Abstract
Ubiquitination of histones plays a critical role in the regulation of several processes within the nucleus, including maintenance of genome stability and transcriptional regulation. The only known ubiquitination site on histones is represented by a conserved Lys residue located at the C terminus of the protein. Here, we describe a novel ubiquitin mark at the N-terminal tail of histone H2As consisting of two Lys residues at positions 13 and 15 (K13/K15). This “bidentate” site is a target of the DNA damage response (DDR) ubiquitin ligases RNF8 and RNF168. Histone mutants lacking the K13/K15 site impair RNF168- and DNA damage-dependent ubiquitination. Conversely, inactivation of the canonical C-terminal site prevents the constitutive monoubiquitination of histone H2As but does not abolish the ubiquitination induced by RNF168. A ubiquitination-defective mutant is obtained by inactivating both the N- and the C-terminal sites, suggesting that these are unique, non-redundant acceptors of ubiquitination on histone H2As. This unprecedented result implies that RNF168 generates a qualitatively different Ub mark on chromatin.
Keywords: chromatin remodeling, DNA damage response, epigenetics, histone ubiquitination, RNF168 ubiquitin ligase
Introduction
Posttranslational modifications (PTMs) are critical to ensure the fine orchestration of the molecular events that govern almost all cellular processes. Histones, the backbone of chromatin, undergo different types of PTMs, including acetylation, methylation, phosphorylation, ADP-ribosylation, sumoylation and ubiquitination. The dynamic pattern of these modifications affects the intrinsic properties of histones and drives the interactions between DNA and proteins, marking different functional regions on chromatin and increasing its accessibility to a number of regulatory factors that govern transcription, DNA repair, DNA replication and recombination.1-4
Ubiquitination, one of the most abundant PTMs occurring on histones, is a versatile regulatory process that takes advantage of the combined action of specific enzymes (E1-activating enzyme, E2-conjugating enzyme and E3 ligase), resulting in the attachment of ubiquitin (Ub) moiety on a substrate protein. Ub contains seven lysine residues that can themselves be substrate of ubiquitination, giving rise to poly-Ub chains that are differentially decoded by the cell.5 In the last decade, the canonical view of ubiquitination as a device to mark proteins for degradation has been evolved to a more multifaceted set of functions, including DNA repair, transcription, cell cycle control, signaling, stress response, viral budding, endocytosis and membrane traffic.5,6
Specific cellular events, such as the formation of DNA double-strand breaks (DSBs) occurring upon genotoxic agents induce additional ubiquitination of core histones.7-10 In this case, DNA damage-induced ubiquitination is initiated by the E3 Ub ligase RNF8, which targets histones H2A and H2A.X and is sustained by the action of RNF168, which promotes the formation of K63-linked Ub chains.11-13 RNF8/RNF168-mediated ubiquitination is critical for the assembly of multi-protein complexes on DSB-flanking chromatin, which initiates downstream signaling pathways.14-18 RNF168-dependent ubiquitination of histones exerts two main functions: it generates docking sites for the tandem UIM domain of Rap80, thereby allowing the recruitment of BRCA1-containing complexes,15-17 and it induces the chromatin relaxation required for 53BP1 recruitment through the binding of its Tudor domain to methylated histones H3 and H4.18,19
So far, it has been shown that histones H2A and H2B are modified by Ub at a conserved Lys residue in the C-terminal tail (K119 for H2A and H2A.X, K120 for H2B). We investigated the possibility that additional PTMs of histones might exist in specific cellular contexts, such as genotoxic stress. Here, we identify the first Ub mark laying on the N-terminal tail of histones H2A and H2A.X. This site is composed by K13 and K15 and is targeted by the DDR ligase RNF8 and RNF168. Indeed, we show that inactivation of K13 and K15 reduces RNF8/RNF168- and DNA damage-dependent ubiquitination of histones H2As, while inactivation of both N- and C-terminal sites completely abolishes histone ubiquitination.
