Showing convergence with budding yeast mitotic exit network signaling, the LATS1/WARTS kinase phosphorylates the MYPT1 phosphatase to control PLK1 at the G2 DNA damage checkpoint.
Abstract
In the mitotic exit network of budding yeast, Dbf2 kinase phosphorylates and regulates Cdc14 phosphatase. In contrast, no phosphatase substrates of LATS1/WARTS kinase, the mammalian equivalent of Dbf2, has been reported. To address this discrepancy, we performed phosphoproteomic screening using LATS1 kinase. Screening identified MYPT1 (myosin phosphatase–targeting subunit 1) as a new substrate for LATS1. LATS1 directly and preferentially phosphorylated serine 445 (S445) of MYPT1. An MYPT1 mutant (S445A) failed to dephosphorylate Thr 210 of PLK1 (pololike kinase 1), thereby activating PLK1. This suggests that LATS1 promotes MYPT1 to antagonize PLK1 activity. Consistent with this, LATS1-depleted HeLa cells or fibroblasts from LATS1 knockout mice showed increased PLK1 activity. We also found deoxyribonucleic acid (DNA) damage–induced LATS1 activation caused PLK1 suppression via the phosphorylation of MYPT1 S445. Furthermore, LATS1 knockdown cells showed reduced G2 checkpoint arrest after DNA damage. These results indicate that LATS1 phosphorylates a phosphatase as does the yeast Dbf2 and demonstrate a novel role of LATS1 in controlling PLK1 at the G2 DNA damage checkpoint.
Introduction
Mitotic entry and many early mitotic events are regulated by mitotic kinases (Nigg, 2001). Of the many known mitotic kinases, Cdk1, together with Cyclin B, plays a major role in mitotic entry of animal cells (Lindqvist et al., 2009). In turn, exit from mitosis requires the inactivation of Cdk1 and the removal of phosphates from Cdk1 substrates (Sullivan and Morgan, 2007).
In budding yeast, the Cdc14 protein controls mitotic exit by means of a mitotic Cdk-counteracting phosphatase (Stegmeier and Amon, 2004). Cdc14 activity can be detected during late mitosis and is regulated by two distinct pathways: the Cdc14 early anaphase release network and the mitotic exit network (MEN). At early anaphase, the Cdc14 early anaphase release network leads to partial activation of Cdc14. From late anaphase, the MEN promotes complete activation of Cdc14, resulting in mitotic exit. The MEN pathway is initiated by the small Ras-like GTPase, Tem1. Activated Tem1 triggers signaling cascades involving Cdc15 and Dbf2 kinases to target Cdc14 for activation (Sullivan and Morgan, 2007; Mohl et al., 2009). Consistent with the yeast pathway, LATS1 (also known as WARTS) kinase, the mammalian equivalent of Dbf2 kinase, plays a role during mitotic exit (Bothos et al., 2005). However, to date, no phosphatase has been identified as the substrate for LATS1. Furthermore, mitotic exit in animal cells is mainly regulated by PP2A (protein phosphatase 2A), not Cdc14 (Manchado et al., 2010; Schmitz et al., 2010). Therefore, although some MEN pathway components are conserved throughout evolution, the cellular function of MEN signaling in mammals remains poorly understood (Stegmeier and Amon, 2004).
LATS1 forms part of the Hippo (Hpo) pathway, which is best characterized in Drosophila melanogaster (Pan, 2010; Zhao et al., 2010). Hpo (MST2 in mammals) phosphorylates LATS/WARTS (LATS1/2 in mammals) to regulate cell proliferation and cell death to ensure appropriate organ size is maintained (Pan, 2010; Zhao et al., 2010). Genetic analysis of Drosophila shows that the Hpo pathway is regulated by Merlin and Expanded, both of which contain a membrane-associated module known as a FERM (4.1, ezrin, radixin, and moesin) domain, suggesting some contribution by extracellular cues to Hpo pathway control (Hamaratoglu et al., 2006). Furthermore, accumulating evidence indicates that many signaling pathways also impinge on the Hpo cascade. Upon DNA damage, ataxia telangiectasia mutated (ATM), the main sensor of double-strand DNA breaks, causes MST2/LATS1 activation by phosphorylating RASSF1A (Ras association domain-containing family 1A) followed by the induction of proapoptotic genes (Hamilton et al., 2009). Consistent with this, LATS1-overexpressing cells undergo apoptosis or cell cycle arrest at G2/M (Yang et al., 2001). However, the role of LATS1 in DNA damage–induced cell cycle control remains obscure.
There is a link between Dbf2 kinase and Cdc14 phosphatase in budding yeast. Thus, in the present study, phosphoproteomic screening was undertaken to search for candidate phosphatases as substrates for LATS1. Screening identified MYPT1 (myosin phosphatase [MP]–targeting subunit 1), the regulatory subunit of PP1C, as a novel target for LATS1. LATS1 directly phosphorylated MYPT1 to promote PP1C activity toward PLK1 (pololike kinase 1). Furthermore, DNA damage–induced activation of LATS1 caused PLK1 suppression via MYPT1 phosphorylation and induced a G2 checkpoint response. Collectively, these results indicate a previously unknown role of LATS1 on PP1C regulation and the G2 DNA damage response.
Results
Identification of MYPT1 as a novel target for LATS1/WARTS
To identify previously unknown targets for LATS1, proteome-wide screening of LATS1 substrates was performed using phosphopeptide enrichment techniques coupled with high-accuracy mass spectrometry (MS; Sugiyama et al., 2007). HeLa cell lysates were dephosphorylated by thermosensitive alkaline phosphatase and used as a substrate for an in vitro kinase assay incorporating recombinant active LATS1 kinase (see Materials and methods). The screening process identified YAP1 (Yes-associated protein 1), a well-known target for LATS1 (Huang et al., 2005), with high frequency (Table 1), thereby confirming the reliability of the assay. Based on the finding that Dbf2 phosphorylates Cdc14 phosphatase in yeast (Mohl et al., 2009), attention was focused on phosphatases phosphorylated by LATS1. Mass spectrometric analysis identified six peptides from phosphatases as candidate targets for LATS1 (Table 1). Of these, MYPT1 and SAPS3 were significantly phosphorylated by LATS1 in our assay. The phosphopeptides showed a remarkably high ratio in the assays (calculated by comparing the phosphorylation status in the presence or absence of the kinase and based on quantitative analysis using stable isotope labeling). In contrast, phosphopeptides derived from PP1RB, PPBN, and PTEN were expected to be the inherent ones not efficiently dephosphorylated by alkaline phosphatase because they were detected regardless of LATS1 spiking (Table 1).
