Abstract
We examined genotoxic signaling and cell fate decisions in response to a potent DNA-protein crosslinker formaldehyde (FA). DNA-protein crosslinks (DPC) are poorly understood lesions produced by bifunctional carcinogens and several cancer drugs. FA-treated human cells showed a rapid activation of ATR kinase that preferentially targeted the p53 transcription factor at low doses and CHK1 kinase at more severe damage, producing bell-shaped and sublinear responses, respectively. CHK1 phosphorylation was transient, and its loss was accompanied by increased p53 accumulation and Ser15 phosphorylation. Activation of p53 was insensitive to inhibition of mismatch repair and nucleotide and base excision repair, excluding the role of small DNA adducts in this response. The p53-targeted signaling was transcription-independent, absent in quiescent cells and specific to S-phase in cycling populations. Unlike other S-phase stressors, FA-activated p53 was functional transcriptionally, promoted apoptosis in lung epithelial cells and caused senescence in normal lung fibroblasts. FA did not induce ATR, RAD1 or RPA foci, and p53 phosphorylation was TopBP1-independent, indicating a noncanonical mode of ATR activation. Replication arrest by FA caused a dissociation of ATR from a chromatin-loaded MCM helicase but no PCNA monoubiquitination associated with stalled polymerases. These results suggest that unlike typical DNA adducts that stall DNA polymerases, replication inhibition by bulkier DPC largely results from blocking upstream MCM helicase, which prevents accumulation of ssDNA. Overall, our findings indicate that S-phase-specific, TopBP1-independent activation of the ATR-p53 axis is a critical stress response to FA-DPC, which has implications for understanding of FA carcinogenesis.
Keywords: ATR, DNA-protein crosslink, apoptosis, cancer, formaldehyde, p53, senescence
Introduction
DNA damage is a main cause of human cancer and can also lead to neurodegeneration and premature aging.1,2 Genotoxic agents cause a broad variety of DNA lesions, among which small base modifications, duplex-distorting DNA adducts, strand breaks and interstrand DNA crosslinks have been studied the most extensively. Genome protection against these forms of DNA damage involves several evolutionary conserved DNA repair processes. For example, carcinogen-DNA adducts and UV-DNA damage are removed by nucleotide excision repair (NER),3 whereas DNA double-strand breaks are repaired by homologous recombination and nonhomologous end joining.4,5 DNA damage also activates signal transduction pathways that orchestrate multiple cellular activities from DNA repair to cell fate decisions.6,7 A common yet poorly understood form of DNA damage are DNA-protein crosslinks (DPC), which are produced by many important chemotherapeutic agents, such as DNA crosslinkers8,9 and DNA demethylating agents.10 DPC are also generated by oxidative DNA lesions,11,12 carcinogenic metals and aldehydes.13-15 NER, the principal mechanism for repair of large DNA adducts, was inactive toward the majority of DPC due to their excessive bulkiness.16-21 For some crosslinking agents, however, NER can diminish the formation of DPC by removing precursor DNA adducts.21 Inhibition of DPC repair by lactacystin in normal and NER-deficient human cells led to a model of DPC removal via proteasomal digestion of crosslinked proteins.16 Similar effects were also found for another proteasome inhibitor MG132;18,21 however, cellular DPC tested negative for polyubiquitinated proteins.20
Formaldehyde (FA) is a well-known DPC-forming chemical that has been recently recognized as a multi-tissue human carcinogen.22 In addition to numerous sources of occupational and indoor exposure, FA is also produced endogenously during normal metabolism23 and in the vicinity of DNA by the Jumanji-type histone demethylases.24 The average concentration of FA in human blood is ~0.1 mM,22,23 suggesting that it could be a significant source of endogenous DNA damage for all tissues. Even low levels of FA can apparently elicit carcinogenic effects, as suggested by elevated risks of internal cancers (leukemia) among occupationally exposed individuals.25,26 A high carcinogenic potential of aldehydes in the bone marrow is supported by spontaneous leukemia development in mice lacking the aldehyde-detoxifying enzyme Aldh2.27 The incidence of FA-induced nasal tumors in rodents has shown a very good correlation with the formation of DPC, leading to the use of DPC as a biomarker of carcinogenic damage in risk assessment.28 The carcinogenic role of DPC is not necessarily linked to mutagenesis, as FA is a relatively weak mutagen in mammalian cells.29 It is also unclear how mammalian cells, particularly human cells, respond to the presence of DPC. Cytotoxicity studies with FA in mutant collections of transformed chicken DT40 and hamster CHO and V79 cells yielded conflicting results,20,30,31 which could reflect major species and/or transformation-related differences in FA-activated signaling.
In this work, we examined how normal human cells respond to FA and activate cell fate decisions. We determined that human cells were able to sense FA-induced DPC only in S-phase, where a rapid activation of ATR kinase preferentially targeted the p53 transcription factor but not its downstream kinase CHK1. ATR activation by DPC occurred via a unique mechanism lacking canonical requirements for ssDNA and TopBP1. Activated p53 played a major role in the engagement of cell death programs, which has implications for understanding of the origin of mutated p53 in FA-induced tumors. Our findings also point to a variable degree of p53 deficiency as a potential factor contributing to the contradictory findings on FA tolerance mechanisms in the previously employed cellular models.20,30-32
Results
p53 is a highly sensitive target of FA-induced stress signaling
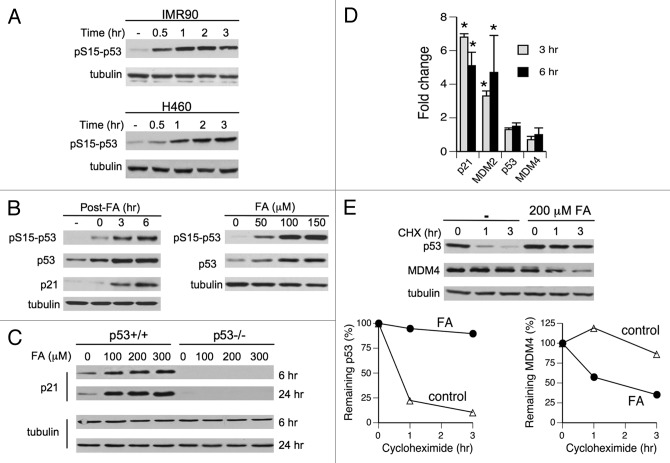
The p53 tumor suppressor is a major regulator of stress responses, which integrates signals from numerous signal transduction pathways.33,34 Therefore, in order to characterize genotoxic signaling by FA, we decided to assess p53 activation and then search upstream to identify apical stress sensors using p53-based readouts. We chose two human cell lines as our main cellular models, primary IMR90 lung fibroblasts and H460 lung epithelial cells. H460 cells have previously been found to accurately reproduce p53 responses of primary cells to genotoxic stress by ionizing radiation35 and small DNA crosslinks.36 We treated cells with FA in the presence of serum, which is physiologically relevant and avoids acute cellular injury. We found that p53 was a very sensitive target of genotoxic signaling elicited by FA in H460 and primary IMR90 cells. Stress-responsive phosphorylation of p53 at Ser15 was already detectable after a 30-min-long FA treatment and increased further at longer 1–3 h exposures (Fig. 1A). Ser15 phosphorylation and p53 protein levels continued to increase for several hours after FA removal (Fig. 1B, left panel). At 6 h post-exposure, a strong phosphorylation at Ser15 was observed with as low as 50 μM FA (Fig. 1B, right panel), which is within its physiological concentration range in human serum.22 Accumulated p53 was transcriptionally active, as evidenced by a strong induction of its target protein, the CDK inhibitor p21 (Fig. 1B and C). RT-qPCR confirmed a transcriptional mechanism of p21 upregulation and another p53-inducible gene MDM2 (Fig. 1D). Blocking of protein synthesis by cycloheximide revealed a dramatically increased stability of p53 protein in FA-treated H460 cells with t1/2 = 18.8 h vs. t1/2 = 0.7 h in controls (Fig. 1E). This result along with the absence of changes in p53 mRNA levels (Fig. 1D) demonstrates that FA-induced accumulation of p53 was primarily due to protein stabilization effects. Enhanced transactivation ability of p53 was promoted by a pronounced destabilization of its inhibitor MDM4 (HDMX), which showed a severely shortened half-life of 1.6 h in FA-treated cells vs. 14.3 h in controls but no changes in mRNA levels (Fig. 1D and E).