Results
C-terminal K118/K119 is not strictly required for the ubiquitination of histones H2A and H2A.X.
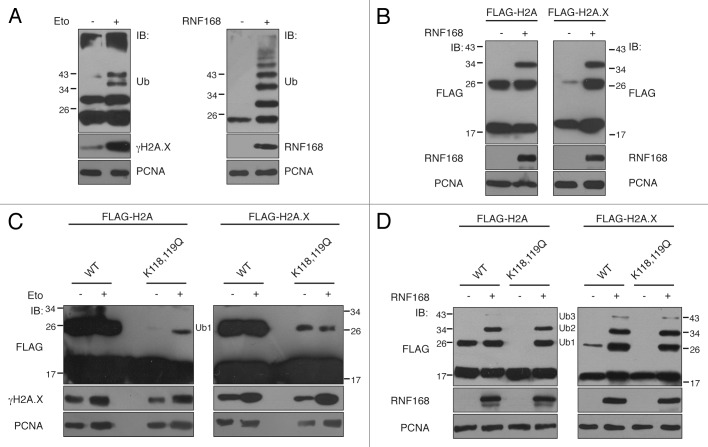
Ubiquitination of histones is a crucial step for the activation of the downstream signaling pathways activated upon formation of DNA DSBs. We observed that the sole ectopic expression of RNF168 is sufficient to induce poly-ubiquitination of chromatin, similarly to what happens upon DSBs formation (Fig. 1A), and to specifically target histone H2A and H2A.X (Fig. 1B). Although it has been demonstrated that RNF168 acts mainly by amplifying the ubiquitination signal originated by RNF8,11-13 we investigated whether RNF168 is able to target other Lys residues on histones, alternative to the conventional K119.
Figure 1. K118/K119 is not the sole ubiquitination site on histones H2A and H2A.X. (A) 293T cells were either treated with etoposide (30 μM) for one hour (left panel) or transfected with cDNA encoding RNF168 or the vector alone (right panel). Three hours after etoposide treatment or 48 h post-transfection, cells were subjected to acid extraction and the histone component was analyzed by SDS-PAGE and immunodecorated with anti-Ub antibody. The induction of DNA damage and expression of RNF168 were verified by phospho-H2A.X (γH2A.X) and RNF168 immunoblotting (IB), respectively; (B) 293T cells were co-transfected with a vector coding RNF168 or the empty vector together with FLAG-tagged histones H2A (left panel) and H2A.X (right panel). Lysates were subjected to acid extraction, analyzed by SDS-PAGE and immunodecorated with anti-FLAG antibody. IB with RNF168 revealed the ectopic expression of the E3 ligase. (C and D) 293T cells expressing the FLAG-tagged forms of histones H2A, H2A.X and their C-terminal mutants K118,119Q were either treated with etoposide (as described in A) or co-transfected with empty vector or RNF168 and immunodecorated with anti-FLAG antibody to evaluate the level of histone ubiquitination. In all cases, an aliquot of cells (1/10) was lysed in LAEMMLI buffer and immunoblotted with PCNA antibody to verify equal loading (lower panels). (Ub1, Ub2, Ub3) indicates the number of Ub moieties appended to histones.
To unveil the existence of putative new ubiquitination sites on histones H2A and H2A.X, we inactivated both K118 and K119 (K118,119Q mutants) to exclude any contribution of the adjacent lysine when the canonical site is absent, and we evaluated the ubiquitination status of the mutants upon induction of DNA DSBs (Fig. 1C) or following ectopic expression of RNF168 (Fig. 1D). Strikingly, we found that ubiquitination of K118,119Q mutants clearly persists after genotoxic treatment (Fig. 1C), suggesting that additional Lys residues are ubiquitinated following DNA damage. Moreover, upon overexpression of RNF168, we observed similar patterns of ubiquitination on wild-type and K118,119Q mutants, consisting of mono-, di- and tri-ubiquitinated species (Fig. 1D). As expected, in the absence of exogenous stimuli (i.e., etoposide treatment and RNF168 overexpression) the K118, 119Q mutants did not show detectable basal ubiquitination (Fig. 1C and D), indicating that this is the major ubiquitination site on histone H2As under normal conditions.