Table 1.
Proteins phosphorylated by LATS1
| UniProt ID | Protein description | Phosphopeptides | Corresponding phosphorylated positions | Ratio (kinase ±) |
| PP1RB_HUMAN | Protein phosphatase 1 regulatory subunit 11 | AFGESSTESDEEEEEGCGHTHCVR | PP1RB_HUMAN; T75 | 1.3 |
| AFGESSTESDEEEEEGCGHTHCVR | PP1RB_HUMAN; S77, PP1RB_HUMAN; S73 or S74 | 1.6 | ||
| SAPS3_HUMAN | Serine/threonine-protein phosphatase 6 regulatory subunit 3 | FADQDDIGNVSFDR | SAPS3_HUMAN; S579 | 9.7 |
| PPBN_HUMAN | Alkaline phosphatase, placental-like | HVPDSGATATAYLCGVK | PPB1_HUMAN; S114, PPBN_HUMAN; S111 | 1.5 |
| MYPT1_HUMAN | Protein phosphatase 1 regulatory subunit 12A | RSTQGVTLTDLQEAEK | MYPT1_HUMAN; S695 or T696 | 14.2 |
| PTEN_HUMAN | Phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase (PTEN) | YSDTTDSDPENEPFDEDQHTQITKV | PTEN_HUMAN; S380, PTEN_HUMAN; T383 | 1.1 |
| YAP1_HUMAN | Yorkie homologue | DESTDSGLSMSSYSVPR | YAP1_HUMAN; S400 | 36.4 |
| DESTDSGLSMSSYSVPR | YAP1_HUMAN; S397 or T398 | 75.2 | ||
| QASTDAGTAGALTPQHVR | YAP1_HUMAN; S109 or T110 | 110.3 |
Phosphoproteomic screening was performed as described in the Materials and methods. Phosphorylated residue of each peptide is indicated by bold character or underline (it was not possible to locate the exact phosphorylation site between two residues). Ratio is increased with the reliability of phosphorylation event; lower ratio (<2) indicates the residual phosphorylation sites before kinase reaction.
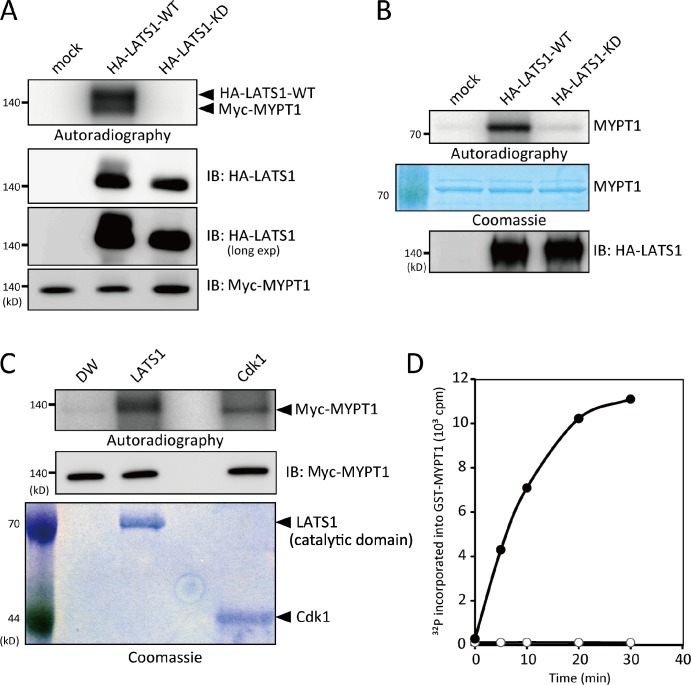
Next, an in vitro kinase assay was performed to confirm whether MYPT1 was a target for LATS1. 293T cells were transfected with a HA-tagged wild-type or kinase-dead (KD) mutant of LATS1 and subsequently treated with okadaic acid to activate the exogenous kinase (Chan et al., 2005). LATS1 was then immunoprecipitated using anti-HA antibodies and allowed to react with immunopurified or recombinant MYPT1. As shown in Fig. 1 (A and B), wild-type, but not the KD mutant, LATS1 phosphorylated both immunopurified (full length) and recombinant (amino acids 345–653) MYPT1. Similarly, active recombinant LATS1 (amino acids 589–1,130; Carna Biosciences, Inc.) efficiently phosphorylated immunopurified MYPT1 (Fig. 1 C). MYPT1 is specifically phosphorylated during mitosis, mainly by Cyclin B/Cdk1 (Totsukawa et al., 1999; Yamashiro et al., 2008). Thus, it was confirmed that MYPT1 was also phosphorylated by a recombinant Cyclin B–Cdk1 complex (Fig. 1 C).
Figure 1.
LATS1 phosphorylated MYPT1 in vitro. (A and B) Immunopurified LATS1 phosphorylated MYPT1. Wild-type (WT) or kinase-dead (KD) HA-LATS1 (full length) immunoprecipitated from 293T cells treated with 100 nM okadaic acid (for 2 h) was incubated with immunopurified Myc-MYPT1 (full length; A) or GST-MYPT1 (amino acids 345–653; B) for 30 min at 30°C, and samples were subjected to SDS-PAGE followed by autoradiography. Arrowheads indicate trans-phosphorylated MYPT1 and autophosphorylated LATS1. Immunoprecipitated proteins were also subjected to immunoblot analysis with the indicated antibodies. (C) Recombinant LATS1 phosphorylated MYPT1. Full-length MYPT1 immunoprecipitated from 293T cells was incubated with 400 ng GST-LATS1 or 100 ng GST-Cdk1–Cyclin B for 30 min. As a control, an equivalent volume of distilled water (DW) was added instead of kinases. The reaction mixtures and immunopurified MYPT1 were analyzed as in A. SDS-PAGE gels were stained with Coomassie blue. (D) Time course analysis of LATS1 kinase activity. Recombinant 2 µg GST-MYPT1 (amino acids 345–653) was incubated with (•) or without (○) 200 ng GST-LATS1 for the indicated times. The amount of 32P incorporated into MYPT1 was determined by Cerenkov counting. This experiment was completed once. IB, immunoblot; exp, exposure.
To rule out possible contamination of the coimmunoprecipitated kinases, an in vitro kinase assay was performed using recombinant LATS1 and MYPT1. As shown in Fig. 1 D, active LATS1 phosphorylated the recombinant MYPT1 fragment in a time-dependent manner. These results indicate that LATS1 is a novel and relevant kinase for MYPT1.