Figure 1. Activation of p53 by FA in human cells. (A) Ser15-p53 phosphorylation in IMR90 and H460 cells treated with 200 μM FA for different periods of time. (B) Left panel: p53 responses in IMR90 cells treated with 150 μM FA for 3 h and collected at different post-exposure times. Right panel: dose dependence of p53 responses in IMR90 cells treated with FA for 3 h and collected 6 h later. (C) Induction of p21 by FA in p53−/− and p53+/+ HCT116 cells. (D) Gene expression in H460 cells collected at 3 and 6 h post-FA (means ± SD, n = 3, * - p < 0.05). (E) Stability of p53 and MDM4 in H460 cells treated with 200 μM FA for 3 h and then incubated in the presence of 100 μg/ml cycloheximide (CHX). A representative western blot and means from two independent protein extracts are shown.
S-phase specificity of FA-induced stress signaling
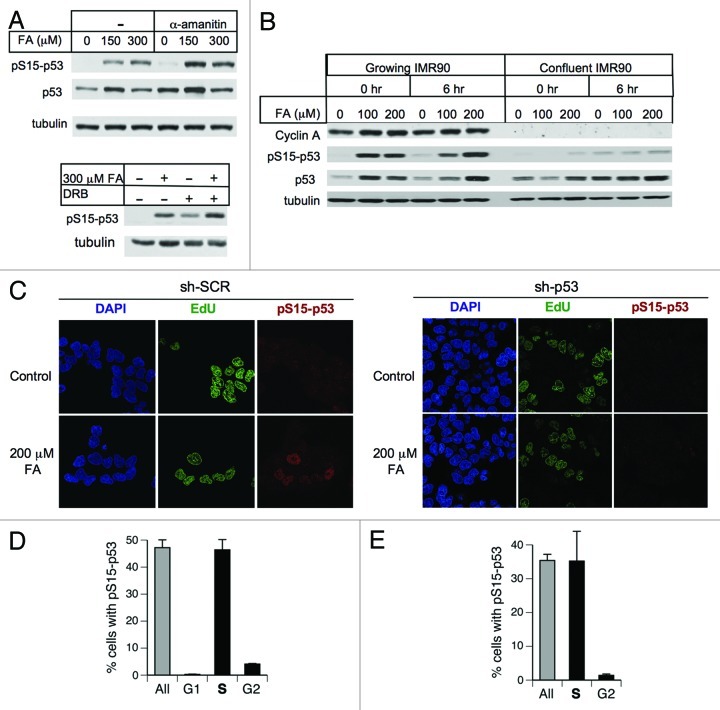
Activation of genotoxic signaling by FA-DNA damage could result from their ability to impede ongoing DNA-based processes, such as transcription or replication. Stalling of RNA pol II with subsequent p53 activation occurs even in response to weakly DNA helix-distorting pyrimidine dimers.37 However, we found that blocking of RNA polymerase II elongation by α-amanitin or 5,6-dichloro-1β-D-ribofuranosylbenzimidazole (DRB) did not inhibit p53 activation by FA (Fig. 2A), arguing against the possibility that a collision of transcriptional complexes with DPC activated p53-targeting stress signaling.
Figure 2. Impact of transcription and cell cycle specificity of p53 activation by FA. (A) Transcription inhibitors α-amanitin (20 μg/ml) and DRB (100 μM) do not prevent p53 activation by FA in H460 cells (0 h post-FA collection). (B) Absence of p53 activation by FA in growth-arrested IMR90 cells. (C) Representative images of H460 cells labeled with EdU for 15 min prior to FA addition and immunostained for phosphorylated Ser15-p53 immediately after FA exposure. sh-SCR – nospecific shRNA, sh-p53 - p53-targeting shRNA. (D) Cell cycle specificity of Ser15-p53 phosphorylation in FA-treated H460 and (E) IMR90 cells. S-phase cells were identified by EdU incorporation (15 min labeling before FA), G1 by CDT1 and G2 by cyclin B1 immunostaining. Cells were treated with 200 μM FA for 3 h and fixed immediately after exposure. Data are means ± SD from three slides with at least 100 cells scored per each slide.
To test the role of cell cycle in responses to FA, we first examined Ser15-p53 phosphorylation using quiescent primary cells that were arrested at confluence in the presence of low serum. Nonreplicating IMR90 populations, evidenced by their lack of the S-phase cyclin A, showed no phospho-p53 induction by FA despite high background levels of p53 protein (Fig. 2B). Next, we analyzed the presence of phospho-p53 in different cell cycle phases by co-staining for CDT1, cyclin B1 and EdU incorporation as markers of G1, G2 and S-phases, respectively. Specificity of the phospho-Ser15-p53 detection was demonstrated by the absence of immunostaining in cells with p53 knockdown (Fig. 2C). We found that FA-induced Ser15-p53 phosphorylation occurred almost exclusively in the S-phase of both H460 and IMR90 cells (Fig. 2C–E). A small percentage of cyclin B1-positive cells containing phospho-p53 likely represented a population of late S-phase cells that progressed into G2 during 3-h long treatments with FA.