Identification of a new ubiquitination mark on histone H2As.
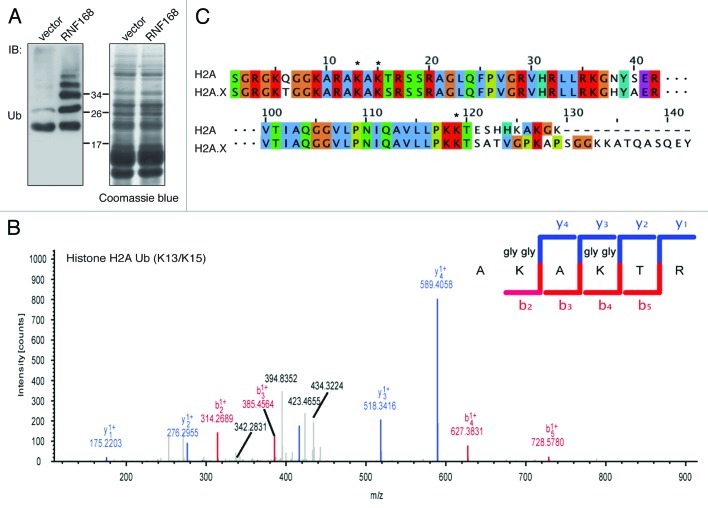
The sum of the above results suggests the intriguing possibility that RNF168 might target histones on Lys residues different from the canonical K118/119, thereby appending a peculiar Ub mark on chromatin to signal the presence of harmful DSBs. To formally prove this point and to map these novel putative sites, we performed a mass spectrometry analysis on chromatin extracts from cells ectopically expressing RNF168 Ub ligase (Fig. 2A and B). In addition to the ubiquitinated peptide matching the K119 site of histone H2A family (Fig. S1A and B), we discovered an additional peptide (AKAKTR) located at the N terminus of histone H2A (Fig. 2B). It is interesting to notice that this novel Ub mark is composed by two Lys residues, which are both modified by Ub. This unprecedented result reveals for the first time the existence of alternative sites of ubiquitination on histone H2As. Importantly, this “bidentate” site is conserved among different members of the H2A family, including H2A.X (Fig. 2C) and throughout evolution (Fig. S2).
Figure 2. Identification of new ubiquitination mark on histone H2As. (A) Acidic extraction of 293T cells, transfected with a vector encoding RNF168 or with the empty vector, were analyzed by SDS-PAGE followed by either Ub immunoblotting (left panel) or Coomassie brilliant blue staining (right panel). Protein bands corresponding to ubiquitinated histones were excised, subjected to trypsin digestion and processed as described in Material and Methods. (B) Mass spectrometry analysis revealed the presence of ubiquitinated peptides corresponding to the N-terminal sequence of histone H2A. The peptide sequence encompassing the ubiquitinated Lys residues in position 13 and 15 are indicated. (C) Sequence alignment of the N-terminal and C-terminal tails of histones H2A and H2A.X, using the Clustalw program and edited with the Jalview software, is shown. ClustalX colors (reflecting the chemical properties of the amino acids) are applied.
K13/K15 site is required for RNF8/RNF168- and etoposide-dependent ubiquitination of histone H2As.