Interaction of MYPT1 with LATS1
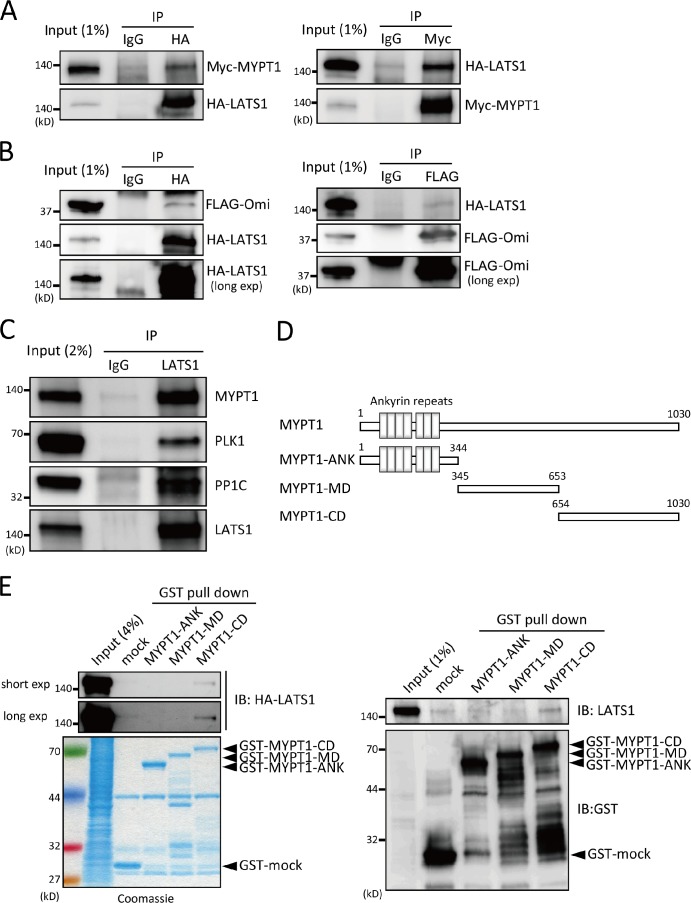
Next, the binding between LATS1 and MYPT1 was examined to further evaluate whether LATS1 targets MYPT1 in cells. Myc-tagged MYPT1 and HA-tagged LATS1 were cotransfected into 293T cells, and HA-LATS1 was subsequently immunoprecipitated with anti-HA antibodies. Myc-MYPT1 was coprecipitated with HA-LATS1 (Fig. 2 A, left). Conversely, HA-LATS1 was detected in immunoprecipitates using anti-Myc antibodies (Fig. 2 A, right). As a positive control, a mature form of Omi/HtrA2 protease was coexpressed with LATS1, and the interaction of these proteins was confirmed (Fig. 2 B; Kuninaka et al., 2005).
Figure 2.
Interaction of MYPT1 with LATS1. (A) Interaction of MYPT1 with LATS1 in 293T cells. 293T cells were cotransfected with HA-LATS1 (full length) and Myc-MYPT1 (full length) expression vectors. Lysates from the transfected cells were immunoprecipitated with control IgG, anti-HA (left), or anti-Myc (right) antibodies. The immunoprecipitates and 1% of the input fractions were analyzed by immunoblotting with anti-Myc and anti-HA antibodies. (B) Interaction of LATS1 with a mature (protease inactive) form of Omi was examined as in A. (C) Endogenous interaction between LATS1 and MYPT1. The lysate from asynchronously growing HeLa cells was immunoprecipitated with control IgG or an anti-LATS1 antibody. The immunoprecipitates and 2% of the input fractions were probed with anti-MYPT1, anti-PLK1, anti-PP1C, or anti-LATS1 antibodies as indicated. (D) Schematic diagram of human MYPT1 and the derived mutants. Rectangles indicate ankyrin repeats. ANK, ankyrin repeats domain; MD, middle domain; CD, C-terminal domain. (E) LATS1-binding region within MYPT1. HA-LATS1 KD (left) or endogenous LATS1 (right) immunoprecipitated from cells treated with 100 nM okadaic acid (2 h) were incubated with either GST alone or GST-MYPT1- ANK, GST-MYPT1-MD, or GST-MYPT1-CD (4 µg of each protein) bound to glutathione–Sepharose beads. Bead-bound proteins (and 4 or 1% of the input fraction) were then subjected to immunoblot analysis with an antibody to HA, GST, LATS1, or Coomassie blue staining. (left, second row). Bead-bound protein was detected after long exposure (exp). IP, immunoprecipitation; IB, immunoblot.
To examine the physiological interaction between these proteins, endogenous LATS1 was immunoprecipitated using a specific antibody, and MYPT1 was identified in the immunoprecipitates (Fig. 2 C). The known binding partners of MYPT1, PP1C and PLK1, also coprecipitated with LATS1 (Ito et al., 2004; Yamashiro et al., 2008). To identify the exact region within MYPT1 that interacted with LATS1, a GST pull-down assay was performed. MYPT1 was divided into three fragments: an N-terminal fragment comprising amino acids 1–344 (designated as ankyrin repeats domain [ANK]), a central fragment comprising amino acids 345–653 (designated as middle domain [MD]), and a C-terminal fragment comprising amino acids 654–1,030 (designated as C-terminal domain [CD]; Fig. 2 D). GST fusion proteins of each fragment were purified and used in GST pull-down assays. As shown in Fig. 2 E, both exogenous and endogenous LATS1 preferentially interacted with the C-terminal region of MYPT1 (MYPT1-CD), which encompasses the previously reported binding sites for several regulatory molecules, such as cGMP-dependent protein kinase I α and RhoA (Ito et al., 2004). These results indicate that LATS1 was complexed with the C terminus of MYPT1.
