Abstract
Phytopathogens can manipulate plant hormone signaling to access nutrients and counteract defense responses. Pseudomonas syringae produces coronatine, a toxin that mimics the plant hormone jasmonic acid isoleucine and promotes opening of stomata for bacterial entry, bacterial growth in the apoplast, systemic susceptibility and disease symptoms. We examined the mechanisms underlying coronatine-mediated virulence and show that coronatine activates three homologous NAC transcription factor (TF) genes, ANAC019, ANAC055 and ANAC072, through direct activity of the TF, MYC2. Genetic characterization of NAC TF mutants demonstrates that these TFs mediate coronatine-induced stomatal reopening and bacterial propagation in both local and systemic tissues by inhibiting the accumulation of the key plant immune signal salicylic acid (SA). These NAC TFs exert this inhibitory effect by repressing ICS1 and activating BSMT1, genes involved in SA biosynthesis and metabolism, respectively. Thus, a signaling cascade by which coronatine confers its multiple virulence activities has been elucidated.
INTRODUCTION
In the constant battle for survival, pathogens evolved diverse mechanisms to facilitate their virulence in the hosts. Many phytopathogens are able to manipulate plant hormone signaling pathways to gain access to nutrients or to counteract plant defense responses. The bacterial pathogen Agrobacterium tumefaciens, through T-DNA-mediated production of the plant hormones auxin and cytokinin, is known to induce the formation of crown galls, where the plant primary metabolites are used to produce opines serving as carbon and nitrogen sources for the pathogen (Escobar and Dandekar, 2003). Another well-known example of hormone manipulation involves the phytotoxin coronatine (COR), which is produced by various strains of Pseudomonas syringae (Mittal and Davis, 1995). COR enhances bacterial growth and disease symptom development and promotes systemic susceptibility (Brooks et al., 2005; Cui et al., 2005; Mittal and Davis, 1995). Structurally similar to the plant hormone jasmonic acid isoleucine (JA-Ile), it has been used as a JA analog in making many essential discoveries in the JA signaling pathway (Browse, 2009).
The Arabidopsis JA perception mutant coi1 (coronatine insensitive 1) was initially identified in a screen for insensitivity to COR (Feys et al., 1994). The coi1 mutant is defective in many JA-induced responses such as root elongation inhibition and wound response. COI1 encodes an F-box protein of the Skp/Cullin/F-box complex (SCFCOI1), which is involved in ubiquitin-mediated degradation of substrates, the JAZ (jasmonate ZIM-domain) proteins (Chini et al., 2007; Thines et al., 2007; Xie et al., 1998). The COI1-JAZ receptor-substrate complex formation is facilitated by JA-Ile or COR (Katsir et al., 2008; Yan et al., 2009). Since JAZ proteins are normally associated with the JA signaling transcription factors (TFs) such as MYC2, degradation of JAZ proteins leads to the release of these TFs and transcription activation of JA/COR-responsive genes (Katsir et al., 2008; Melotto et al., 2008).
For defense against Pseudomonas syringae, the plant immune signal salicylic acid (SA) plays an essential role. However, some plant hormones, such as JA, can counteract the SA-mediated defense to fine-tune the immune response through signaling crosstalk (Grant and Jones, 2009; Kunkel and Brooks, 2002; Spoel and Dong, 2008). In the COR-insensitive coi1 mutant, SA levels are elevated and resistance to Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) and Pseudomonas syringae pv. maculicola ES4326 (Psm ES4326) is enhanced (Kloek et al., 2001). The JA signaling mutant, myc2/jin1, which is less sensitive to COR, also exhibits a similar phenotype (Laurie-Berry et al., 2006; Nickstadt et al., 2004). Since COR can activate the JA signaling pathway, it is hypothesized to enhance bacterial virulence by suppressing SA-mediated defense through crosstalk. Supporting this hypothesis, virulence can be restored to the COR-deficient Pst DC3000 mutant (Pst DC3000 cor−) when inoculated into the Arabidopsis mutant plants defective in SA production (Brooks et al., 2005). However, the downstream signal transduction pathway by which COR suppresses SA-mediated immunity remains largely unknown. Block et al. showed that wild-type Arabidopsis plants did not accumulate more SA in response to Pst DC3000 cor− compared to Pst DC3000 (Block et al., 2005), whereas in other studies COR was found to suppress SA accumulation (de Torres Zabala et al., 2009; Uppalapati et al., 2007). Moreover, COR targets more than one cellular function. Melotto et al. discovered that COR was essential for the reopening of the stomata, which were closed upon Pst DC3000 infection, to facilitate bacterial entry into the apoplast (Melotto et al., 2006). However, even when infiltrated directly into the apoplast, the Pst DC3000 cor− mutant still exhibited a reduced growth rate compared to the toxin-producing strain, revealing a role of COR in inhibiting plant defense in the apoplast (Brooks et al., 2004; Zeng and He, 2010). Besides affecting bacterial growth, COR contributes to the chlorotic (yellow) disease symptom development that is independent of bacterial growth (Mittal and Davis, 1995). While Pst DC3000 cor− grew to a similar level as Pst DC3000 in the SA-deficient mutant, the former induced a less chlorotic symptom than the latter (Brooks et al., 2005). Moreover, COR not only functions locally in the infected tissue but also enhances bacterial virulence systemically (Cui et al., 2005). Despite these studies, it is still largely unknown what downstream host signaling components are targeted by the COR-COI1-JAZ-MYC2 module and whether there is a common signaling pathway leading to these various COR effects.
Here we report the identification of a signaling cascade by which COR suppresses SA accumulation. We discovered three homologous NAC (petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2) TFs, the expression of which is induced by COR directly through MYC2. These three NAC TFs function not only in COR-mediated stomatal reopening, but also in promoting bacterial growth in the apoplast and systemic susceptibility. We further demonstrate that these TFs suppress SA accumulation by regulating genes involved in SA synthesis and metabolism. We propose that the same signaling cascade is responsible for the virulence effects of COR in stomata, apoplast, as well as systemic tissue.
EXPERIMENTAL PROCEDURES
Plants and bacterial strains
Arabidopsis plants were grown on soil in 22 °C under 16/8 hour day/night cycle. The myc2 (SALK_017005), anac019 (SALK_096295), anac055 (SALK_014331) and anac072 (SALK_083756) mutants were obtained from the Arabidopsis Biological Resource Center. Pst DC3000, Psm ES4326 and a COR deficient strain, Psm ES4326 cor−, were grown on King’s medium B plates with corresponding antibiotics at 30 °C for 2 days (Cui et al., 2005; Dong et al., 1991).
Gene expression analysis
For chemical treatment, plants were grown on vertical MS plates for 12 days and then transferred to MS plates with either 5 µM ABA (Sigma), 1 µM COR (Sigma), or no additional chemicals as a control. The rosette leaves were collected for RNA extraction after 24 hours. For bacterial infection, three-week-old leaves were pressure-infiltrated with either 10 mM MgCl2, Psm ES4326, or Psm ES4326 cor−. OD600nm = 0.01 of bacteria was inoculated unless specified. Statistical analyses were performed using Student’s t test of the differences between two means (*, p<0.05; **, p<0.01; ***, p<0.001). Primers used for qRT-PCR are listed in supplemental information.