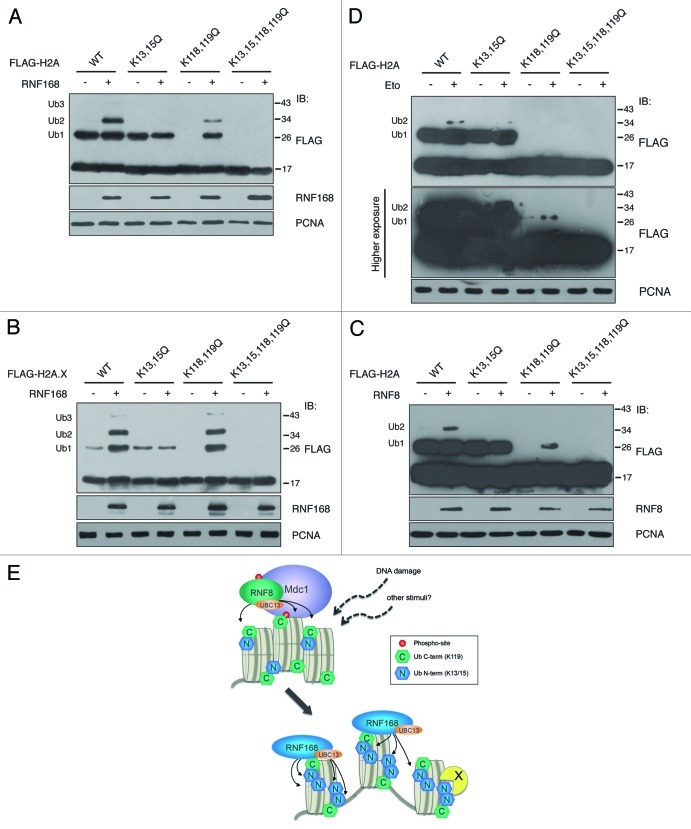
To further characterize this novel Ub mark, we tested whether its inactivation affected the ubiquitination profile of histone H2As. We observed that point mutations on K13 and K15 (K13,15Q mutants) almost blocked the ability of H2A and H2A.X to undergo RNF168-dependent poly-ubiquitination, while they did not alter the formation of mono-ubiquitinated species (Fig. 3A and B). These results demonstrate that, when the N-terminal ubiquitination site is inactivated, RNF168-induced ubiquitination is impaired, suggesting that K13/K15 is the preferential site targeted by this Ub ligase. Furthermore, we found that the amino acid substitution of both sites (K13,15,118,119Q mutants) generates proteins that are unable to undergo ubiquitination (Fig. 3A and B).
Figure 3. Ubiquitination of histones H2A and H2A.X depends on K13 and K15. (A) and (B) In vivo ubiquitination of histones H2A and H2A.X and their mutant forms were evaluated in cells co-transfected with a vector encoding RNF168 or with the empty vector together with FLAG-tagged histones, as indicated. After acidic extraction, samples were analyzed by SDS-PAGE. Immunoblot with anti-FLAG antibodies revealed the presence of higher molecular weight proteins compatible with mono- (Ub1), di- (Ub2) and tri- (Ub3) ubiquitinated forms of the histones. (C) Cells have been co-transfected with cDNA coding for FLAG-RNF8, together with the indicated histone mutants. The experimental procedure is the same as in A. (D) 293T cells expressing the indicated forms of FLAG-tagged histone H2A were treated with etoposide (30 μM) for one hour, and three hours later processed as in A and B. Cell loading was normalized by PCNA immunoblotting as described in Figure 1. (E) Model representing the RNF8 and RNF168-dependent ubiquitination on histone H2As. In addition to amplification of histone ubiquitination initiated by RNF8, RNF168 promotes multi-ubiquitination on different sites, thereby translating upstream post-translational modifications into specific DNA damage-response signals. X represents possible K13/15-specific binding protein.
Since the function of RNF8 and RNF168 in the context of DNA damage is strictly correlated, we asked whether RNF8 targets the same sites on histones. Indeed, we found that RNF8 is still able to ubiquitinate histone H2A when the C-terminal site is absent, and it failed to form di-Ub forms on the mutant K13,15Q (Fig. 3C). Overall, RNF8 recapitulated the results obtained with RNF168, suggesting that both Ub ligases, either directly or indirectly, are able to ubiquitinate the N-terminal tail of histones.