LATS1 preferentially phosphorylated the middle region (MD) of MYPT1
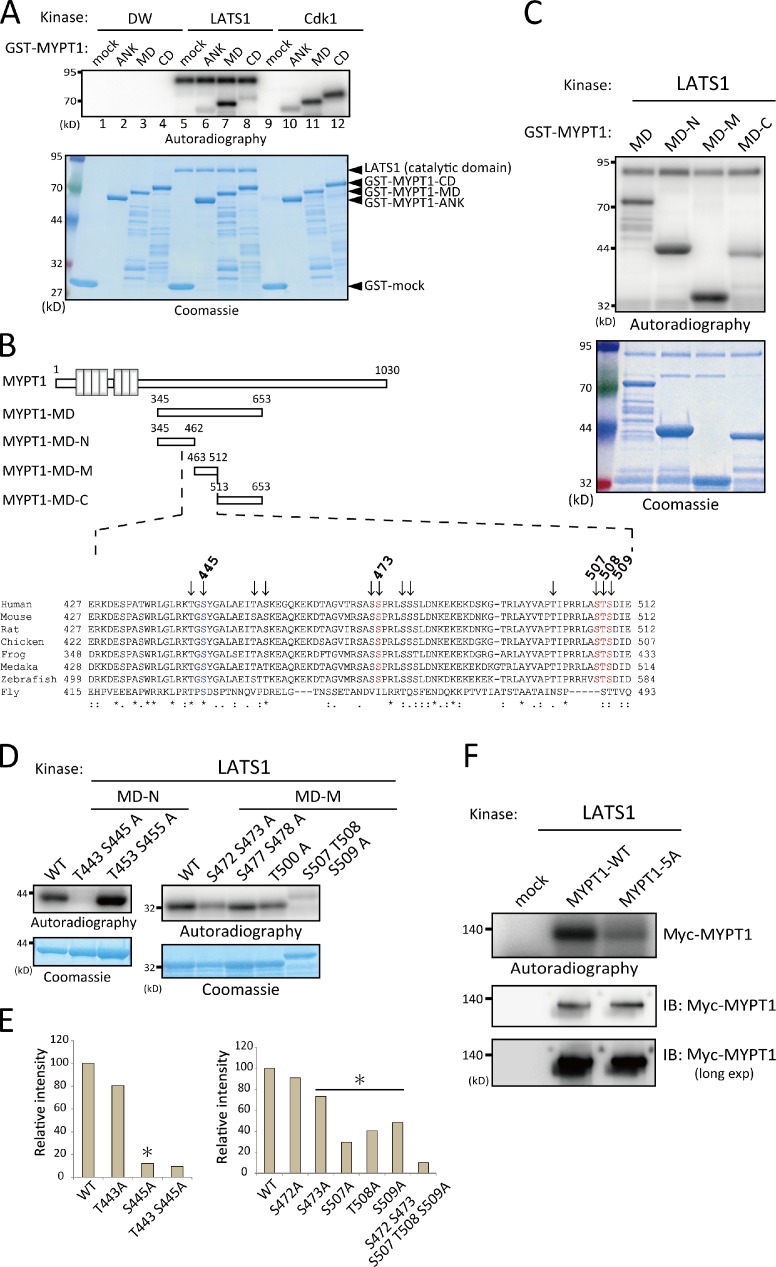
Initial screens identified the C terminus of MYPT1 (Serine [S] 695 or Threonine [T] 696) as a candidate phosphorylation site for LATS1 kinase (Table 1). To reexamine this result, we performed mass spectrometric analysis of the immunopurified MYPT1 from cells transfected with or without an siRNA oligonucleotide (oligo) against LATS1. Purification of full-length MYPT1 protein provided an opportunity to obtain the missing information of the LATS1-mediated phosphorylation sites. There were several candidate sites, including S695 and T696 (Table S1). To determine the major sites for the LATS1-mediated phosphorylation, an in vitro kinase assay was performed using deletion mutants (Fig. 2 D) of MYPT1. Consistent with the screening results, LATS1 phosphorylated the C terminus of MYPT1 (amino acids 654–1,030); however, the middle region of MYPT1 (amino acids 345–653, designated as MYPT1-MD) was more efficiently phosphorylated by LATS1 than the C terminus (Fig. 3 A, lanes 5–8). In contrast to LATS1, Cdk1–Cyclin B phosphorylated the C-terminal (amino acids 654–1,030) and middle regions of MYPT1 to the same extent (Fig. 3 A, lanes 9–12), suggesting the physiological significance of the middle region of MYPT1 for LATS1 signaling.
Figure 3.
Determination of LATS1 phosphorylation sites within MYPT1 in vitro. (A) Mapping of the LATS1 phosphorylation region within MYPT1. 400 ng active GST-LATS1 or 60 ng GST-Cdk1–Cyclin B was incubated with either GST alone or GST-MYPT1-ANK, GST-MYPT1-MD, or GST-MYPT1-CD (2 µg of each protein) for 30 min. Samples were then subjected to SDS-PAGE followed by autoradiography and Coomassie blue staining. As a control, an equivalent volume of distilled water (DW) was added instead of the kinases (lanes 1–4). (B) A schematic representation of human MYPT1 and the derived mutants and ClustalW alignment of the LATS1 phosphorylation sites within the middle domain (MD) of MYPT1. Consensus sequences are indicated by asterisks. The phosphorylation sites identified by MS are indicated by arrows. Residues validated in the kinase assay are shown in blue or orange (top numbers indicate amino acids). Double and single dots indicate the “strong” and “weaker” groups of amino acids, all of which positively scored in the PAM250 matrix (Gonnet). The strong and weak groups are defined as strong score >0.5 and weak score <0.5, respectively. (C) 200 ng active GST-LATS1 was incubated with either GST-MD, GST-MD-N, GST-MD-M, or GST-MD-C (details of each mutant are shown in Fig. 3 B) for 30 min, and the reaction mixtures were analyzed as in A. (D) MYPT1 mutation analysis. 200 ng active GST-LATS1 was incubated with the indicated GST-MD-N or GST-MD-M mutants for 30 min, and the reaction mixtures were analyzed as in A. (E) The intensity of each band on autoradiography was quantified and normalized according to the intensity of the bands in the Coomassie blue–stained gels. Relative intensity to that of the wild type (WT) is shown (percentage of wild type). Asterisks indicate the identified target sites. This experiment was repeated at least once with similar results. (F) 200 ng active GST-LATS1 was incubated with immunopurified full-length wild-type or 5A MYPT1 for 30 min, and the reaction mixtures were subjected to SDS-PAGE followed by autoradiography. Immunoprecipitated proteins were also subjected to immunoblot analysis with the anti-Myc antibody. IB, immunoblot; exp, exposure.
Next, the phosphopeptides in the in vitro kinase reaction mixture of MYPT1-MD were enriched and then subjected to mass spectrometric analysis. 20 phosphorylation sites were identified within the MD (Fig. 3 B and not depicted). Of these, S445, S472, S478, and S507 partially matched the His-x-Arg/His/Lys-x-x-Ser/Thr consensus motif for LATS1 substrates (in which x denotes any amino acid; Pearce et al., 2010). Stoichiometric analysis of the phosphorylated peptides was also performed by analyzing the reaction mixture before and after phosphopeptide enrichment (Olsen et al., 2010). However, it was difficult to collect all the corresponding peptides and definitively compare the frequency of phosphorylation; therefore, it was not possible to identify the significant phosphorylation sites (unpublished data).