Chromatin immuno-precipitation
For MYC2-GFP ChIP, untreated three-week-old leaves were harvested. For ANAC019-GFP ChIP, leaves were pressure-infiltrated with Psm ES4326 (OD600nm = 0.01) and harvested after 24 hours. ChIP was performed as previously described (Busch et al., 2010) and the samples were analyzed by qPCR. The primers for qPCR are listed in supplemental information. Input samples were first used to normalize the results. Fold difference was then calculated by taking ratios between normalized results from wild-type plants and from MYC2-GFP or ANAC019-GFP plants. Finally, the fold enrichment was calculated as the ratio between the probes and the corresponding negative control.
Stomatal analysis
Stomatal assay was performed as described previously (Zeng and He, 2010). For chemical treatment, 15 µM of abscisic acid (Sigma) and 1 ng/µl of COR (purchased from C. Bender, Oklahoma State University) were used.
SA, SAG and MeSA measurement
Three-week-old leaves were pressure-infiltrated with Psm ES4326 or Psm ES4326 cor− (OD600nm = 0.01) and harvested at specified time points. SA and SAG were extracted and measured by HPLC as previously described (Liu et al., 2010). MeSA was extracted and measured using GC/MS as previously described (Liu et al., 2010). Three replicates were taken for each data point. Statistical analyses were performed using Student’s t test of the differences between two means.
Pseudomonas syringae infection
For pressure infiltration, three-week-old plants were pressure-infiltrated using a needleless syringe with Psm ES4326 (OD600nm = 0.001) and eight plants were assayed for each data point. Bacterial growth was measured after three days as described previously (Durrant et al., 2007). For dip-inoculation, four-week-old plants were dipped into a solution of Psm ES4326 (OD600nm = 0.2) with 0.02% Silwet L-77 and covered with clear plastic lids for one day. The infected leaves were surface-sterilized with 15% H2O2 and washed twice before homogenized to assay the bacterial growth. Statistical analyses were performed on means of log-transformed data using one-way ANOVA and Bonferroni's multiple comparison tests or Student’s t test (*, p<0.05; **, p<0.01; ***, p<0.001).
For the COR-induced systemic susceptibility assay, two lower leaves were pressure-infiltrated with Psm ES4326 (OD600nm = 0.2) or Psm ES4326 cor− (OD600nm = 0.2). Three days later, the infected leaves were removed and two upper leaves were pressure-infiltrated with Psm ES4326 (OD600nm = 0.0002). Colony forming unit was determined three days later. Student’s t test was used to compare the means of log-transformed data (*, p<0.05; **, p<0.01; ***, p<0.001).
RESULTS
Pseudomonas syringae induces the expression of three NAC TF genes through the JA and ABA signaling pathways and through COR production
To identify the signaling cascade responsible for Pseudomonas virulence promoted by the phytotoxin COR, we first searched for TF genes that are inducible by both bacterial infection and treatment of the plant hormone JA, as COR is a known mimic of JA-Ile. We found three homologous NAC family TF genes, ANAC019, ANAC055 and ANAC072, as plausible candidates since they were induced by Pst DC3000 infection as well as by methyl-JA treatment (de Torres-Zabala et al., 2007; Ooka et al., 2003; Tran et al., 2004). These NAC genes were also induced in our “Pseudomonas half leaf injection” microarray experiment (Figure S1A; NASCARRAYS-168) where the virulent Psm ES4326 bacteria was infiltrated into one half of the Arabidopsis leaf and gene expression was examined in the uninfected half. Since COR can trigger systemic susceptibility in plants (Cui et al., 2005), the induction of NAC TFs in the uninfected tissue suggests that they are possible host targets for this COR function. Interestingly, treatment with another plant hormone, abscisic acid (ABA), can also increase the expression of these three NAC TF genes (Tran et al., 2004). Biosynthesis of ABA and ABA-mediated gene expression are rapidly induced by Pst DC3000 through effector proteins delivered into the host cells by the type III secretion system (de Torres-Zabala et al., 2007), suggesting that the three NAC TF genes could be targets of not only COR but also type III effectors.
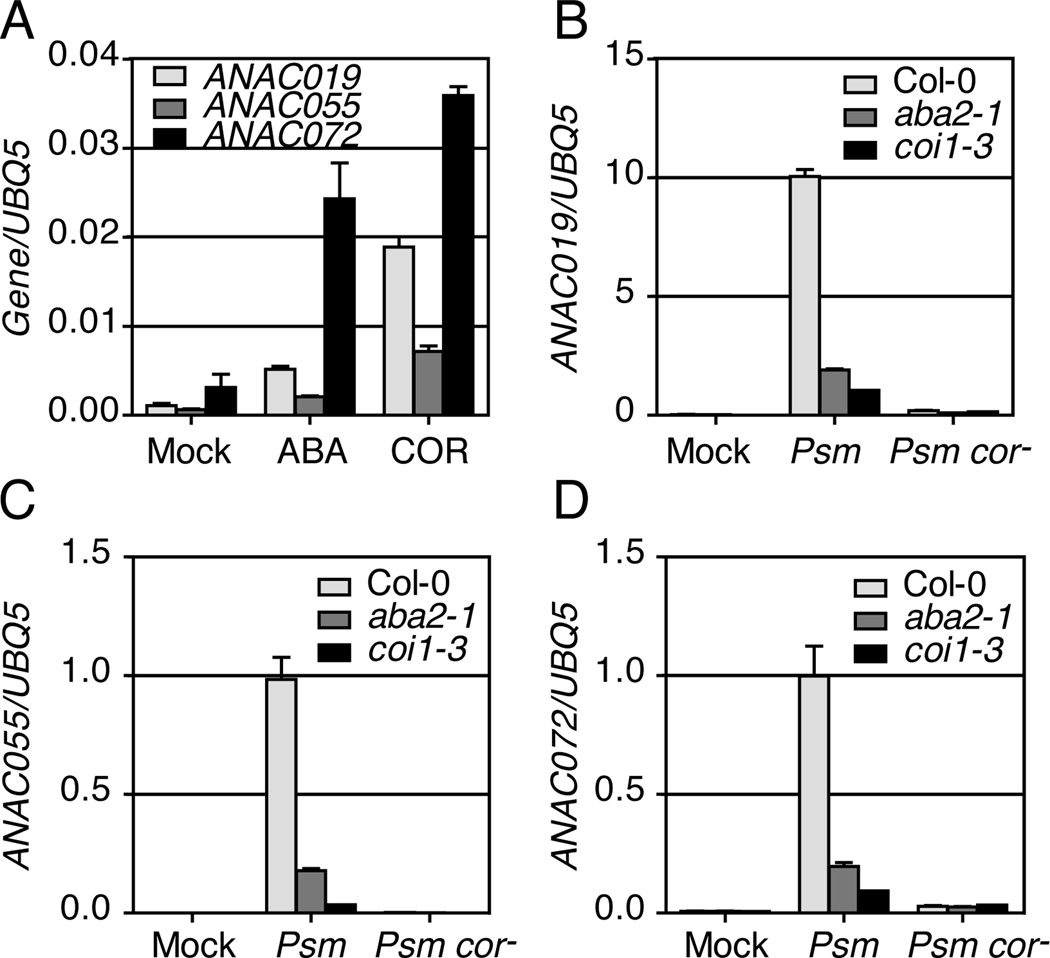
The microarray data were further supported by qRT-PCR analysis. As shown in Figure 1, COR and ABA treatment as well as Psm ES4326 infection all increased the expression of these NAC genes. The Psm ES4326-induced expression was diminished in the JA receptor mutant coi1-3 and the ABA biosynthesis mutant aba2-1 (Figure 1B–D), indicating that Psm ES4326 induces the three NAC genes through the JA and ABA signaling pathways. In wild-type plants, the COR-deficient mutant Psm ES4326 cor− (Cui et al., 2005) caused little increase in transcription of the NAC genes, suggesting that Psm ES4326 induces these genes mainly through COR.