Even though the data we obtained are significant, they might be restricted to the experimental condition used, namely the overexpression of RNF8 and RNF168. We then tested our mutants upon induction of DNA DSBs by etoposide. Consistent with our observations, we found that the K13,15Q mutant displayed a reduction of the di-Ub forms (Fig. 3D), and that the K118,119Q mutant was still able to undergo ubiquitination, albeit to a lesser extent compared to the wild type, since we could not detect significant di-Ub signal. This can be explained by the fact that the increase in histone ubiquitination following DNA damage is typically very low, thereby reducing the probability that K13 and K15 are simultaneously ubiquitinated. Conversely, when histone ubiquitination is enhanced by RNF168 ectopic expression (Fig. 3A), di-Ub and tri-Ub forms are clearly detectable. We conclude that inactivation of both N- and C-terminal sites (K13,15,118,119Q) completely abrogated H2A ubiquitination (Fig. 3D), likely indicating that these are unique, non-redundant acceptors of Ub on histone H2As in response to genotoxic lesions.
Discussion
A remarkable number of histones are constantly mono-ubiquitinated within cells, indicating an important role of this modification for cell homeostasis. When the stability of genetic information is undermined by the formation of DNA DSBs, histones undergo further ubiquitination by the action of the E3 Ub ligases RNF8 and RNF168, which form K63-linked Ub chains.
Ubiquitination events are known to occur on a unique Lys residue at the C-terminal tail of histone H2As, namely K119. Strikingly, we found that mutants of H2A and H2A.X lacking this canonical site are still able to undergo extensive RNF168- and DNA damage-dependent ubiquitination, which raises two important issues. First, the K119 is not strictly required for the attachment of multiple Ub on histones, and, under particular conditions, different sites might be acceptor of ubiquitination. Second, since we observed that multiple Ub moieties are appended to histone mutants, we can envision that either a single lysine is poly-ubiquitinated, or multiple lysines can be targeted by monoubiquitination. To elucidate these points, we exploited mass spectrometry to better characterize histone ubiquitination, and we adopted experimental conditions (i.e., RNF168 overexpression) to intentionally shift the balance toward this novel, preferential ubiquitination site of RNF168 in order to increase the probability of finding modified peptides.
By using this approach, in this study, we described for the first time a novel Ub tag on histone H2As, targeted by the E3 ligase RNF168 and composed by the residues K13 and K15 within the peptide AKAKTR. This exceptional finding might suggest that, when the genome is injured by formation of DSBs, cells induce unconventional PTMs on chromatin, exemplified by this peculiar Ub signal on the N-terminal tail of histone H2As. The relevance of this new site is corroborated by the fact that histone mutants lacking K13/K15 display reduced ubiquitination upon DNA damage, and that the simultaneous inactivation of both sites (K13,15,118,119Q) abolishes it (Fig. 3D). Consistently, these results are recapitulated upon RNF8 and RNF168 overexpression (Fig. 3A–C).
These data do not exclude that, in different cellular contexts and upon different stimuli, other Ub ligases might target K13/K15 site on H2As, as also suggested by the fact that depletion of RNF168 did not significantly alter the ubiquitination status of histone H2A (data not shown). In addition, we assumed that the effect of RNF168 on K13/K15 ubiquitination is directly depending on its Ub ligase activity, since it has been reported that the integrity of RNF168’s RING finger domain is required to ubiquitinate histone H2A.11,12 Nevertheless, we should consider the possibility that RNF168, and RNF8, by virtue of their protein-protein interaction domains, might also act as scaffolds, mediating chromatin recruitment of additional Ub ligases or other regulatory proteins involved in this process.