To narrow down the relevant sites for LATS1-mediated phosphorylation, MYPT1-MD was divided into three regions: MD-N (amino acids 345–462), MD-M (amino acids 463–512), and MD-C (amino acids 513–653; Fig. 3 B). An in vitro kinase assay using these mutants showed that MD-N and MD-M were mainly responsible for LATS1-mediated phosphorylation (Fig. 3 C). Combining this result with those from the mass spectrometric analysis showed that MD-N contained four candidate phosphorylation sites, and MD-M contained eight (Fig. 3 B, residues are indicated by arrows).
We next categorized these putative target sites into several groups based on their vicinity. After introducing an alanine mutation into each group, GST–MD-N or -M carrying these mutations was used in the in vitro kinase assay. The results showed that two alanine substitutions in MD-N (T443 and S445) and five in MD-M (S472, S473, S507, T508, and S509) significantly attenuated LATS1-mediated phosphorylation (Fig. 3 D). Further mutational analysis revealed that five residues within MD were the major sites at which LATS1 phosphorylates MYPT1 in vitro (Fig. 3 B and Fig. S1). Mutation of these residues (S445, S473, S507, T508, and S509) decreased the phosphorylation of each mutant by >20% compared with that of the corresponding protein (Fig. 3 E). Furthermore, alanine substitution of all phosphorylation sites (5A) significantly reduced LATS1-mediated phosphorylation in vitro (Fig. 3 F). These results indicate that LATS1 directly and preferentially phosphorylates the middle region of MYPT1 (Fig. 3 B, residues colored in blue or orange).
LATS1-regulated PLK1 activity
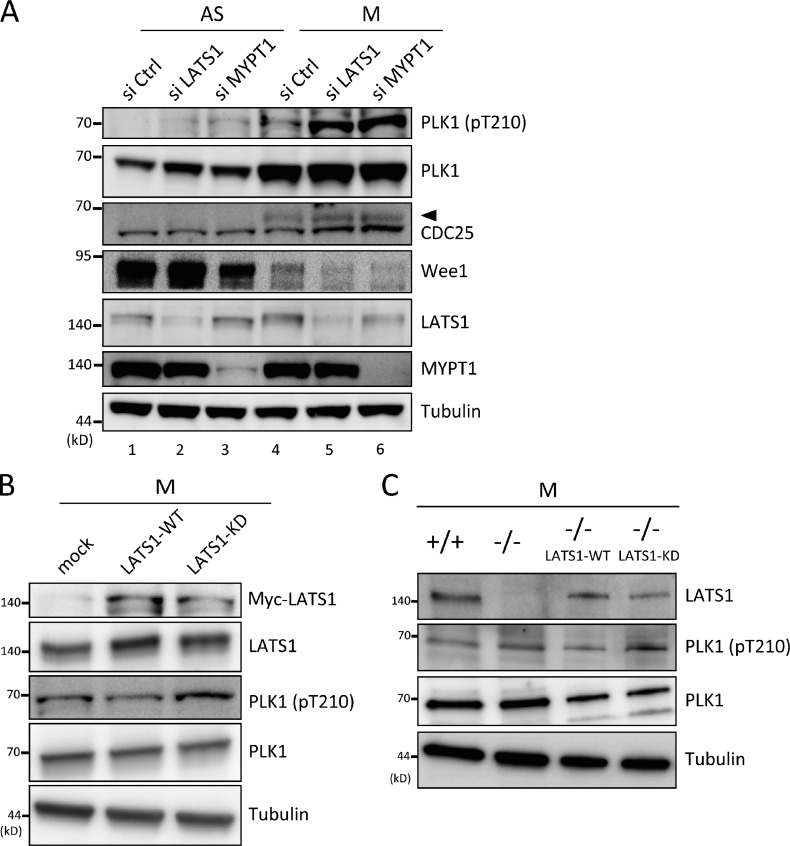
Phosphorylation of MYPT1 regulates MYPT1–PP1C activity toward several targets (Matsumura and Hartshorne, 2008). Recent data indicate that MYPT1–PP1C antagonizes PLK1 activity by dephosphorylating Thr 210 within its T loop (Yamashiro et al., 2008). Because both LATS1 and PLK1 are mitotic kinases (Nigg, 2001), the effects of LATS1 depletion on PLK1 activity were examined. As PLK1 is activated during mitosis (Nigg, 2001), T210 within PLK1 was phosphorylated when cells were synchronized at mitosis using nocodazole (Noc; Fig. 4 A, first row, compare lane 4 with lane 1; Jang et al., 2002). Phosphorylation was further increased by the depletion of LATS1 and MYPT1 (Fig. 4 A, first row, lanes 5 and 6), implying that LATS1 has an inhibitory effect on PLK1 via the MYPT1–PP1C pathway. Up-regulated PLK1 activity was also confirmed by increase in the phosphorylated form of Cdc25 and the decreased abundance of Wee1, both of which are the significant downstream targets of PLK1 (Fig. 4 A, third row [arrowhead] and fourth row; Kumagai and Dunphy, 1996; Watanabe et al., 2004). To confirm the result obtained using RNAi, HeLa cell lines stably expressing the mock, wild-type, or KD mutant of LATS1 were established. The KD mutant form of LATS1 has dominant-negative effects on endogenous LATS1 (Iida et al., 2004). When cells were synchronized at mitosis, a band corresponding to pT210 PLK1 was more evident in KD mutant cells than wild-type LATS1-expressing cells (Fig. 4 B, third row), further indicating the negative effects of LATS1 on PLK1.
Figure 4.
LATS1 regulated PLK1 activity. (A) LATS1 depletion promoted PLK1 activity. HeLa cells were transfected with control (Ctrl; GL2), LATS1, or MYPT1 siRNA, after which cells were grown asynchronously (AS) or treated with Noc subsequent to mitotic shake-off (M). The resultant cells were lysed and then subjected to immunoblot analysis with the indicated antibodies. Arrowhead indicates the phosphorylated form of Cdc25. (B) Stable expression of a LATS1 dominant-negative mutant (KD) led to PLK1 activation. HeLa cells stably expressing mock, wild-type (WT), or KD LATS1 were collected as described for mitotic shake-off (M) and analyzed as in A. (C) PLK1 activation in LATS1−/− MEFs. Primary MEFs derived from LATS1−/− mouse embryos were subjected to serum starvation for 72 h and, after release into complete medium for 12 h, arrested at mitosis by treatment with 10 µM K858 and collected 12 h later. For the add-back experiments, MEFs transfected with wild-type or KD HA-LATS1 were released from serum starvation and collected as described in A. The cells were analyzed as in A.