Figure 1.
Psm ES4326 induces the expression of ANAC019, ANAC055 and ANAC072 through the ABA- and JA-signaling pathways as well as COR. (A) ANAC019, ANAC055 and ANAC072 transcripts were measured in wild type (Col-0) treated with either ABA, COR or mock. (B-D) Col-0 plants, the ABA-deficient mutant aba2-1 and the JA-signaling mutant coi1-3 were pressure-infiltrated with the mock solution (Mock), Psm ES4326 (Psm) or Psm ES4326 cor− (Psm cor−) and samples were collected 24 hours later. Error bars represent the standard deviation of three technical replicates. Each experiment was repeated at least three times with similar results. See also Figure S1.
Pathogen-produced COR up-regulates the expression of ANAC019, ANAC055 and ANAC072 directly through MYC2
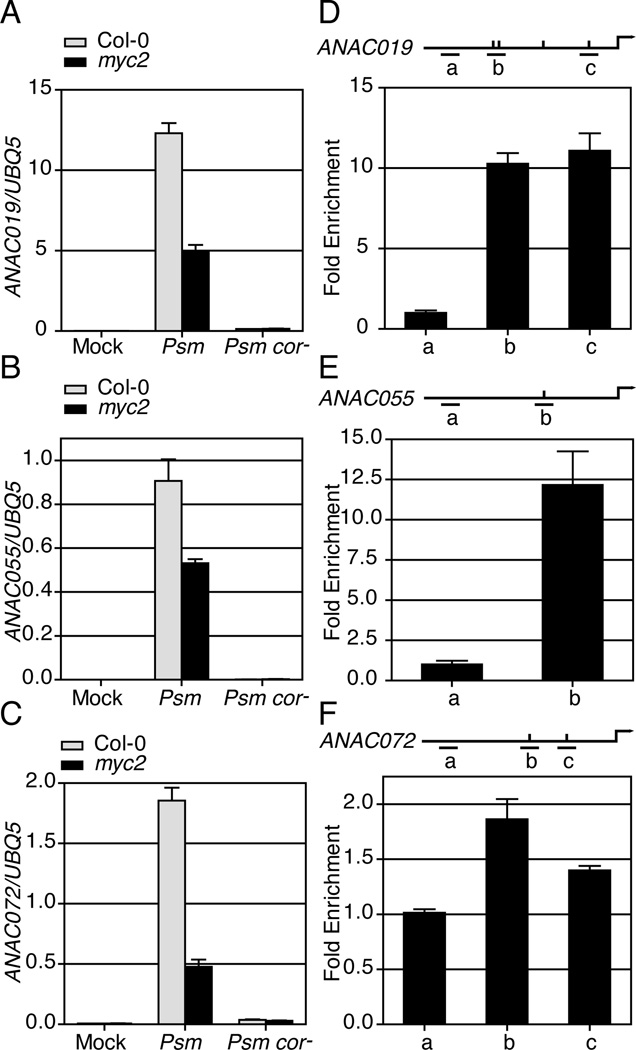
To study the mechanism by which Psm ES4326-generated COR induces ANAC019, ANAC055 and ANAC072, we analyzed the time-course expression patterns of these three genes upon Psm ES4326 infection and found them to be highly correlated using linear regression (Figure S1B–C). This suggests a common regulatory mechanism that controls the three NAC genes. The promoters of these three NAC genes were then searched for conserved cis-elements. Based on the Athena online analysis tool (O'Connor et al., 2005), the CACGTG G-box element was enriched in these three promoters (p-value = 0.0061). Since this G-box element is the preferential binding site of the TF MYC2 (Dombrecht et al., 2007), the three NACs might be regulated by MYC2. This is consistent with the observation that induction of ANAC019 and ANAC055 by both MeJA and ABA was dependent on MYC2 (Bu et al., 2008). To test whether induction of ANAC019, ANAC055 and ANAC072 by Psm ES4326 is also MYC2-dependent, we performed the infection experiment in the myc2 mutant and found significant reduction in expression of all three genes (Figure 2A–C). These results suggest that MYC2, a known TF released after COR-triggered degradation of JAZ, is involved in Psm ES4326-induced ANAC019, ANAC055 and ANAC072 expression. However, this induction was not completely abolished in the myc2 mutant, probably due to the intact functions of MYC3 and MYC4, which are also JAZ-bound TFs that are activated in response to JA (Fernandez-Calvo et al., 2011).
Figure 2.
MYC2 induces the expression of ANAC019, ANAC055 and ANAC072 by directly interacting with their promoters. (A-C) Transcript levels of ANAC019, ANAC055 and ANAC072 were measured in Col-0 and myc2 in response to mock, Psm ES4326 or Psm ES4326 cor−. Error bars represent standard deviation of three technical replicates. (D-F) 35S:MYC2-GFP transgenic plants were used for ChIP experiments. Long horizontal lines represent the promoters. Ticks above the lines represent putative MYC2 binding sites. Short horizontal lines show the regions where different qPCR primers amplify. Error bars represent standard error of three technical replicates. Each experiment was repeated at least three times with similar results. See also Figure S2 and Table S1.
We next tested whether MYC2 TF directly control transcription of the three NAC genes using chromatin immuno-precipitation (ChIP) experiments performed in a transgenic Arabidopsis line constitutively expressing the MYC2-GFP fusion protein (Figure S2). For the promoter of each NAC gene, several DNA regions were analyzed to cover all, but one (1234bp upstream of the transcription start site of ANAC019), potential MYC2-binding sites (Table S1). In addition, promoter regions without the binding sites were used as negative controls. As shown in Figure 2D and 2E, all DNA segments containing the potential MYC2-binding sites were enriched in the ANAC019 and ANAC055 promoters. For the ANAC072 promoter, enrichment was found in one of the two potential binding sites (Figure 2F). These results show that MYC2 activates expression of the three NAC genes by direct interaction.
ANAC019, ANAC055 and ANAC072 are required for COR-regulated stomatal reopening, but not ABA-mediated stomatal closure
To further investigate the functions of ANAC019, ANAC055 and ANAC072 in plant defense, homologous T-DNA insertion mutants for each gene were isolated. The anac019 mutant (SALK_096295) has an insertion in the second exon of ANAC019, anac055 (SALK_014331) in the third exon of ANAC055 and anac072 (SALK_083756) in the first exon of ANAC072. Since ANAC019, ANAC055 and ANAC072 have high amino acid sequence similarities and are able to bind to the same cis-element (Tran et al., 2004), they are likely to have functional redundancy. In anticipation of this possibility, we generated double mutants of all combinations and a triple mutant by crossing. The anac019, anac055, anac072 triple mutant (the nac triple mutant) was verified by measuring the transcripts of these three genes (Figure S3A).