Role of p53 in the fate determination of FA-treated cells
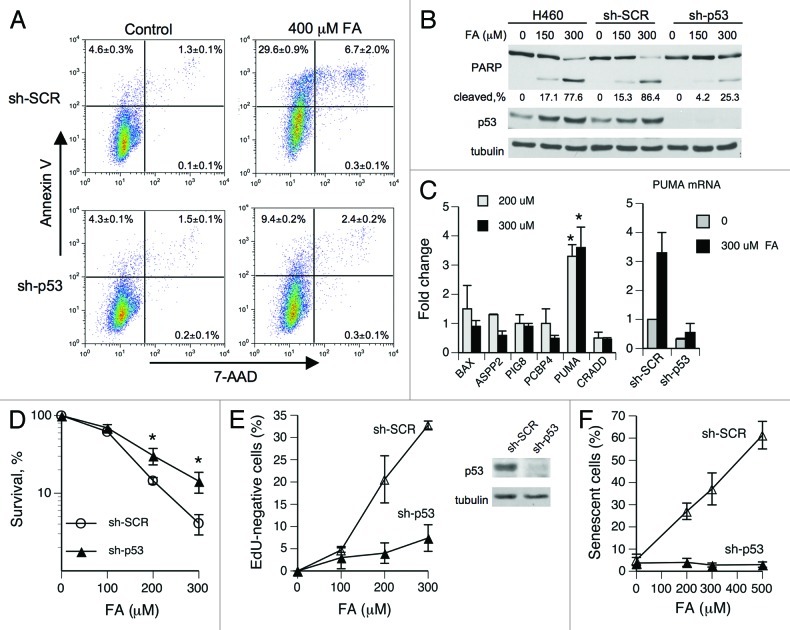
The consequences of S-phase-specific p53 responses typically do not include replication checkpoint and are generally less understood than for other cell cycle phases.33,34 Studies with DNA synthesis inhibitors found that despite its accumulation, p53 was functionally impaired in cells experiencing replication stress.38,39 In contrast, we found a robust induction of p53 targets p21 and MDM2 in FA-treated cells (Fig. 1D), even though p53 activation was also S-phase-specific. To further explore the functionality of p53 in FA-treated cells, we examined its role in cell fate decisions. FACS analyses of H460 cells co-stained with Annexin V and the DNA-binding dye 7-aminoactinomycin D (7-AAD) showed that FA caused apoptosis (detected as Annexin V-positive cells) and no necrosis (< 0.5% Annexin V-negative/7-AAD-positive cells) (Fig. 3A). Numbers of both early (Annexin V-positive/7-AAD-negative) and late (Annexin V-positive/7-AAD-positive) apoptotic cells were strongly decreased by p53 knockdown. The importance of p53 in FA-induced apoptosis was also evident from a strongly diminished PARP cleavage in p53 knockdown cells (Fig. 3B). To identify pro-apoptotic genes upregulated via p53, we performed RT-qPCR measurements of mRNA levels for apoptosis-associated genes in H460 cells with normal and shRNA-depleted p53 levels. PUMA (BBC3) showed the most robust transactivation by p53 in response to FA (Fig. 3C). Inhibition of apoptosis in p53-depleted cells did not appear to activate alternative forms of cell death, because p53-deficient cells also had a higher clonogenic survival (Fig. 3D).
Figure 3. Role of p53 in FA-induced apoptosis and senescence. (A) Representative FACS profiles of H460 cells stained with Annexin V and 7-AAD at 24 h post-FA treatment. Values are means ± SD for three cell samples. (B) PARP cleavage in H460 cells collected at 24 h post-FA. (C) Expression of pro-apoptotic genes in H460 cells at 6 h after FA exposures (means ± SD from three experiments, *- p < 0.05). (D) Clonogenic survival of H460 cells expressing nonspecific (sh-SCR) and targeting (sh-p53) shRNA (means ± SD from three experiments, * - p < 0.05). (E) Frequency of replication-defective IMR90 cells in populations expressing control and p53-targeting shRNA (means ± SD from three slides with > 100 cells scored per each slide). EdU was added at 24 h after FA exposure and cells were labeled for 48 h. (F) Abrogation of FA-induced senescence in IMR90 cells by p53 knockdown (means ± SD from three slides with > 100 cells scored per each slide). Senescent cells were identified by SA-β-Gal staining at 6 d after FA exposure.
Senescence is another form of programmed cell death that can be induced by DNA damage. Primary fibroblasts express low levels of proapototic factors and are generally prone to engaging senescence programs rather than apoptosis. Consistent with this general trend, FA did not cause activation of apoptosis in primary IMR90 fibroblasts. The main cytotoxic response in these cells was senescence, which was assessed by the failure to incorporate EdU during prolonged labeling and the appearance of a common senescence marker SA-β-Gal staining. We found that the ability of FA-treated IMR90 cells to enter into stable replication arrest, and senescence was completely dependent on p53 (Fig. 3E and F). Collectively, our cell survival studies determined that p53 was highly functional in FA-treated human cells.
ATR kinase is responsible for p53 activation
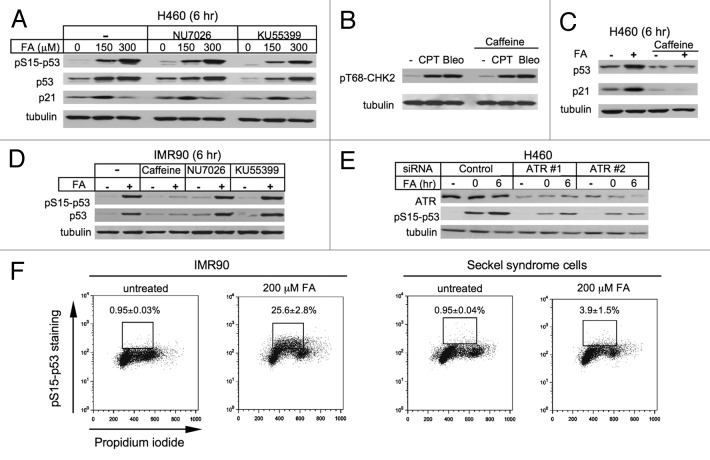
Given the importance of p53 in FA-induced cell death, we sought to identify the upstream stress sensors targeting this critical regulator of cell fate decisions. Activation of p53 is accomplished via a range of its posttranslational modifications, including phosphorylation by multiple kinases.34,35 Ser15, a site that was extensively phosphorylated in response to FA (Fig. 1A and B), can be targeted by several stress-activated kinases, including three DNA damage-responsive kinases DNA-PK, ATM and ATR. The addition of specific inhibitors of DNA-PK (NU7026) and ATM (KU55399) produced no effect on Ser15 phosphorylation, p53 accumulation and p21 induction in FA-treated H460 cells (Fig. 4A). The same concentration of KU55399 completely blocked the ATM-specific Thr68-CHK2 and Ser15-p53 phosphorylation in H460 cells treated with the radiomimetic bleomycin (not shown). The effectiveness of NU7026 was confirmed by a diminished clonogenic survival of bleomycin-treated H460 cells (not shown).
Figure 4. Role of DNA damage-induced kinases in p53 activation. (A) Inhibitors of DNAPK (10 μM NU7026) and ATM (10 μM KU55399) had no effect on p53 responses in FA-treated H460 cells. (B) Caffeine (4 mM) does not inhibit Thr68-CHK2 phosphorylation by ATM in H460 cells treated for 4 h with 1 μM CPT or 20 μg/ml bleomycin (Bleo). (C) Caffeine (3 mM) blocks p53 accumulation and p21 induction in H460 cells treated with 200 μM FA for 3 h and collected 6 h later. (D) Caffeine (3 mM) but not NU7026 or KU55399 abolishes Ser15-p53 phosphorylation and p53 accumulation in IMR90 cells treated with 150 μM FA (6 h post-FA collection). (E) ATR knockdown inhibits Ser15-p53 phosphorylation in H460 cells treated with 200 μM FA (0 and 6 h post-FA collection). (F) FACS profiles of IMR90 and Seckel syndrome fibroblasts immunostained for phosphorylated Ser15-p53. Cells were collected immediately after FA exposures.