Next, mouse embryonic fibroblasts (MEFs) derived from LATS1 knockout (KO) mice (Fig. S2) were examined. Homozygous KO (LATS1−/−) MEFs were synchronized at mitosis using the Eg5 inhibitor K858 (Nakai et al., 2009), and the activation status of PLK1 (pT210) was evaluated by immunoblotting. Consistent with the aforementioned results, LATS1−/− MEFs showed an increase in the density of pT210 (Fig. 4 C, second row). To analyze whether LATS1 kinase activity was responsible for PLK1 regulation, wild-type or KD mutant LATS1 was added back into LATS1−/− MEFs, and PLK1 activity was examined after mitotic synchronization. As shown in Fig. 4 C, PLK1 activity was effectively suppressed in wild-type LATS1-expressing cells but not in KD mutant cells (Fig. 4 C, second row, lanes 2–4). These results support the notion that LATS1-mediated phosphorylation is involved in MYPT1–PP1C-mediated PLK1 suppression.
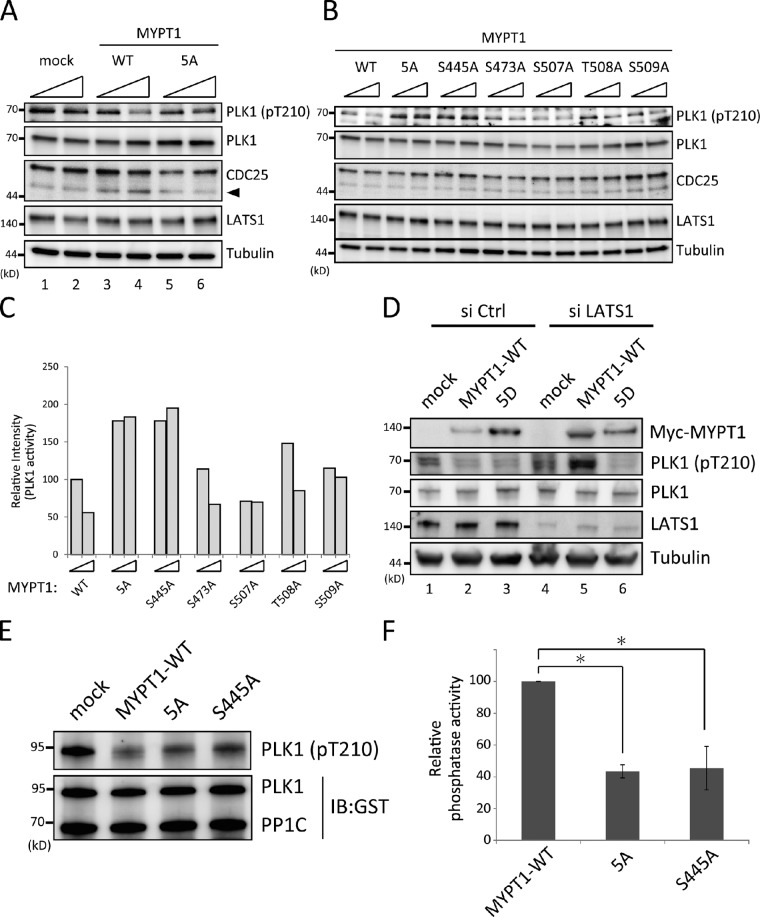
LATS1 activated MYPT1–PP1C to counteract PLK1
To explore the molecular link between LATS1 and PLK1, we next examined whether LATS1-mediated phosphorylation of MYPT1 participates in PLK1 regulation. To this end, wild-type or mutant forms of MYPT1 were overexpressed in cells and PLK1 activity was subsequently analyzed via T210 phosphorylation status. To exclude any possible effects on the cell cycle or cell survival by transient overexpression of MYPT1, 293T cells were used for transfection (Wu et al., 2005) and arrested at prometaphase to compare PLK1 activity. Overexpression of wild-type MYPT1 caused a marked decrease in T210-phosphorylated PLK1 protein expression in a dose-dependent manner compared with that in the control (Fig. 5 A, first row, lanes 1–4), which is consistent with a previous study indicating the antagonistic effects of MYPT1 on PLK1 (Yamashiro et al., 2008). Consistently, the unphosphorylated form of Cdc25, downstream targets of PLK1, was increased in wild-type MYPT1-expressing cells (Fig. 5 A, third row, arrowhead). However, the observed inhibition of wild-type MYPT1 to PLK1 was abrogated by the 5A mutation (Fig. 5 A, first row, lanes 5 and 6), suggesting that these MYPT1 phosphorylation sites are important for its activity toward PLK1. To further analyze the regulatory mechanisms of MYPT1 in more detail, various MYPT1 mutants were introduced into 293T cells, and PLK1 activity was examined via T210 phosphorylation status. Of the five phosphorylation sites within MYPT1, only the S445A mutant showed the same inhibitory effect against PLK1 T210 as the 5A mutant (Fig. 5, B and C). Although it has been reported that phosphorylation of MYPT1 S473 generates the binding motif for the polo box domain of PLK1 to inhibit its activity (Yamashiro et al., 2008), the S473A mutant alone had little effect on PLK1 T210 phosphorylation (Fig. 5, B and C). These results imply that an interaction via phosphorylated S473 is not enough to modulate PLK1 activity.
Figure 5.
LATS1-mediated phosphorylation of MYPT1 promoted MYPT1–PP1C activity toward PLK1. (A) The MYPT1 5A mutant showed reduced activity against PLK1. 293T cells were transfected with increasing amounts (1 or 2 µg) of the mock, Myc-MYPT1-wild type (WT), or Myc-MYPT1-5A mutant expression vectors for 48 h and then treated with Noc for 12 h. Cells were collected, lysed, and subjected to immunoblot analysis with the indicated antibodies. Arrowhead indicates the unphosphorylated form of Cdc25. (B) Determination of the responsible phosphorylation site for phosphatase activity. Myc-MYPT1 with the indicated mutations was introduced into cells and analyzed as in A. (C) Quantification of the pT210 signals. Relative intensity to that of 1 µg MYPT1-wild type is shown (percentage of control; wild type was set as 100%). The data shown are from a single representative experiment out of three repeats. (D) Phosphomimic mutant (5D) induced PLK1 inactivation in the absence of LATS1. 293T cells were transfected with siRNA against LATS1 or control for 48 h and subsequently with the indicated vectors. The cells were treated with Noc for 12 h before harvest and analyzed as in A. (E) In vitro phosphatase assay. 293T cells were transfected with mock, Myc-MYPT1-wild type, Myc-MYPT1-5A mutant, or Myc-MYPT1-S445A mutant and treated with Noc for 14 h. Proteins precipitated by the anti-Myc antibody were subjected to an in vitro phosphatase assay as described in the Materials and methods. (F) Quantification of phosphatase activity. The intensity of the bands precipitated with the anti-pT210 antibody was quantified and normalized according to the intensity of PLK1 bands (anti-GST antibody). Data show the relative intensity compared with the wild type (percentage of wild type) and represent the mean of three independent experiments (*, P < 0.05, Student’s two-tailed t test). Error bars indicate the SDs. Ctrl, control; IB, immunoblot.