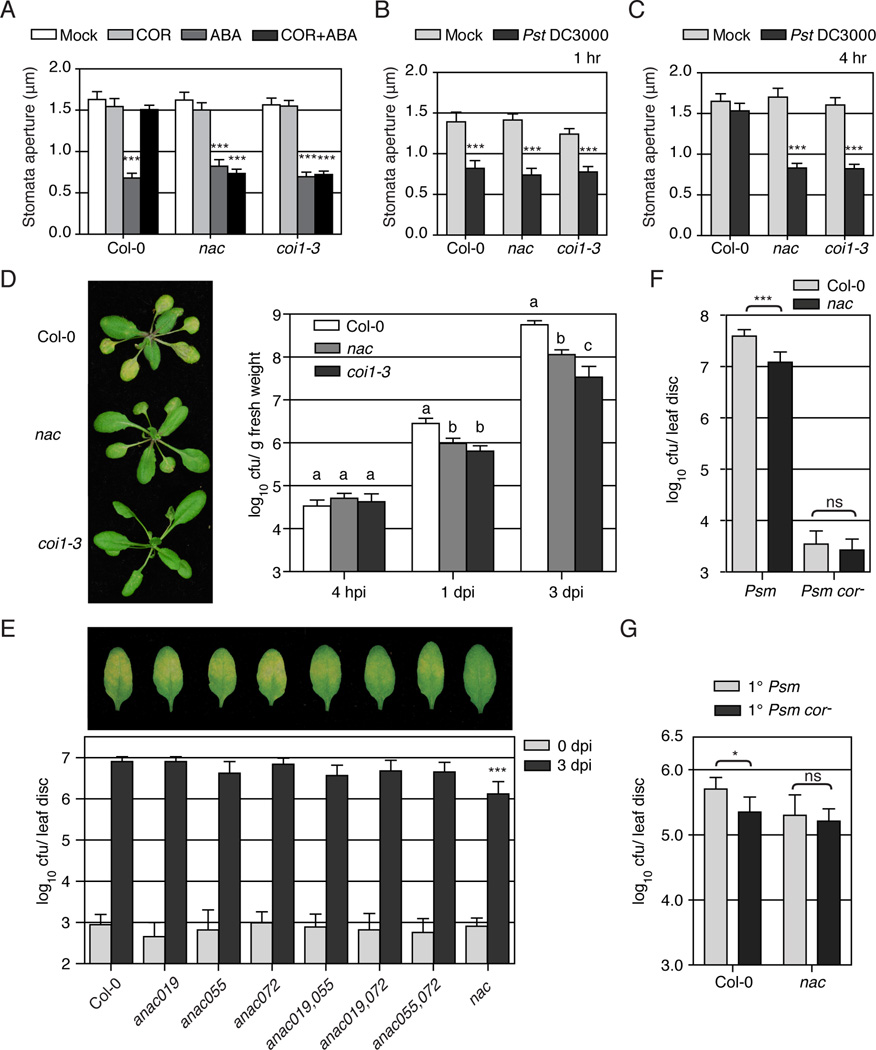
It is known that ABA signaling is responsible for pathogen-triggered stomatal closure, a key step in plant immune response against Pseudomonas syringae, while COR produced by this pathogen can reopen stomata to facilitate pathogen entry into the plant (Melotto et al., 2006). Since ANAC019, ANAC055 and ANAC072 are induced by both ABA and COR, we were interested in their roles in ABA-mediated stomatal closure and COR-mediated stomatal reopening. As shown in Figure 3A, in wild-type plants (Col-0), ABA treatment induced stomatal closure and COR inhibited this ABA effect (Melotto et al., 2006). In response to ABA, the nac triple mutant closed the stomata, demonstrating that these three NACs are not required for ABA-mediated stomatal closure. However, COR could not inhibit ABA-induced stomatal closure in the nac triple mutant, suggesting the requirement of ANAC019, ANAC055 and ANAC072 for this COR function.
Figure 3.
ANAC019, ANAC055 and ANAC072 are involved in COR-triggered virulence in stomata, apoplast and systemic tissue. (A) Stomata aperture from leaf peels of Col-0, the nac triple mutant and the coi1-3 mutant treated with mock, COR, ABA or combination of COR and ABA for 1 hour. (B–C) Stomatal closure response in leaf peels of Col-0, the nac triple mutant and the coi1-3 mutant after 1-hour incubation (B) or 4-hour incubation (C) with either mock or Pst DC3000. Error bars represent standard error. Each experiment was repeated at least twice with similar results. Student’s t test was used to compare means between mock and specific treatment of the same genotype. (D) Col-0, the nac triple mutant and the coi1-3 mutant were dip-inoculated with Psm ES4326. Pictures of the disease symptoms were taken at 3 dpi and bacterial growth was measured at 4 hpi, 1 and 3 dpi. Student’s t test was used to compare the log-transformed data of different genotypes at the same time point. (E) Col-0, the single, double mutants of ANAC019, ANAC055 and ANAC072 and the nac triple mutant were pressure-infiltrated with Psm ES4326. Photographs of the disease symptoms were taken 3 dpi. Bacterial growth was measured at the same time. Bonferroni's multiple comparison test was preformed to compare the log-transformed data of each mutant to that of Col-0. (F) Col-0 and the nac triple mutant were pressure-infiltrated with Psm ES4326 or Psm ES4326 cor− and assayed 3 dpi. (G) Psm ES4326 growth in systemic leaves with pre-inoculation with Psm ES4326 or Psm ES4326 cor−. Error bars represent 95% confidence intervals. Experiments were repeated for at least twice with similar results. ns, no significant difference. See also Figure S3.
To further confirm the function of ANAC019, ANAC055 and ANAC072 in stomatal regulation, we incubated Pst DC3000 with leaf peels from wild-type plants, the nac triple mutant and the coi1-3 mutant. Previous studies have shown that Pst DC3000 causes a reduction in the stomatal aperture in wild-type plants one hour after bacterial incubation. However, stomata reopen 4 hours after bacterial incubation and this reopening is dependent on COR (Melotto et al., 2006; Zeng and He, 2010). We found that the presence of Pst DC3000 caused stomatal closure in all three genotypes (Figure 3B). But the coi1-3 mutant and the nac triple mutant were compromised in the ability to reopen the stomata after 4 hours and the stomatal apertures remained small (Figure 3C). These results suggest that the nac triple mutant, like the coi1 mutant, is defective in COR-triggered stomatal reopening.
To examine the effect of the nac triple mutant on bacterial entry, we dip-inoculated wild type, the nac triple mutant and the coi1-3 mutant plant with Psm ES4326 and observed symptom development and measured the bacterial growth (Figure 3D). At 1 day post inoculation (dpi), the bacterial numbers present in the nac triple mutant and the coi1-3 mutant were similarly lower than that in wild-type plants, probably due to the fact that these mutants are equally defective in early infection steps including reopening of stomata (Figure 3A and C). Interestingly, at 3 dpi, the nac triple mutant exhibited a bacterial titer that was lower than wild type, but higher than the coi1-3 mutant, suggesting that the three NACs may play a partial role in COR-mediated Psm ES4326 growth in apoplast later in the infection.
COR promotes virulence in the apoplast partly through ANAC019, ANAC055 and ANAC072
To validate our hypothesis that the NAC TFs play a role in the COR-mediated virulence in the apoplast, we pressure-infiltrated Psm ES4326 into the nac single, double, and triple mutants to bypass the stomatal regulation. Though the single and double mutants developed similar symptoms as those observed in wild-type plants, the nac triple mutant was more resistant than the wild type (Figure 3E). Furthermore, disease susceptibility could be restored in the nac triple mutant by expressing any one of the three wild-type NAC genes (Figure S3B). These results demonstrate that the three NACs are required for Psm ES4326 virulence in the apoplast. When the COR-deficient Psm ES4326 cor− was pressure-infiltrated, wild-type plants and the nac triple mutant exhibited similar levels of bacterial growth (Figure 3F), supporting the notion that the virulence effect of COR is mediated by the three NACs. However, the growth difference between Psm ES4326 and Psm ES4326 cor− in wild-type plants was much greater than the growth difference of Psm ES4326 in wild-type plants and the nac triple mutant, suggesting the existence of other TFs mediating the COR activity in the apoplast.