Notably, we found that both lysines of the novel site are marked by the di-glycine signature. This finding allows us to make some considerations. Although we do not know if this is a frequent event, it indicates that these two residues can be simultaneously ubiquitinated within cells, giving rise to a peculiar bifunctional docking site for Ub receptors. Nevertheless, we should take into consideration that histone sequences are highly enriched in Lys and Arg residues, making the proteomic analysis following trypsin digestion intrinsically problematic. As a consequence, peptides that are contemporary modified on both sites have more chances to be identified compared with peptides in which a single lysine is modified by Ub.
Generation of K13/K15 Ub mark on histone H2As represents a potentially new docking site for proteins involved in the DDR (and in other) processes. It will be of interest to understand how this novel signal is interpreted by the downstream mediators of DDR, and whether the N- and C-terminal ubiquitination sites of H2As recruit common Ub receptors (e.g., Rap80), or if they show different specificity. Among other DDR proteins, there is RNF168 itself. We previously demonstrated that RNF168 is endowed with three Ub binding domains (UBDs), namely UMI, MIU1 and MIU2,20,21 which are all required for the proper localization to DDR foci, through the binding to ubiquitinated histones.11,13 This apparent redundancy of UBDs’ functions can be unraveled by the discovery of this novel ubiquitination site at the N terminus of histones. Thus, we can conceive that the three UBDs recognize distinct Ub marks on nucleosomes, and cooperate to allow proper recruitment of RNF168 to chromatin structures. This hypothesis is substantiated by our observation that MIU2 has a major role in RNF168 recruitment to chromatin, mainly in the absence of DNA damage,12 likely suggesting that MIU2 is responsible for the constitutive binding of RNF168 to chromatin structures, while UMI and MIU1 might stabilize it upon DSBs formation through the binding of K13/K15 site.
By promoting multi-ubiquitination of histones at the N-terminal tail, RNF8 and RNF168 generate a qualitatively different Ub signal on chromatin, revealing for the first time that additional Ub marks can be appended to histones to transmit specific signals (i.e., genotoxic stress).
Overall, these results open new perspectives in the field of histone modifications, also suggesting that there may be other Ub ligases, addressing either the same K13/K15 or additional sites, acting in cellular contexts different from the genomic insults. Development of new experimental tools, such as generation of modification specific antibodies (ubK13-H2A, ubK15-H2A) will be fundamental for future studies to better analyze the dynamics of this ubiquitination event and to assess whether it is a specific cellular response to genomic lesions.
Materials and Methods
Cell culture and antibodies.
293T cells were grown in Dulbecco’s modified Eagle’s medium (SIGMA) supplemented with 10% fetal bovine serum (GIBCO) and 2 mM L-Glutamine (SIGMA). Cells were transfected with calcium phosphate method. Antibodies used in this study included mouse monoclonal anti-Ub P4D1 (Santa Cruz), mouse monoclonal anti-FLAG (M2, SIGMA), anti-phospho-Histone H2AX (Ser139; Upstate), mouse monoclonal anti-PCNA (Santa Cruz). Antibody against RNF168 was generated as previously described.20
Construct design.
The full-length human RNF168 cDNA was cloned into pcDNA3 (Invitrogen). The cDNA of FLAG-RNF8 is a gift of Dr. Lukas. Plasmid encoding FLAG-H2A and FLAG-H2A.X were already described in reference 12. Point mutations were introduced by site-directed mutagenesis using the following oligonucleotides: H2A K13,15Q_for GGC AAA GCT CGC GCC CAG GCC CAG ACC CGC TCT TCT CGG; H2A K13,15Q_rev CCG AGA AGA GCG GGT CTG GGC CTG GGC GCG AGC TTT GCC; H2AK118,119Q_for GGC CGT GCT ACT GCC CCA GCA GAC CGA GAG CCA CCA CAA GG; H2AK118,119Q_rev CCT TGT GGT GGC TCT CGG TCT GCT GGG GCA GTA GCA CGG CC; H2A.X K13,15Q_for CGG CAA GGC CCG CGC CCA GGC CCA GTC GCG CTC GTC GCG C; H2A.X K13,15Q_rev GCG CGA CGA GCG CGA CTG GGC CTG GGC GCG GGC CTT GCC G; H2A.X K118,119Q_for GCC GTG CTG CTG CCC CAG CAG ACC AGC GCC ACC GTG; H2A.X K118,119Q_rev CAC GGT GGC GCT GGT CTG CTG GGG CAG CAG CAC GGC. All the constructs were sequence verified.