To further evaluate whether LATS1-mediated phosphorylation is important for MYPT1 regulation, LATS1 was depleted in cells overexpressing MYPT1. In the absence of LATS1, wild-type MYPT1 lost its ability to decrease T210-phosphorylated PLK1 protein (Fig. 5 D, second row, lanes 4 and 5), whereas phosphomimic mutant of MYPT1 (5× D) efficiently inhibited T210 phosphorylation (Fig. 5 D, lanes 4 and 6), indicating that the identified phosphorylation sites are important for MYPT1 activity.
To confirm the results of the in vivo overexpression experiments, in vitro phosphatase assays were performed. Wild-type or mutant MYPT1 was overexpressed in 293T cells, which were arrested at prometaphase using Noc and subsequently subjected to immunoprecipitation. Immunopurified MYPT1 was combined with recombinant GST-PP1C to reconstitute the MYPT1–PP1C complex and then used in a phosphatase assay incorporating an active form of GST-fused full-length PLK1 as a substrate. MYPT1–PP1C activity toward PLK1 was analyzed by immunoblotting with an anti-PLK1 pT210 antibody. As shown in Fig. 5 (E and F), PP1C plus the 5A or S445A mutant of MYPT1 showed significantly reduced levels of phosphatase activity toward PLK1 compared with wild-type MYPT1, which was consistent with the in vivo assay results (Fig. 5, A–D). This strongly suggested that LATS1-mediated phosphorylation of MYPT1 promotes MYPT1–PP1C activity toward PLK1.
LATS1–MYPT1 pathway was required for DNA damage–induced PLK1 inactivation
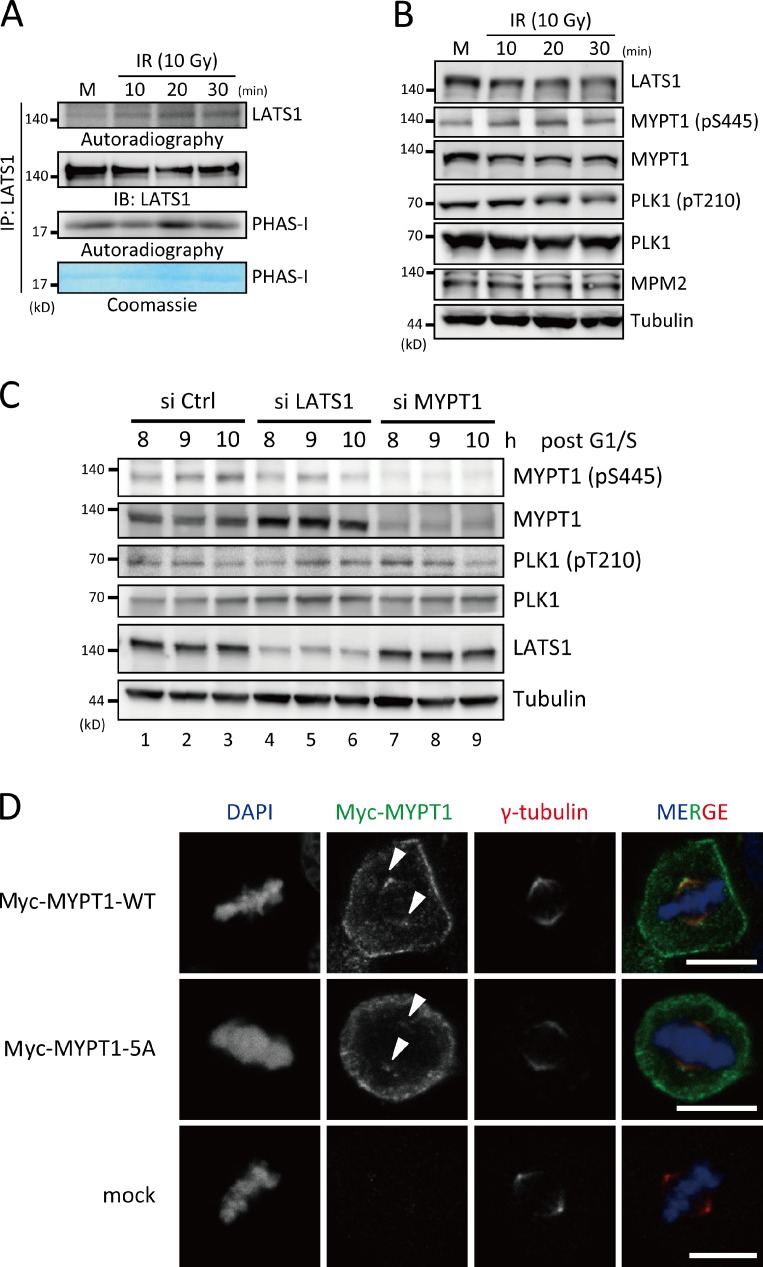
LATS1 is a mitotic kinase but its kinase activity during mitosis is subtle (Iida et al., 2004). It has been reported that the RASSF1A–MST2–LATS1 signaling cascade is activated upon DNA damage (Hamilton et al., 2009). In contrast, PLK1 is inactivated upon DNA damage (Smits et al., 2000). These reciprocal effects of DNA damage on the activity of each kinase prompted us to examine whether MYPT1–PP1C links these two different functional ends. First, we confirmed that LATS1 activity was up-regulated after DNA damage, which was analyzed by autophosphorylation or the phosphorylation of an in vitro substrate, PHAS-I (phosphorylated heat–acid stabled protein I; Fig. 6 A and Fig. S3 A). Consistent with a previous study (Tsvetkov and Stern, 2005), DNA damage–induced attenuation of PLK1 activity was detected via its phosphorylation of T210 in U2OS cells (Fig. 6 B, fourth row). Importantly, the phosphorylation status of MYPT1 at S445 was reciprocally correlated with PLK1 activity (T210 phosphorylation; Fig. 6 B, second and fourth rows). Phosphorylation of S445 within MYPT1 was still detected in mitotic arrested U2OS cells using an MYPT1 phospho-S445–specific antibody, possibly reflecting the existence of other S445 kinases, such as NUAK1 (Zagórska et al., 2010), but the phosphorylation level increased approximately twofold and peaked 20 min after ionizing radiation (IR; Fig. 6 B, second row), which correlated well with the timing of PLK1 suppression (Fig. 6 B, fourth row). To determine whether LATS1 was responsible for DNA damage–induced MYPT1 S445 phosphorylation, HeLa cells synchronized at G1/S boundary by double-thymidine block were released and transfected with siRNA against LATS1, MYPT1, or control. The G2/M cells were treated with 0.5 µM doxorubicin (DRB). Under these conditions, S445 phosphorylation was reduced in LATS1-depleted cells (Fig. 6 C, first row, lanes 4–6), but it was conversely increased in control cells (Fig. 6 C, first row, lanes 1–3). The decline in PLK1 activity (T210 phosphorylation) was observed in control cells, whereas the activity was relatively retained in both LATS1- and MYPT1-depleted cells (Fig. 6 C, third row), indicating that LATS1-mediated phosphorylation of MYPT1 leads to PLK1 inhibition after DNA damage.