ANAC019, ANAC055 and ANAC072 are involved in COR-triggered systemic susceptibility
Another known function of COR in counteracting plant defense is the promotion of plant systemic susceptibility (Cui et al., 2005). The systemic induction of ANAC019, ANAC055 and ANAC072 by Psm ES4326 (Figure S1A) suggests that they play a role. As shown in Figure 3G, wild-type plants pre-inoculated with Psm ES4326 exhibited significantly enhanced susceptibility than those pre-inoculated with Psm ES4326 cor−, displaying COR-mediated systemic susceptibility. However, in the nac triple mutant, this induced systemic susceptibility was abolished, validating our hypothesis.
COR suppresses SA accumulation through ANAC019, ANAC055 and ANAC072
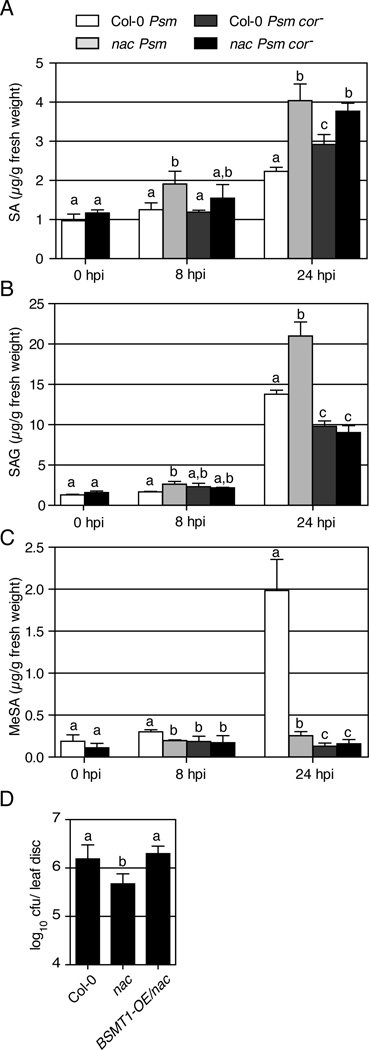
To understand how COR promotes Psm ES4326 virulence through induction of ANAC019, ANAC055 and ANAC072, we sought for their downstream targets. Since SA is an essential signal for resistance against Pseudomonas infection and SA levels in the COR-insensitive coi1 mutant are elevated (Kloek et al., 2001), we first measured the SA levels in wild type and the nac triple mutant in response to Psm ES4326 and Psm ES4326 cor− infection. As shown in Figure 4A, in wild-type plants, Psm ES4326 cor− infection induced a higher level of SA compared to Psm ES4326 infection at 24 hours post inoculation (hpi), displaying the suppressive effect of COR on SA accumulation. In the nac triple mutant, the SA level was dramatically higher than that in the wild type after Psm ES4326 inoculation at both 8 hpi and 24 hpi, indicating that the NAC TFs are principally responsible for COR-mediated repression of SA accumulation. Supporting this conclusion, in the nac triple mutant, the wild type Psm ES4326 and the COR-deficient Psm ES4326 cor− strain triggered comparable levels of SA accumulation.
Figure 4.
COR suppresses SA accumulation through ANAC019, ANAC055 and ANAC072 to promote virulence. Col-0 and the nac triple mutant were pressure-infiltrated with Psm ES4326 or Psm ES4326 cor−. Samples were collected at specified time points to measure the levels of SA (A), SAG (B) and MeSA (C). Error bars represent standard deviation of three replicates. Student’s t test was used to compare two means at the same time point. (D) Psm ES4326 growth was measured in Col-0, the nac triple mutant, and the nac triple mutant overexpressing BSMT1 (BSMT1-OE/nac) as described in Figure 3E. Error bars represent 95% confidence intervals. Experiments were repeated at least twice with similar results. See also Figure S4 and Table S2.
ANAC019, ANAC055 and ANAC072 directly regulate SA biosynthesis and metabolism genes
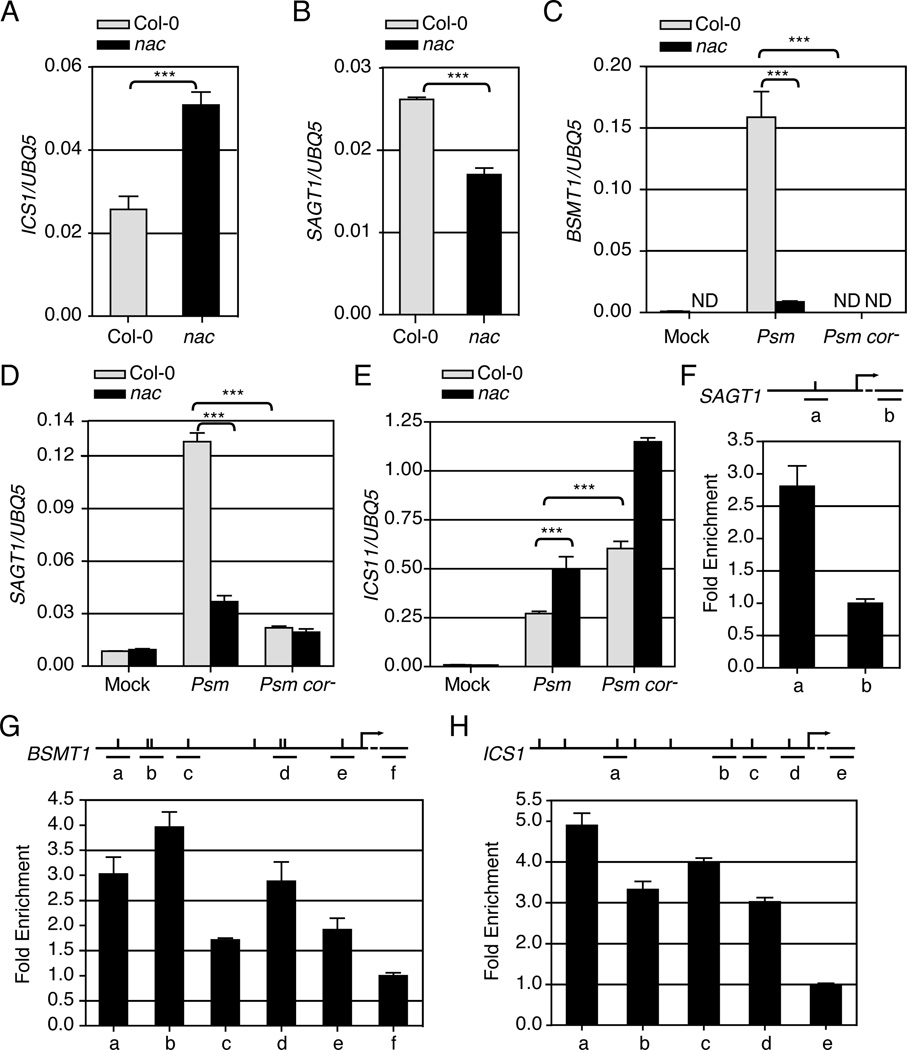
Several Arabidopsis genes are known to affect SA biosynthesis and metabolism (Table S2)(Vlot et al., 2009). We tested the possibility that these genes are direct targets of the three NACs. First, we found six of these SA-related genes to contain at least one copy of the known NAC core-binding site (CACG tetramer) (Tran et al., 2004) in their promoters (Table S2). To determine whether these genes could be regulated by ANAC019, ANAC055 and ANAC072, we analyzed their expression. We found that the basal transcript level of the SA synthesis enzyme isochorismate synthase gene 1 (ICS1) was higher in the nac triple mutant than in the wild type (Figure 5A). In contrast, the SA glucosyl transferase gene 1 (SAGT1), which converts SA to the inactive storage forms SA glucose ester (SGE) and SA O-β-glucoside (SAG)(Dean and Delaney, 2008), had a lower transcript level in the nac triple mutant (Figure 5B). These results suggest the three NAC TFs may decrease SA synthesis, but increase SA storage through transcriptional repression of ICS1 and transcriptional activation of SAGT1, respectively. The basal expression levels of other SA biogenesis genes were not significantly different between the wild type and the nac triple mutant, implying that they are not regulated by the three NACs (Figure S5A–C). The basal transcript level of the SA methyl transferase 1 (BSMT1), which converts SA to the inactive, volatile methyl SA (MeSA)(Chen et al., 2003), was too low for accurate measurement.
