constraints on phosphoproteomes. Trends Genet. 2009;25:193–197. doi: 10.1016/j.tig.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Hogues H, Mallick J, Sellam A, Nantel A, Whiteway M. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 2010;8:e1000329. doi: 10.1371/journal.pbio.1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Iakoucheva LM, Mooney SD, Radivojac P. Loss of post-translational modification sites in disease. Pac Symp Biocomput. 2010:337–347. doi: 10.1142/9789814295291_0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienhard GE. Non-functional phosphorylations? Trends Biochem Sci. 2008;33:351–352. doi: 10.1016/j.tibs.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V. A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein-protein interactions in living cells. Methods Enzymol. 2010;470:335–368. doi: 10.1016/S0076-6879(10)70014-8. [DOI] [PubMed] [Google Scholar]
- Moses AM, Landry CR. Moving from transcriptional to phospho-evolution: generalizing regulatory evolution? Trends Genet. 2010;26:462–467. doi: 10.1016/j.tig.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Moses AM, Liku ME, Li JJ, Durbin R. Regulatory evolution in proteins by turnover and lineage-specific changes of cyclin-dependent kinase consensus sites. Proc Natl Acad Sci U S A. 2007;104:17713–17718. doi: 10.1073/pnas.0700997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan A, Jacobson MP. Computational studies of protein regulation by post-translational phosphorylation. Curr Opin Struct Biol. 2009;19:156–163. doi: 10.1016/j.sbi.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Neduva V, Russell RB. Linear motifs: evolutionary interaction switches. FEBS Lett. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Paolinelli R, Mendoza-Maldonado R, Cereseto A, Giacca M. Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat Struct Mol Biol. 2009;16:412–420. doi: 10.1038/nsmb.1583. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8:R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Soroka J, Wandinger SK, Mausbacher N, Schreiber T, Richter K, Daub H, Buchner J. Conformational Switching of the Molecular Chaperone Hsp90 via Regulated Phosphorylation. Mol Cell. 2012;45:517–528. doi: 10.1016/j.molcel.2011.12.031. [DOI] [PubMed] [Google Scholar]
- Soufi B, Kelstrup CD, Stoehr G, Frohlich F, Walther TC, Olsen JV. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol Biosyst. 2009;5:1337–1346. doi: 10.1039/b902256b. [DOI] [PubMed] [Google Scholar]
- Stark C, Su TC, Breitkreutz A, Lourenco P, Dahabieh M, Breitkreutz BJ, Tyers M, Sadowski I. PhosphoGRID: a database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database (Oxford) 2010;2010:bap026. doi: 10.1093/database/bap026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Ceol A, Aloy P. 3did: identification and classification of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2011;39:D718–723. doi: 10.1093/nar/gkq962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Tan C, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner M, Jørgensen C, Bader G, Aebersold R, Pawson T, Linding R. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci Signal. 2009;2:ra39. doi: 10.1126/scisignal.2000316. [DOI] [PubMed] [Google Scholar]
- Tsong AE, Tuch BB, Li H, Johnson AD. Evolution of alternative transcriptional circuits with identical logic. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- van Dam TJ, Snel B. Protein complex evolution does not involve extensive network rewiring. PLoS Comput Biol. 2008;4:e1000132. doi: 10.1371/journal.pcbi.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Neutralism and selectionism: a network-based reconciliation. Nat Rev Genet. 2008;9:965–974. doi: 10.1038/nrg2473. [DOI] [PubMed] [Google Scholar]
- Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111 013284. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandinger SK, Suhre MH, Wegele H, Buchner J. The phosphatase Ppt1 is a dedicated regulator of the molecular chaperone Hsp90. EMBO J. 2006;25:367–376. doi: 10.1038/sj.emboj.7600930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Wagner SA, Horn H, Henriksen P, Liu WR, Olsen JV, Jensen LJ, Choudhary C. Proteome-wide mapping of the Drosophila acetylome demonstrates a high degree of conservation of lysine acetylation. Sci Signal. 2011;4:ra48. doi: 10.1126/scisignal.2001902. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Gregoire S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol Cell. 2006;23:779–786. doi: 10.1016/j.molcel.2006.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.