ATR kinase lacks highly specific inhibitors, leading us to employ caffeine, which preferentially inhibits ATR but can also diminish ATM activity at high concentrations. We found that ATM kinase activity was not affected by 4 mM caffeine, as measured by the phosphorylation levels of the ATM target Thr68-CHK2 in response to DNA damage induced by bleomycin or the topoisomerase I poison camptothecin (CPT) (Fig. 4B). In FA-treated H460 cells, the addition of 3 mM caffeine suppressed p53 stabilization and p21 induction (Fig. 4C). Similar to H460 cells, caffeine completely blocked p53 accumulation and Ser15-p53 phosphorylation by FA in primary IMR90 cells, whereas DNA-PK and ATM inhibitors had no effects (Fig. 4D). In agreement with caffeine experiments, transfection of H460 cells with two ATR-targeted siRNAs suppressed FA-induced Ser15-p53 phosphorylation proportionally to the decrease in ATR levels (Fig. 4E). FACS analyses did not find significant changes in cell cycle distribution of ATR-depleted cells (siControl: G1 = 51.8%, S = 38.0%, G2/M = 9.0%; siATR-1: G1 = 48.0%, S = 37.3%, G2/M = 14.2%; siATR-2: G1 = 47.2%, S = 39.4%, G2/M = 12.9%). Finally, we examined Ser15-p53 phosphorylation in response to FA in Seckel syndrome fibroblasts that contain biallelic hypomorphic mutations in the ATR gene.40 We found that treatments with 200 μM FA resulted in the presence of Ser15-phosphorylated p53 in 25.6% IMR90 cells, but the same genotoxic insult in Seckel syndrome fibroblasts produced almost seven times fewer cells with detectable phospho-p53 (Fig. 4F).
Small FA-DNA adducts do not contribute to p53 activation
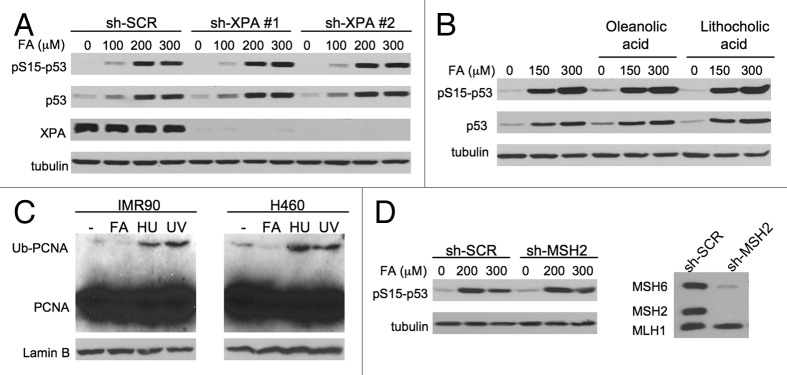
In addition to its predominant genetic damage DPC, FA also forms smaller DNA adducts and crosslinks.20,41 The small DNA-peptide crosslinks are very good substrates for human NER,20 which does not operate on FA-induced16 and other forms of DPC.17,20,21 Small DNA-peptide crosslinks did not show a significant contribution to p53 activation, as evidenced by a normal accumulation of p53 protein and its Ser15 phosphorylation in IMR90 cells with two shRNA knockdowns of the NER protein XPA (Fig. 5A). XPA is required for both transcription-coupled and global genome branches of NER.3 Small DNA base modifications can also be removed by base excision repair, which recruits DNA polymerase β for filling in single-nucleotide gaps. We found that inhibition of DNA polymerase β activity with oleanolic42 or lythocholic acid43 had no effect on p53 accumulation and Ser15-p53 phosphorylation by FA (Fig. 5B). Similar results with both inhibitors were also obtained in H460 cells (not shown). Replication stress induced by non-DPC adducts promotes monoubiquitination of the replication factor PCNA, which results from stalling of DNA polymerases at the sites of DNA damage.44,45 We found no increases in PCNA monoubiquitination in FA-treated IMR90 or H460 cells, which showed the expected positive responses with hydroxyurea and UV-B (Fig. 5C). In the case of highly mutagenic base modifications, genotoxic signaling can be initiated through the detection of the adduct-containing mispairs by mismatch repair proteins.46 Mismatch repair did not play a detectable role in the activation of p53 as evidenced by normal levels of Ser15-p53 phosphorylation in IMR90 cells with MSH2 knockdown (Fig. 5D). MSH2, one of the essential mismatch repair proteins, forms DNA-binding complexes and its loss also results in a destabilization (co-depletion) of its dimerization partner MSH6 but not the upstream-acting MLH1 (Fig. 5D, right panel). A similar clonogenic survival of human HCT116 cells lacking and containing MLH147 further argues against a role of mismatch repair in cytotoxic signaling by FA. Overall, non-DPC adducts do not appear to make a significant contribution to the activation of the ATR-p53 axis by FA either directly or through sensing by mismatch repair.
Figure 5. Activation of p53 in cells with downregulated repair of small DNA adducts. (A) Normal activation of p53 in IMR90 cells with stable XPA knockdowns (6 h post-FA collection). (B) No effect of DNA polymerase β inhibitors oleanolic and lithocholic acids (50 μM each) on p53 activation (6 h post-FA collection). (C) PCNA monoubiquitination in chromatin after 3-h long treatments with 200 μM FA and 10 mM hydroxyurea or 3 h after 100 J/m2 UV-B (302 nm). Soluble proteins were removed by a PBS-0.2% Triton X-100 extraction for 5 min on ice. (D) Normal Ser15-p53 phosphorylation in IMR90 with knockdown of the mismatch repair protein MSH2.
Noncanonical features of ATR activation by FA
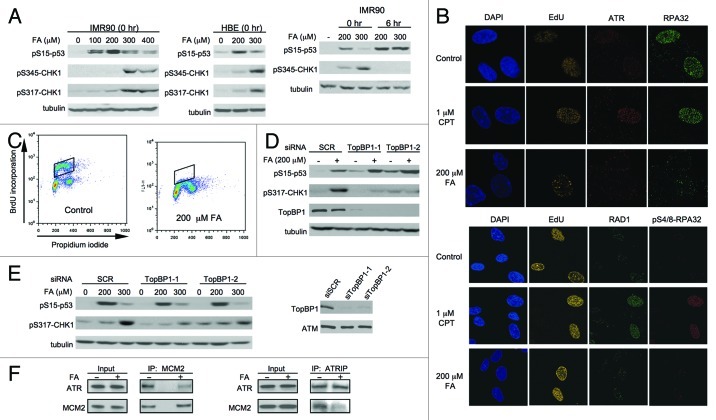
A common model of ATR activation is based on the generation of single-stranded (ss)DNA, which is rapidly coated with RPA, resulting in ATR recruitment via its binding partner ATRIP.48 The ssDNA also attracts the RAD9-RAD1-HUS1 trimer, TopBP1 and other adaptor proteins, and the fully assembled checkpoint complex stimulates ATR-mediated phosphorylation of its downstream kinase CHK1.49-51 We found that CHK1 phosphorylation at two ATR target sites Ser317 and Ser345 became apparent only in IMR90 cells treated with high FA doses (Fig. 6A). In contrast, Ser15-p53 phosphorylation was readily detectable at low FA concentrations and exhibited a bell-shaped dose response, with its inhibitory trend coinciding with the appearance of phosphorylated CHK1. The diminished Ser15-p53 phosphorylation and the appearance of CHK1 phosphorylation at higher FA dosages were also observed in primary human bronchial epithelial cells (Fig. 6A, middle panel), further demonstrating that ATR activity was redirected from p53 to other substrates at severe damage levels. CHK1 phosphorylation was lost at 6 h post-exposure, whereas Ser15-p53 phosphorylation continued to increase, which was particularly noticeable for the highest dose (Fig. 6A, right panel). Confocal microscopy showed that the induction of DPC in IMR90 cells did not lead to the formation of ATR or RPA foci (Fig. 6B, top panel), which occurs during the canonical ATR activation by ssDNA. FA exposures visually appeared to diminish the amounts of chromatin-bound ATR and RPA32 in the S-phase cells. FA-treated IMR90 cells also lacked foci of RAD1 and phospho-Ser4/8-RPA32, which are formed on ssDNA during a canonical ATR activation (Fig. 6B, low panel). RPA32 phosphorylation is a specific biochemical marker of replication-associated DSB.52 Parallel experiments with the topoisomerase I poison camptothecin (CPT) showed numerous ATR, RPA, RAD1 and phospho-RPA32 foci (Fig. 6B), arguing against technical reasons for our negative findings in FA-exposed cells. The absence of significant amounts of ssDNA in FA-treated cells could have potentially resulted from the inefficient blockage of DNA replication, which would limit the production of ssDNA through uncoupling of helicase and polymerase activities. However, measurements of BrdU incorporation by FACS found that our most frequently employed dose of 200 μM FA completely blocked replicative DNA synthesis (Fig. 6C). TopBP1 is required for activation of ATR by the canonical ssDNA-mediated mechanism.49-51 We determined that TopBP1 depletion by two independent siRNA resulted in the expected inhibition of CHK1 phosphorylation but not Ser15-p53 phosphorylation by FA in H460 and primary IMR90 cells (Fig. 6D and E). The ATR dimerization partner ATRIP mediates association of ATR with the replication helicase complex MCM.53 Since the MCM helicase represents a leading edge of the replication complexes, it may activate/release ATR via a collision with a particularly bulky DNA damage, such as DPC. We found that the replication arrest by FA did lead to a dissociation of ATR from the chromatin-bound MCM helicase, as evidenced by a strongly diminished co-immunoprecipitation of ATR/ATRIP and the MCM2 subunit (Fig. 6F).