Figure 5.
ANAC019, ANAC055 and ANAC072 differentially regulate ICS1, BSMT1 and SAGT1 through direct interaction with their promoters. Basal transcript levels of ICS1 (A) and SAGT1 (B) were measured in three-week-old leaves. Col-0 and the nac triple mutant were pressure-infiltrated with 10 mM MgCl2, Psm ES4326 or Psm ES4326 cor− and were collected after 24 hours to measure the expression of BSMT1 (C), SAGT1 (D), and ICS1 (E). For ICS1, a higher dose of pathogen (OD600nm = 0.02) was used. ND: no detectable signal. Error bars represent standard deviation of three technical replicates. (F-H) ChIP experiments were performed using the Psm ES4326-infected 35S:ANAC019-GFP transgenic plants. Long horizontal lines represent the promoters of SAGT1 (F), BSMT1 (G) and ICS1 (H). Ticks above the lines represent NAC core-binding sites. Short horizontal lines show the regions where different qPCR primers amplify. Error bars represent standard error. Experiments were repeated three times with similar results. See also Figure S5 and Table S3.
We then analyzed the potential target gene expression after Psm ES4326 induction to determine whether they are regulated by COR through the NACs. As shown in Figure 5C, while BSMT1 was induced by Psm ES4326 in wild type, this induction was dramatically compromised in the nac triple mutant. Similarly, induction of SAGT1 by Psm ES4326 was also blocked in the nac triple mutant (Figure 5D). Consistent with our hypothesis that COR represses SA accumulation through NACs, transcript reductions were observed for SAGT1 and BSMT1 in wild-type plants when the CORdeficient Psm ES4326 cor− was inoculated instead of Psm ES4326 (Figure 5C–D). In contrast to the SA metabolic enzyme genes, the expression of the SA synthesis gene ICS1 was further induced in the nac triple mutant and by Psm ES4326 cor− compared to Psm ES4326 (Figure 5E), consistent with our hypothesis that COR suppresses SA accumulation through NAC-mediated repression of ICS1 expression. However, we could only consistently observe elevated ICS1 expression in the nac triple mutant when a higher number of bacteria were infiltrated. This was probably due to the fact that Psm ES4326 not only produces COR, which negatively regulates the ICS1 gene expression, but also carries other signals that positively regulate the plant defense machinery, including the ICS1 gene.
To demonstrate that the three NACs regulate SAGT1, BSMT1 and ICS1 through direct interaction, ChIP experiments were performed. We constructed a transgenic Arabidopsis line that constitutively expresses the ANAC019-GFP fusion protein (Figure S5D) for these experiments. For each promoter, primers were designed to cover the NAC core-binding sites, except for regions where no appropriate primers could be found. Since the NAC core-binding sites are scattered in the entire promoters (Table S3), a region in the coding sequence where there is no NAC core-binding site within a 500-bp stretch of sequence was included to measure the background binding. As shown in Figure 5F–H, the DNA samples precipitated by ANAC019-GFP were enriched in some NAC core-binding sites in the SAGT1, BSMT1 and ICS1 promoters, suggesting a direct role of ANAC019 in regulating expression of these genes. Since ANAC019, ANAC055 and ANAC072 bound to the same cis-element in an Arabidopsis protoplast assay (Tran et al., 2004), it is reasonable to extrapolate that ANAC055 and ANAC072 could also bind to the promoters of SAGT1, BSMT1 and ICS1, and regulate their expression. The binding of these three NACs might lead to induction of the SA metabolic genes SAGT1 and BSMT1 and repression of the SA synthesis gene ICS1, resulting in an overall reduction of the defense signal SA accumulation and enhancement of bacterial virulence.
To further validate the aforementioned hypothesis, we measured the SAG and MeSA levels (the products of SAGT1 and BSMT1 enzymatic activities, respectively) in wild type and the nac triple mutant. In wild-type plant, Psm ES4326 cor− induced lower SAG and MeSA levels than Psm ES4326 at 24 hpi (Figure 4B–C), consistent with the lower SAGT1 and BSMT1 transcript levels found in the wild type in response to Psm ES4326 cor− (Figure 5C–D). However, the nac triple mutant accumulated more SAG than the wild type in response to Psm ES4326 (Figure 4B), despite of the lower SAGT1 transcript level in the nac triple mutant (Figure 5D). This result indicates that the amount of SAGT1 transcript or enzyme might not be a limiting factor in the nac triple mutant in converting SA to SAG and SAGT1 expression is not a significant contributor to the inhibitory effect of the NAC TFs on SA accumulation. In contrast, the MeSA levels in the nac triple mutant were significantly lower than those in the wild type at both 8 hpi and 24 hpi (Figure 4C), suggesting a more significant role of BSMT1 expression regulation in affecting SA level. This conclusion is further supported by the observation that overexpression of BSMT1 in the nac triple mutant (BSMT1-OE/nac) abolished the enhanced resistance of the nac triple mutant (Figure 4D) and restored symptom development (Figure S4). Therefore, COR suppresses SA level through the three NAC TFs mainly by activation of BSMT1 and repression of ICS1 (Figure 6).
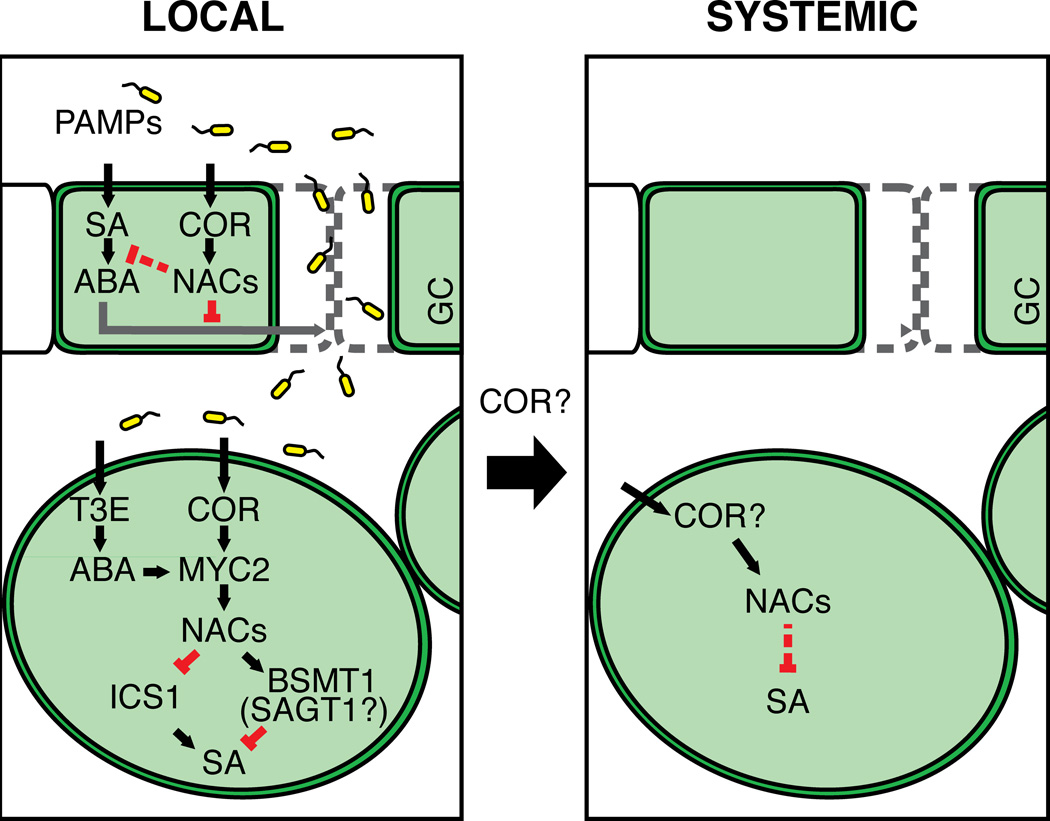
Figure 6.