In vivo detection of ubiquitinated histones.
Forty-eight hours after transfection, 293T cells were collected in Phosphate Saline Buffer (PBS) containing protease inhibitor cocktail (SIGMA), 1 mM PMSF and 20 ∝M NEM. 1/10 of the samples were separately processed for protein normalization, while the remaining cell pellets were subjected to acid extraction of chromatin proteins as previously described.12
Gel separation of proteins, in-gel digestion and LC-MS/MS analysis.
Proteins were extracted in SDS-PAGE sample buffer and separated by one-dimensional electrophoresis. Protocols for protein processing and peptide desalting and concentration are described.22 Peptides were analyzed by liquid chromatography on an Agilent 1100 LC system (Agilent Technologies Inc.) coupled to LTQ-FT ultra (Thermo Fisher Scientific). Mass spectrometric data were analyzed for protein identification and presence of digly signature using Mascot Deamon and Proteome Discoverer 1.1 (1.1.0.263 Thermo Fisher Scientific Inc.) considering the following parameters: GlyGly (K) +114.043 Da, LeuArgGlyGly (K) +383,228 Da, peptide tolerance 10 ppm, MS/MS tolerance 0.5 Da). All Mascot or Sequest ubiquitination spectra were manually validated.
Supplementary Material
Acknowledgements
This work was funded by grants from Associazione Italiana per la Ricerca sul Cancro (IG11979) and Regione Piemonte RSF to L.P. Research in the Polo laboratory is supported by Associazione Italiana per la Ricerca sul Cancro (IG11627) and by the EMBO Young Investigator Program. M.G. was supported by Fondazione CRT (Lagrange Fellowship) and S.P. was supported by the FIRC (Federazione Italiana per la Ricerca sul Cancro).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here:
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/20919
References
- 1.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–99. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 3.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–63. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, Price BD. Chromatin dynamics and the repair of DNA double-strand breaks. Cell Cycle. 2011;10:261–7. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woelk T, Sigismund S, Penengo L, Polo S. The ubiquitination code: a signalling problem. Cell Div. 2007;2:11. doi: 10.1186/1747-1028-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–86. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 8.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–40. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–14. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan Q, Dutt S, Xu R, Graves K, Juszczynski P, Manis JP, et al. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol Cell. 2009;36:110–20. doi: 10.1016/j.molcel.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart GS, Panier S, Townsend K, Al-Hakim AK, Kolas NK, Miller ES, et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 2009;136:420–34. doi: 10.1016/j.cell.2008.12.042. [DOI] [PubMed] [Google Scholar]
- 12.Pinato S, Scandiuzzi C, Arnaudo N, Citterio E, Gaudino G, Penengo L. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol Biol. 2009;10:55. doi: 10.1186/1471-2199-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–46. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 14.Bekker-Jensen S, Mailand N. The ubiquitin- and SUMO-dependent signaling response to DNA double-strand breaks. FEBS Lett. 2011;585:2914–9. doi: 10.1016/j.febslet.2011.05.056. [DOI] [PubMed] [Google Scholar]
- 15.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, Fukai S. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J. 2009;28:2461–8. doi: 10.1038/emboj.2009.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–83. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huyen Y, Zgheib O, Ditullio RA, Jr., Gorgoulis VG, Zacharatos P, Petty TJ, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–11. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 19.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinato S, Gatti M, Scandiuzzi C, Confalonieri S, Penengo L. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol Cell Biol. 2011;31:118–26. doi: 10.1128/MCB.00818-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–95. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 22.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.