Figure 6. ATR-associated responses in FA-treated cells. (A) Dose-dependent phosphorylation of p53 after FA exposure for 3 h (0 and 6 h post-FA collection times). HBE – human bronchial epithelial cells. (B) ATR, RPA32, RAD1 and phospho-RPA32 foci in the S-phase of IMR90 cells processed immediately after 3 h-long exposures with FA or CPT. S-phase cells were labeled by EdU incorporation for 15 min immediately before FA exposure. (C) FACS profiles of IMR90 cells stained for DNA content and BrdU incorporated during the last 30 min of mock/FA exposures. (D) Impact of TopBP1 depletion on p53 and CHK1 phosphorylation in H460 and (E) IMR90 cells. Cells were collected immediately after 3-h-long FA exposures. (F) Loss of ATR-MCM association in chromatin of FA-treated IMR90 cells (200 μM FA for 3 h, immediate collection).
Discussion
FA-DPC are activators of the ATR-p53 axis controlling fate of FA-treated cells
We found that FA caused a rapid, potent activation of p53-targeted stress signaling that was initiated by ATR kinase in S-phase. The dose dependence profile of ATR activation and p53 phosphorylation contained bell-shaped and biphasic components, which has not been found for other genotoxic agents. The diminished p53 activation at high FA doses appeared to result from a shift of ATR activity toward other targets, such as CHK1 kinase. In addition to its predominant lesion DPC,22 FA also forms less stable small DNA-peptide crosslinks and base adducts,20,41 which play a minor role in cytotoxic effects of FA in human16 and avian cells.30,31 Unaltered p53 activation in cells with suppressed mismatch repair, NER and base excision repair suggests that non-DPC lesions did not make a significant contribution to ATR-p53 signaling. The near-absence of phospho-CHK1 at FA doses causing maximal p53 phosphorylation is also inconsistent with the profile of ATR activity induced by the non-DPC type adducts, which produce ssDNA and strong CHK1 activation.54 The ATR-p53 axis played a major role in determining cell fate in response to FA, as p53 knockdown led to a suppressed apoptosis, higher clonogenic survival and a complete inability to establish permanent growth arrest (senescence). Unlike other S-phase stressors inhibiting DNA synthesis,38,39 DPC-induced ATR signaling did not impair the ability of p53 to transactivate its target genes p21, MDM2 (Fig. 1) or GADD45 (not shown). Suppressed transactivation of these genes was linked to the inhibitory effect of activated CHK1 on mRNA elongation,39 which would not occur in FA-treated cells due to their weak CHK1 activation. Thus, our findings demonstrate that replication stress-activated p53 can be functional transcriptionally and regulate critical cell fate decisions.
Noncanonical mechanism of ATR activation by FA-DPC
A canonical model of ATR activation by DNA damage in S-phase is based on the presence of RPA-coated ssDNA that recruits ATR via its interacting partner ATRIP.48 Activation of ssDNA-bound ATR requires loading of additional factors, such as RAD17, RAD9-RAD1-HUS1 (9-1-1) clamp and TopBP1.49-51 Recruitment of these factors is ATR-independent and is initiated by the recognition of ssDNA-dsDNA junctions by the RAD17-RFC complex, which then loads the 9-1-1 trimer.55,56 TopBP1 plays a role of a scaffold that links two sets of independently loaded complexes through interactions with both RAD9 and ATR.49,50,57 Loss of TopBP1 leads to impaired phosphorylation of CHK1 and many other ATR targets.51 However, activation of ATR by DPC did not fit into the canonical model, as DPC did not cause characteristic features of ssDNA-dependent ATR activation, such as the formation of chromatin foci by ATR, RAD1, RPA or phospho-RPA32. TopBP1 knockdown failed to inhibit ATR-mediated Ser15-p53 phosphorylation, further supporting the engagement of a noncanonical mechanism of ATR activation in response to DPC.
DNA damage-induced formation of ssDNA stretches, detectable in S-phase as nuclear foci by immunostaining, can originate from ongoing duplex unwinding despite damage-induced stalling of the leading-strand polymerase, reinitiation of replication upstream from the arresting lesion and exonuclease digestion of DNA double-strand breaks producing 3′-ssDNA tails.54 FA-treated cells lacked phospho Ser4/8-RPA32, indicating the absence of DNA break-originated ssDNA tails.52 DPC were found to cause a complete arrest of elongating DNA polymerase during in vitro replication,8 which is fully expected, since even small DNA adducts frequently act as potent replication blockers. However, unlike polymerase-blocking DNA adducts,54 replication blockage by cellular DPC did not appear to cause uncoupling of helicase and polymerase activities, since ssDNA regions were not detected by RPA immunofluorescence. CHK1 phosphorylation54 and PCNA monoubiquitination,44,45 two other biochemical markers associated with the formation of ssDNA through uncoupling of polymerase and helicase activities, were also absent at FA doses causing a maximal p53 phosphorylation by ATR.