A model of the signaling cascade mediating the virulence effects of COR. In stomata, PAMPs from the pathogen trigger stomatal closure through SA and ABA. COR produced by Pseudomonas inhibits this effect and reopens the stomata through ANAC019, ANAC055 and ANAC072 (NACs). In apoplast, COR induces the ANAC019, ANAC055 and ANAC072 by the MYC2 TF. Type III effectors (T3E) might also induce the NACs through ABA signaling. These TFs then repress ICS1 and induce BSMT1 and possibly SAGT1 to inhibit SA accumulation and promote bacterial virulence. In systemic tissue, ANAC019, ANAC055 and ANAC072 are induced possibly by translocated COR or a COR-generated signal. GC, guard cell.
DISCUSSION
The bacterial toxin COR is known to cause multiple physiological effects in plants (Brooks et al., 2005; Cui et al., 2005; Melotto et al., 2006; Mittal and Davis, 1995). Our study identified three NAC TFs as targets of COR by which COR exerts its virulence in stomata, apoplast and systemic tissue with different levels of dependency. The three NACs appear to be essential for COR to function in reopening stomata and systemic induced susceptibility, but only partially responsible in COR-mediated growth in the apoplast. Even though we did not specifically examine the effect of the NAC TFs on COR-dependent chlorosis symptom development, we found that restoration of chlorotic symptom in the BSMT1-OE/nac transgenic plant (Figure S4B) correlated with an increase in bacterial growth (Figure 4D). This suggests that NACs may not be required for the symptom promotion function of COR as described by Brook and coworkers (Brooks et al., 2005).
Our study not only identified three TF targets of COR but also elucidated a complete COR-activated signaling cascade. As shown in Figure 6, COR produced by Pseudomonas syringae triggers the SCFCOI1-mediated degradation of the JAZ protein, resulting in the release of the MYC2 TF. MYC2 directly activates the expression of ANAC019, ANAC055 and ANAC072, which then differentially regulate the expression of ICS1, BSMT1 and SAGT1. Repression of the SA biosynthesis gene ICS1 and activation of the SA metabolic gene BSMT1 result in a net reduction in the accumulation of SA and compromised resistance to the pathogen. Though previous studies showed that ANAC019, ANAC055 and ANAC072 had transactivation activity (Bu et al., 2008; Tran et al., 2004), our study suggests that they may also function as transcriptional suppressors, binding to the ICS1 promoter and repressing its expression. This dual transcriptional capability may be achieved by recruiting or interacting with different transcriptional partners.
Our study clearly shows that COR can suppress host defense by inhibiting SA accumulation. We believe that the contradicting data obtained in previous studies resulted from the interplay between pathogen virulence involving COR and induction of host defense during infection (Block et al., 2005). Using dip-inoculation, as reported by Block et al., fewer COR-deficient mutant bacteria could enter the apoplast than the wild type Pst DC3000 to induce SA production, obscuring the effect of COR. Pressure-infiltration used in our study ensured that the same number of wild type and the COR-deficient mutant bacteria were delivered into the apoplast to induce SA production, thus provided a better comparison. This COR-triggered SA suppression, which is mediated by the three NACs, may be the molecular mechanism for COR-mediated virulence in stomata, local and systemic tissue (Figure 6). Zeng et al. has shown that SA biosynthesis is required and acts upstream of ABA signaling to trigger stomatal closure (Zeng and He, 2010). However, it has yet to be determined whether NACs promote stomatal reopening by reducing SA accumulation or through a different mechanism. A method for measuring SA levels specifically in the stomata needs to be developed.
The roles of BSMT1 and SAGT1 in regulating SA level and plant defense have been studied previously. Though single mutants do not exhibit enhanced local resistance, overexpressing either one of them decreases local SA accumulation and increases plant susceptibility to Pseudomonas syringae infection (Liu et al., 2010; Song et al., 2009; Song et al., 2008). Our study further illustrates that BSMT1, and possibly SAGT1, are the plant defense regulatory nodes that can be manipulated by pathogens. With a relatively low Km (16 μM) (Chen et al., 2003), BSMT1 can respond to small increases in SA levels and convert SA to MeSA. Therefore, COR-triggered BSMT1 induction is able to effectively suppress SA accumulation and reduce host defense. Our study, however, does not rule out the possibility that COR also inhibits the expression or the activity of MeSA esterase, which converts MeSA back to SA (Forouhar et al., 2005).
Since COR is a JA-Ile mimic, the signaling cascade we identified for COR signaling may normally mediate the JA-SA crosstalk in plants. The antagonism between JA and SA has been reported, mostly in the direction of SA inhibiting JA (Bostock, 2005). The opposite direction has been shown in a few cases such as the higher SA level observed in the coi1 mutant (Kloek et al., 2001). This JA-SA antagonism is believed to be involved in fine-tuning the plant defense responses based on the types of pathogens encountered (Grant and Jones, 2009; Spoel et al., 2007). Despite its importance, not all signaling components underlying this crosstalk are identified. Since the three NACs can be induced by MeJA treatment, it is reasonable to predict that JA activates the same signaling cascade as COR to suppress SA accumulation, representing one mechanism underlying the JA-SA antagonism.
As mediators of the phytotoxicity of COR, ANAC019, ANAC055 and ANAC072 are hijacked by pathogen to promote virulence. What is the normal physiological function of these TFs? Tran et al. showed that overexpressing any one of them increased drought tolerance (Tran et al., 2004). Fujita et al. found that ANAC072 overexpressing plants are more sensitive to ABA in terms of growth inhibition (Fujita et al., 2004). Consistently, our own study showed that the nac triple mutant was less sensitive in ABA-triggered inhibition on seed germination (data not shown). Moreover, ANAC019 and ANAC055 overexpression lines exhibited higher expression of VSP1 and LOX2 in response to MeJA (Bu et al., 2008), suggesting their function in JA-mediated wound response. Together these results indicate that ANAC019, ANAC055 and ANAC072 are integral parts of the ABA and JA signaling pathways, necessary for regulating plant developmental processes as well as responses to drought stress and wounding. Our study illustrates an example in which the Pseudomonas bacterial pathogen manipulates the plant signaling components, through COR production, to inhibit host defense and promote virulence.
Supplementary Material
HIGHLIGHTS.
-
➢
Coronatine induces NAC transcription factors ANAC019, ANAC055 and ANAC072 through MYC2.