The ssDNA would be absent if DPC were stalling replication by blocking DNA unwinding instead of the downstream DNA polymerases. The central channel of a ring-shaped replicative MCM helicase has 2.5 nm width at its ends but is even wider in the middle,58,59 which allows unobstructed passage of ssDNA containing typical DNA adducts. Consequently, DNA adducts arrest replication by sterically blocking downstream polymerase complexes, which leads to the production of ssDNA regions due to the continuing helicase activity.60 DPC are too bulky to pass through the MCM hexameric ring and would arrest translocation of this helicase, which has recently been observed in vitro with a DPC-like streptovidin-avidin block in the leading strand.61 The presence of DPC in the translocating strand also inhibited in vitro activity of bacterial UvrD helicase.62 A fraction of ATR is normally associated with the MCM helicase through binding of its partner ATRIP to the MCM7 subunit,53 which points to a possibility that the helicase-riding ATR can be directly activated by a collision with DPC. The observed dissociation of ATR from the chromatin-bound MCM complexes is consistent with the release of ATR, allowing it to target nucleoplasmic p53. The ability of ATR to respond to the mechanical stress generated by the collision of MCM with DPC could be related to the presence of numerous HEAT repeats in its structure,63 which can act as mechanosensors, as shown for the HEAT repeat-containing subunit of PP2A.64
Implications for FA carcinogenesis
FA has been initially established as a risk factor for nasal cancers, but it is now recognized as a multi-tissue human carcinogen.22 The formation of nasal tumors in rodents showed a good mathematical fit with FA-induced cytotoxicity,65,66 although a biological mechanism for this association has been unclear. The cell death-based model of carcinogenesis questions the risks for internal cancers from low FA doses, including the leukemia linkage found in epidemiological studies.25,26 Our findings on S-phase specificity of FA-induced genotoxic responses suggest that the rate of cell turnover likely exerts a major impact on its carcinogenic risks due to a relatively quick clearance of DPC from human cells.16 A highly proliferative state of the bone marrow cells is expected to strongly increase their sensitivity to chromosomal damage by FA-derived DPC and, consequently, their susceptibility to leukemogenesis. High-dose FA carcinogenesis at the site of direct contact likely reflects slow turnover rates in the upper respiratory epithelium, where cytotoxicity-induced regenerative proliferation would increase the percentage of cells entering S-phase and allow DPC to manifest their genotoxic properties. Chronic exposure to cytotoxic doses of FA can also select for resistant cells lacking functional p53, which we found to be important for engagement of FA-induced cell death programs. Consistent with this possibility, a large percentage of FA-induced nasal tumors contained mutated p53 gene,67 despite the fact that FA is a weak mutagen.29
Materials and Methods
Cells and treatments
IMR90 and H460 cells were obtained from ATCC. Control and p53−/− HCT116 cells were a gift from B. Vogelstein (Johns Hopkins University). H460, IMR90 and HCT116 derivatives were grown in RPMI-10% serum, DMEM-15% serum and F12/DMEM-10% serum media, respectively. Seckel syndrome fibroblasts (GM18366, Coriell) were propagated in DMEM-15% serum. Primary human bronchial epithelial cells were grown as recommended by a supplier (Clonetics). With the exception of experiments in Figure 1A, cells were treated with FA for 3 h in their complete media. Inhibitors were typically added for 1 h before FA exposure. IMR90 cells were put into a quiescent state by growing them to a full confluency and then maintaining for 2 d in 0.5% serum.
shRNA and siRNA
For stable expression of shRNA, cells were infected with the constructs based on the pSUPER-RETRO vector. The targeting sequences were GACTCCAGTG-GTAATCTAC for p53 and GGTGATATGAAACTCTACT and GACCTGTTATGGAATTTGA for XPA. The shRNA sequence for MSH2 was reported earlier.68 Packaging, infection and selection conditions were used as described previously.69 For ATR knockdown by siRNA, ON-TARGETplus (CGAGACUUCUGCGGAUUGCdTdT; GAACAACACUGCUGGUUUGUU) and control siRNAs were purchased from Dharmacon. H460 cells were seeded to obtain ~25% confluency on the day of transfection. Transfections were performed twice with 50 nM siRNA (final concentration) using Lipofectamine RNAimax (Invitrogen). Cells were treated with FA 48 h after initial transfection.
Western blotting and immunoprecipitation
Cellular extracts were prepared as described earlier.36 Proteins were separated by SDS-PAGE and electrotransferred to ImmunoBlot PVDF membrane (BioRad). Primary antibodies used were: p53 (DO-1, Santa Cruz), ATR (N-19, Santa Cruz), PARP (9542, Cell Signaling), pS15-p53 (9284, Cell Signaling), p21 (SX118, BD Biosciences), CHK1 (2345, Cell Signaling), pS317-CHK1 (2344, Cell Signaling), pS345-CHK1 (2341, Cell Signaling), pT68-CHK2 (2661, Cell Signaling), XPA (556453, BD Biosciences), MDM4 (A300–287A, Bethyl Laboratories) and cyclin A (H-432, Santa Cruz).
In the immunoprecipitation experiments, cells were crosslinked with 1% FA for 3 min followed by quenching with 0.125M glycine in PBS. Cells were then lysed for 5 min on ice in buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, protease inhibitors) containing 0.1% Triton X-100. After centrifugation (3000 g, 4°C, 5 min), pellets were washed with buffer A and then incubated in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, protease inhibitors) on ice for 5 min. After another centrifugation, insoluble chromatin was washed with buffer B and resuspended in a digestion buffer (10 mM TRIS-HCl pH 7.5, 1.5 mM MgCl2, 10 mM NaCl, 1 mM EDTA, 0.34 M sucrose, 10% glycerol, 0.1% Triton X-100, 10 mM caffeine, 1 μM wortmannin, 100 U/ml benzonase (Sigma) and protease inhibitors) followed by 37°C incubation for 25 min. After digestion, equal volume of binding buffer (100 mM Na2HPO4, 2 mM K2HPO4, 137 mM NaCl, 2.7 mM KCl, 1% Triton X-100, and protease inhibitors) was added, and the lysates were then centrifuged (15,000 g, 4°C, 15 min). Supernatants were incubated with primary antibodies for 2 h at 4°C. Subsequently, 20 μl protein G/A beads (Santa Cruz) were added and incubated for another 1 h before washing three times with 1 ml binding buffer. Samples were boiled in SDS loading buffer for 10 min to reverse crosslinks and antibody binding
Immunofluorescence
Cells were grown on coverslips, treated with FA for 3 h and fixed with 4% paraformaldehyde for 15 min followed by PBS-0.2% Triton X-100 permeabilization for 10 min. S-phase cells were labeled with 10 μM 5-ethynyl-2'-deoxyuridine (EdU) for 15 min. For immunostaining of ATR, RPA32, pS4/8-RPA32 and RAD1, PBS-0.5% Triton X-100 pre-permeabilization was applied before fixation for 10 min on ice. Cells were then blocked with 3% BSA for 1 h followed by EdU staining (Alexa Fluor 647 azide or Alexa Fluor 488 azide) using Click-iT Alexa Fluor EdU Imaging kits (Invitrogen). After EdU staining, cells were incubated with primary antibodies overnight at 4°C. The following primary antibodies and dilutions were used: RAD1 (Q-18 from Santa Cruz, 1:100), RPA32 (RPA34–20 from Calbiochem, 1:300), cyclin B1 (H-433 from Santa Cruz, 1:300), ATR (N-19 from Santa Cruz, 1:200), CDT1 (ab83174 from Abcam, 1:300), pS4/8-RPA32 (Bethyl Laboratories, 1:300) and pS15-p53 (16G8 from Cell Signaling, 1:100). Cells were then washed twice and incubated with secondary antibody for 1 h at room temperature. Images were obtained with Zeiss LSM 710 confocal microscope.