-
➢
The nac triple mutant is defective in multiple coronatine-triggered virulence effects.
-
➢
Coronatine suppresses salicylic acid (SA) accumulation through the NAC TFs.
-
➢
The NAC TFs regulate the expression of SA biosynthesis and metabolism genes.
ACKNOWLEDGEMENTS
We thank Dr. Hironaka Tsukagoshi for providing the MYC2-GFP overexpression plants, Dr. Chuanyou Li for sharing the anac019 anac055 double mutant, Wei Wang for assisting the statistical analysis and Musoki Mwimba for assisting the HPLC analysis. This work was supported by a grant from NSF (IOS – 0929226) to X. Dong, a NIH training pre-doctoral training grant to N. Spivey, grants from NIH (R01AI068718) and DOE (the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science; DE–FG02–91ER20021) to S.Y. He, and a grant from NSF (IOS-0820405) to D. F. Klessig. X. Dong and S.Y. He are Howard Hughes Medical Institute-Gordon and Betty Moore Foundation Investigators.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Block A, Schmelz E, Jones JB, Klee HJ. Coronatine and salicylic acid: the battle between Arabidopsis and Pseudomonas for phytohormone control. Mol Plant Pathol. 2005;6:79–83. doi: 10.1111/j.1364-3703.2004.00265.x. [DOI] [PubMed] [Google Scholar]
- Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annual review of phytopathology. 2005;43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- Brooks DM, Bender CL, Kunkel BN. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol Plant Pathol. 2005;6:629–639. doi: 10.1111/j.1364-3703.2005.00311.x. [DOI] [PubMed] [Google Scholar]
- Brooks DM, Hernandez-Guzman G, Kloek AP, Alarcon-Chaidez F, Sreedharan A, Rangaswamy V, Penaloza-Vazquez A, Bender CL, Kunkel BN. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Mol Plant Microbe Interact. 2004;17:162–174. doi: 10.1094/MPMI.2004.17.2.162. [DOI] [PubMed] [Google Scholar]
- Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol. 2009;60:183–205. doi: 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 2008;18:756–767. doi: 10.1038/cr.2008.53. [DOI] [PubMed] [Google Scholar]
- Busch W, Miotk A, Ariel FD, Zhao Z, Forner J, Daum G, Suzaki T, Schuster C, Schultheiss SJ, Leibfried A, et al. Transcriptional control of a plant stem cell niche. Dev Cell. 2010;18:849–861. doi: 10.1016/j.devcel.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Chen F, D'Auria JC, Tholl D, Ross JR, Gershenzon J, Noel JP, Pichersky E. An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J. 2003;36:577–588. doi: 10.1046/j.1365-313x.2003.01902.x. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proc Natl Acad Sci U S A. 2005;102:1791–1796. doi: 10.1073/pnas.0409450102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala M, Bennett MH, Truman WH, Grant MR. Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. 2009;59:375–386. doi: 10.1111/j.1365-313X.2009.03875.x. [DOI] [PubMed] [Google Scholar]
- de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bogre L, Grant M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007;26:1434–1443. doi: 10.1038/sj.emboj.7601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean JV, Delaney SP. Metabolism of salicylic acid in wild-type, ugt74f1 and ugt74f2 glucosyltransferase mutants of Arabidopsis thaliana. Physiol Plant. 2008;132:417–425. doi: 10.1111/j.1399-3054.2007.01041.x. [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19:2225–2245. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM. Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell. 1991;3:61–72. doi: 10.1105/tpc.3.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Wang S, Dong X. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4223–4227. doi: 10.1073/pnas.0609357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar MA, Dandekar AM. Agrobacterium tumefaciens as an agent of disease. Trends in plant science. 2003;8:380–386. doi: 10.1016/S1360-1385(03)00162-6. [DOI] [PubMed] [Google Scholar]
- Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant cell. 2011;23:701–715. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis Mutants Selected for Resistance to the Phytotoxin Coronatine Are Male Sterile, Insensitive to Methyl Jasmonate, and Resistant to a Bacterial Pathogen. The Plant cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouhar F, Yang Y, Kumar D, Chen Y, Fridman E, Park SW, Chiang Y, Acton TB, Montelione GT, Pichersky E, et al. Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1773–1778. doi: 10.1073/pnas.0409227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K. A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. The Plant journal : for cell and molecular biology. 2004;39:863–876. doi: 10.1111/j.1365-313X.2004.02171.x. [DOI] [PubMed] [Google Scholar]
- Grant MR, Jones JD. Hormone (dis)harmony moulds plant health and disease. Science. 2009;324:750–752. doi: 10.1126/science.1173771. [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc Natl Acad Sci U S A. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 2001;26:509–522. doi: 10.1046/j.1365-313x.2001.01050.x. [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Laurie-Berry N, Joardar V, Street IH, Kunkel BN. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact. 2006;19:789–800. doi: 10.1094/MPMI-19-0789. [DOI] [PubMed] [Google Scholar]
- Liu PP, Yang Y, Pichersky E, Klessig DF. Altering expression of benzoic acid/salicylic acid carboxyl methyltransferase 1 compromises systemic acquired resistance and PAMP-triggered immunity in arabidopsis. Mol Plant Microbe Interact. 2010;23:82–90. doi: 10.1094/MPMI-23-1-0082. [DOI] [PubMed] [Google Scholar]
- Melotto M, Mecey C, Niu Y, Chung HS, Katsir L, Yao J, Zeng W, Thines B, Staswick P, Browse J, et al. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. Plant J. 2008;55:979–988. doi: 10.1111/j.1365-313X.2008.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126:969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- Mittal S, Davis KR. Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Molecular plant-microbe interactions : MPMI. 1995;8:165–171. doi: 10.1094/mpmi-8-0165. [DOI] [PubMed] [Google Scholar]
- Nickstadt A, Thomma BP, Feussner I, Kangasjarvi J, Zeier J, Loeffler C, Scheel D, Berger S. The jasmonate-insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Molecular plant pathology. 2004;5:425–434. doi: 10.1111/j.1364-3703.2004.00242.x. [DOI] [PubMed] [Google Scholar]
- O'Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21:4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. doi: 10.1093/dnares/10.6.239. [DOI] [PubMed] [Google Scholar]
- Song JT, Koo YJ, Park JB, Seo YJ, Cho YJ, Seo HS, Choi YD. The expression patterns of AtBSMT1 and AtSAGT1 encoding a salicylic acid (SA) methyltransferase and a SA glucosyltransferase, respectively, in Arabidopsis plants with altered defense responses. Mol Cells. 2009;28:105–109. doi: 10.1007/s10059-009-0108-x. [DOI] [PubMed] [Google Scholar]
- Song JT, Koo YJ, Seo HS, Kim MC, Choi YD, Kim JH. Overexpression of AtSGT1, an Arabidopsis salicylic acid glucosyltransferase. Phytochemistry. 2008;69:1128–1134. doi: 10.1016/j.phytochem.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci U S A. 2007;104:18842–18847. doi: 10.1073/pnas.0708139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448:661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–2498. doi: 10.1105/tpc.104.022699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Wangdi T, Kunkel BN, Anand A, Mysore KS, Bender CL. The phytotoxin coronatine contributes to pathogen fitness and is required for suppression of salicylic acid accumulation in tomato inoculated with Pseudomonas syringae pv. tomato DC3000. Molecular plant-microbe interactions : MPMI. 2007;20:955–965. doi: 10.1094/MPMI-20-8-0955. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. Salicylic Acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, He SY. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 2010;153:1188–1198. doi: 10.1104/pp.110.157016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.