FACS
Cells were trypsinized, spun down and then fixed in 1.5% formaldehyde for 10 min followed by ice-cold methanol permeabilization for 10 min. For BrdU immunostaining, cells were incubated with 2N HCl for 20 min. Subsequently, cells were washed twice with 1% BSA in PBS. Samples were incubated with FITC-conjugated anti-BrdU (BD Biosciences) and mouse anti-pS15-p53 (16G8, Cell Signaling) antibodies for 30 min at room temperature. Secondary antibodies for anti-pS15-p53 were Alexa Fluor 488 goat anti-mouse IgG (1:300, Invitrogen). After immunostaining, cells were washed twice and resuspended in 1 ml TE buffer containing
Forty µg/ml propidium iodide and 40 µg/ml RNase for 30 min at 37°C before FACS analysis (FACSCalibur, BD Biosciences). For Annexin V/7-AAD staining, FITC-Annexin V apoptosis staining kit was utilized (BD Biosciences). Cells were harvested and spun down (250 g, 4°C, 4 min) followed by washing twice with PBS and a binding buffer. Cells were then stained with FITC-Annexin V and 7-AAD for 15 min at room temperature followed by immediate FACS analysis.
RT-qPCR
RNA samples were extracted with Trizol (Invitrogen), purified with RNeasy mini kit (Qiagen) and further treated with DNase I. cDNA was then synthesized by RT First Strand kit (SABioscience) per manufacturer’s protocol. Primers were obtained from SABioscience and amplification reactions were performed using ABI 7900HT Real-Time PCR system (Applied Biosystems). Expression of five housekeeping genes (B2M, HPRT1, RPL13A, GAPDH, ACTB) were used for normalization purposes. Fold differences of gene expression were calculated by the 2-DDCt method.
Senescence
Senescent cells were identified by staining for senescence-associated β–galactosidase activity (SA-β-Gal assay).70 IMR90 cells were seeded in 6-well plates, treated with FA and, 3 d later, were replated to avoid over-confluent cultures. On day 6 post-FA, cells were fixed with 0.5% glutaraldehyde in PBS (pH 6.0) for 5 min, washed twice with PBS and then incubated with a β-galactosidase staining solution adjusted to pH 6.0 (Roche) at 37°C for 16 h. To facilitate counting of cells, 100 ng/ml DAPI was added to stain nuclei. Five fields were randomly selected to count at least 100 cells for each well.
Acknowledgments
This work was supported by grants ES008786 and ES020689.
Glossary
Abbreviations:
- 7-AAD
7-aminoactinomycin D
- CPT
camptothecin
- DPC
DNA-protein crosslink
- DRB
5,6-dichloro-1β-D-ribofuranosylbenzimidazole
- EdU
5-ethynyl-2'-deoxyuridine
- FA
formaldehyde
- NER
nucleotide excision repair
- ssDNA
single-stranded DNA
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20905
References
- 1.Coppedè F, Migliore L. DNA repair in premature aging disorders and neurodegeneration. Curr Aging Sci. 2010;3:3–19. doi: 10.2174/1874609811003010003. [DOI] [PubMed] [Google Scholar]
- 2.Hanawalt PC. Emerging links between premature ageing and defective DNA repair. Mech Ageing Dev. 2008;129:503–5. doi: 10.1016/j.mad.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 4.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–6. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 7.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 8.Novakova O, Kasparkova J, Malina J, Natile G, Brabec V. DNA-protein cross-linking by trans-[PtCl(2)(E-iminoether)(2)]. A concept for activation of the trans geometry in platinum antitumor complexes. Nucleic Acids Res. 2003;31:6450–60. doi: 10.1093/nar/gkg863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeber RL, Michaelson-Richie ED, Codreanu SG, Liebler DC, Campbell CR, Tretyakova NY. Proteomic analysis of DNA-protein cross-linking by antitumor nitrogen mustards. Chem Res Toxicol. 2009;22:1151–62. doi: 10.1021/tx900078y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santi DV, Norment A, Garrett CE. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc Natl Acad Sci USA. 1984;81:6993–7. doi: 10.1073/pnas.81.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc Natl Acad Sci USA. 2010;107:22475–80. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X, Muller JG, Ye Y, Burrows CJ. DNA-protein cross-links between guanine and lysine depend on the mechanism of oxidation for formation of C5 vs C8 guanosine adducts. J Am Chem Soc. 2008;130:703–9. doi: 10.1021/ja077102a. [DOI] [PubMed] [Google Scholar]
- 13.Costa M, Zhitkovich A, Harris M, Paustenbach D, Gargas M. DNA-protein cross-links produced by various chemicals in cultured human lymphoma cells. J Toxicol Environ Health. 1997;50:433–49. doi: 10.1080/00984109708984000. [DOI] [PubMed] [Google Scholar]
- 14.Macfie A, Hagan E, Zhitkovich A. Mechanism of DNA-protein cross-linking by chromium. Chem Res Toxicol. 2010;23:341–7. doi: 10.1021/tx9003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat Res. 2005;589:111–35. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Quievryn G, Zhitkovich A. Loss of DNA-protein crosslinks from formaldehyde-exposed cells occurs through spontaneous hydrolysis and an active repair process linked to proteosome function. Carcinogenesis. 2000;21:1573–80. doi: 10.1093/carcin/21.8.1573. [DOI] [PubMed] [Google Scholar]
- 17.Reardon JT, Sancar A. Repair of DNA-polypeptide crosslinks by human excision nuclease. Proc Natl Acad Sci USA. 2006;103:4056–61. doi: 10.1073/pnas.0600538103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker DJ, Wuenschell G, Xia L, Termini J, Bates SE, Riggs AD, et al. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J Biol Chem. 2007;282:22592–604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 19.de Graaf B, Clore A, McCullough AK. Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair (Amst) 2009;8:1207–14. doi: 10.1016/j.dnarep.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, et al. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J Biol Chem. 2009;284:27065–76. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zecevic A, Hagan E, Reynolds M, Poage G, Johnston T, Zhitkovich A. XPA impacts formation but not proteasome-sensitive repair of DNA-protein cross-links induced by chromate. Mutagenesis. 2010;25:381–8. doi: 10.1093/mutage/geq017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Toxicology Program Final Report on Carcinogens Background Document for Formaldehyde. Rep Carcinog Backgr Doc. 2010;(i-512):10–5981. [PubMed] [Google Scholar]
- 23.IARC Working Group on the Evaluation of Carcinogenic Risks to Humans Formaldehyde, 2-butoxyethanol and 1-tert-butoxypropan-2-ol. IARC Monogr Eval Carcinog Risks Hum. 2006;88:1–478. [PMC free article] [PubMed] [Google Scholar]
- 24.Culhane JC, Cole PA. LSD1 and the chemistry of histone demethylation. Curr Opin Chem Biol. 2007;11:561–8. doi: 10.1016/j.cbpa.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauptmann M, Stewart PA, Lubin JH, Beane Freeman LE, Hornung RW, Herrick RF, et al. Mortality from lymphohematopoietic malignancies and brain cancer among embalmers exposed to formaldehyde. J Natl Cancer Inst. 2009;101:1696–708. doi: 10.1093/jnci/djp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwilk E, Zhang L, Smith MT, Smith AH, Steinmaus C. Formaldehyde and leukemia: an updated meta-analysis and evaluation of bias. J Occup Environ Med. 2010;52:878–86. doi: 10.1097/JOM.0b013e3181ef7e31. [DOI] [PubMed] [Google Scholar]
- 27.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–8. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 28.Subramaniam RP, Chen C, Crump KS, Devoney D, Fox JF, Portier CJ, et al. Uncertainties in biologically-based modeling of formaldehyde-induced respiratory cancer risk: identification of key issues. Risk Anal. 2008;28:907–23. doi: 10.1111/j.1539-6924.2008.01083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heck H, Casanova M. The implausibility of leukemia induction by formaldehyde: a critical review of the biological evidence on distant-site toxicity. Regul Toxicol Pharmacol. 2004;40:92–106. doi: 10.1016/j.yrtph.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Ridpath JR, Nakamura A, Tano K, Luke AM, Sonoda E, Arakawa H, et al. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–22. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 31.Rosado IV, Langevin F, Crossan GP, Takata M, Patel KJ. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol. 2011;18:1432–4. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- 32.Noda T, Takahashi A, Kondo N, Mori E, Okamoto N, Nakagawa Y, et al. Repair pathways independent of the Fanconi anemia nuclear core complex play a predominant role in mitigating formaldehyde-induced DNA damage. Biochem Biophys Res Commun. 2011;404:206–10. doi: 10.1016/j.bbrc.2010.11.094. [DOI] [PubMed] [Google Scholar]
- 33.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D, Zaugg K, Mak TW, Elledge SJ. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell. 2006;126:529–42. doi: 10.1016/j.cell.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 36.Reynolds M, Zhitkovich A. Cellular vitamin C increases chromate toxicity via a death program requiring mismatch repair but not p53. Carcinogenesis. 2007;28:1613–20. doi: 10.1093/carcin/bgm031. [DOI] [PubMed] [Google Scholar]
- 37.Derheimer FA, O’Hagan HM, Krueger HM, Hanasoge S, Paulsen MT, Ljungman M. RPA and ATR link transcriptional stress to p53. Proc Natl Acad Sci USA. 2007;104:12778–83. doi: 10.1073/pnas.0705317104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottifredi V, Shieh S, Taya Y, Prives C. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc Natl Acad Sci USA. 2001;98:1036–41. doi: 10.1073/pnas.021282898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beckerman R, Donner AJ, Mattia M, Peart MJ, Manley JL, Espinosa JM, et al. A role for Chk1 in blocking transcriptional elongation of p21 RNA during the S-phase checkpoint. Genes Dev. 2009;23:1364–77. doi: 10.1101/gad.1795709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 41.Moeller BC, Lu K, Doyle-Eisele M, McDonald J, Gigliotti A, Swenberg JA. Determination of N2-hydroxymethyl-dG adducts in the nasal epithelium and bone marrow of nonhuman primates following 13CD2-formaldehyde inhalation exposure. Chem Res Toxicol. 2011;24:162–4. doi: 10.1021/tx1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Z, Maloney DJ, Dedkova LM, Hecht SM. Inhibitors of DNA polymerase beta: activity and mechanism. Bioorg Med Chem. 2008;16:4331–40. doi: 10.1016/j.bmc.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 43.Stachelek GC, Dalal S, Donigan KA, Campisi Hegan D, Sweasy JB, Glazer PM. Potentiation of temozolomide cytotoxicity by inhibition of DNA polymerase beta is accentuated by BRCA2 mutation. Cancer Res. 2010;70:409–17. doi: 10.1158/0008-5472.CAN-09-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J Biol Chem. 2006;281:32081–8. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- 45.Niimi A, Brown S, Sabbioneda S, Kannouche PL, Scott A, Yasui A, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16125–30. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol Cell. 2006;22:501–10. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peterson-Roth E, Reynolds M, Quievryn G, Zhitkovich A. Mismatch repair proteins are activators of toxic responses to chromium-DNA damage. Mol Cell Biol. 2005;25:3596–607. doi: 10.1128/MCB.25.9.3596-3607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 49.Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–7. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J, Dunphy WG. Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks. Mol Biol Cell. 2010;21:926–35. doi: 10.1091/mbc.E09-11-0958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol. 2006;26:6056–64. doi: 10.1128/MCB.00492-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc Natl Acad Sci USA. 2004;101:10078–83. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J. 2011;436:527–36. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA. 2003;100:13827–32. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu S, Shiotani B, Lahiri M, Maréchal A, Tse A, Leung CC, et al. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pape T, Meka H, Chen S, Vicentini G, van Heel M, Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–83. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, et al. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc Natl Acad Sci USA. 2008;105:20191–6. doi: 10.1073/pnas.0808037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–52. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fu YV, Yardimci H, Long DT, Ho TV, Guainazzi A, Bermudez VP, et al. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–41. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kumari A, Minko IG, Smith RL, Lloyd RS, McCullough AK. Modulation of UvrD helicase activity by covalent DNA-protein cross-links. J Biol Chem. 2010;285:21313–22. doi: 10.1074/jbc.M109.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perry J, Kleckner N. The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell. 2003;112:151–5. doi: 10.1016/S0092-8674(03)00033-3. [DOI] [PubMed] [Google Scholar]
- 64.Grinthal A, Adamovic I, Weiner B, Karplus M, Kleckner N. PR65, the HEAT-repeat scaffold of phosphatase PP2A, is an elastic connector that links force and catalysis. Proc Natl Acad Sci USA. 2010;107:2467–72. doi: 10.1073/pnas.0914073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conolly RB, Kimbell JS, Janszen DB, Miller FJ. Dose response for formaldehyde-induced cytotoxicity in the human respiratory tract. Regul Toxicol Pharmacol. 2002;35:32–43. doi: 10.1006/rtph.2001.1515. [DOI] [PubMed] [Google Scholar]
- 66.Conolly RB, Kimbell JS, Janszen D, Schlosser PM, Kalisak D, Preston J, et al. Human respiratory tract cancer risks of inhaled formaldehyde: dose-response predictions derived from biologically-motivated computational modeling of a combined rodent and human dataset. Toxicol Sci. 2004;82:279–96. doi: 10.1093/toxsci/kfh223. [DOI] [PubMed] [Google Scholar]
- 67.Recio L, Sisk S, Pluta L, Bermudez E, Gross EA, Chen Z, et al. p53 mutations in formaldehyde-induced nasal squamous cell carcinomas in rats. Cancer Res. 1992;52:6113–6. [PubMed] [Google Scholar]
- 68.Reynolds M, Stoddard L, Bespalov I, Zhitkovich A. Ascorbate acts as a highly potent inducer of chromate mutagenesis and clastogenesis: linkage to DNA breaks in G2 phase by mismatch repair. Nucleic Acids Res. 2007;35:465–76. doi: 10.1093/nar/gkl1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reynolds M, Peterson E, Quievryn G, Zhitkovich A. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J Biol Chem. 2004;279:30419–24. doi: 10.1074/jbc.M402486200. [DOI] [PubMed] [Google Scholar]
- 70.Itahana K, Campisi J, Dimri GP. Methods to detect biomarkers of cellular senescence: the senescence-associated beta-galactosidase assay. Methods Mol Biol. 2007;371:21–31. doi: 10.1007/978-1-59745-361-5_3. [DOI] [PubMed] [Google Scholar]