Figure 6.
DNA damage–induced LATS1 activation led to PLK1 suppression. (A) DNA damage–induced LATS1 activation. U2OS cells were treated with 250 nM Noc. Mitotic cells, detached by repeated shaking, were collected at the indicated times after ionizing radiation (IR). Endogenous LATS1 was immunoprecipitated and then subjected to an in vitro kinase assay, in which the phosphorylation of PHAS-I (phosphorylated heat–acid stabled protein I) and autophosphorylation of LATS1 were analyzed. Immunoprecipitated proteins were also subjected to immunoblot analysis with the anti-LATS1 antibody. (B) MYPT1-S445 phosphorylation was increased after IR. U2OS cells were treated and collected as described in A and subjected to immunoblot analysis with the indicated antibodies. (C) MYPT1-S445 phosphorylation was dependent on LATS1 kinase. HeLa cells blocked with thymidine were released and transfected with siRNA against LATS1, MYPT1, or control. The G2/M cells were treated with 0.5 µM DRB. The cells were collected at the indicated times from the release and then subjected to immunoblot analysis with the indicated antibodies. (D) Centrosomal localization of the MYPT1-5A mutant. HeLa cells were transfected with mock, Myc-MYPT1-wild type (WT), or Myc-MYPT1-5A mutant expression vectors for 48 h. Cells were preextracted with digitonin in KHM buffer, fixed with cold methanol, and processed for indirect immunofluorescence staining with anti–γ-tubulin and anti-Myc antibodies. DNA was visualized by DAPI. Arrowheads indicate centrosomes. Bars, 5 µm. IP, immunoprecipitation; IB, immunoblot; Ctrl, control; M, mitotic cells.
Next, HeLa cells were transfected with the 5A mutant MYPT1 expression vector, and their localization was analyzed to determine whether LATS1 modulates MYPT1 localization via its phosphorylation. As shown in Fig. 6 D, both MYPT1-5A and wild-type MYPT1 were localized in centrosomes at metaphase, where PLK1 staining is detected (Nigg, 2001), indicating that S445 phosphorylation of MYPT1 is not necessarily required for colocalization with PLK1. Therefore, together with the in vitro analysis, these results strongly suggest that LATS1-mediated phosphorylation of MYPT1 promotes MYPT1–PP1C activity toward PLK1 in vivo.
Role of LATS1–MYPT1 pathway in the G2 DNA damage response
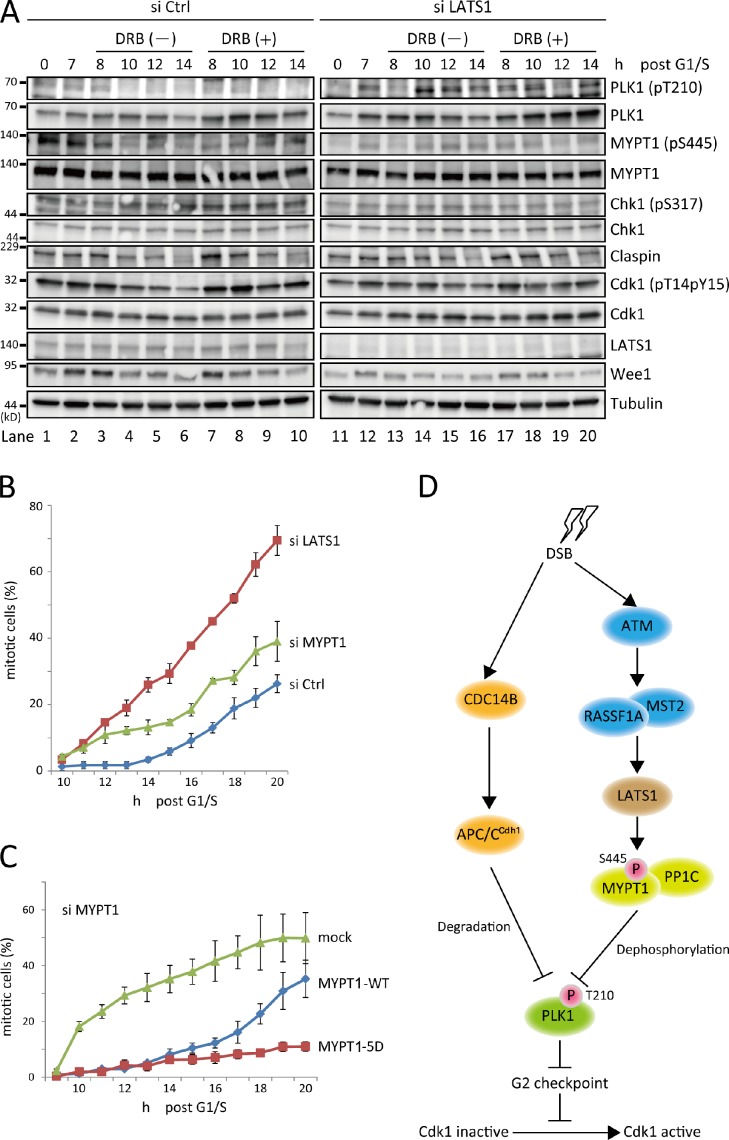
PLK1 suppression is required for an efficient DNA damage–induced G2 checkpoint by inducing the stabilization of Claspin and Wee1 (Bassermann et al., 2008). Therefore, to address the physiological role of LATS1-mediated regulation of PLK1, the G2 DNA damage response in LATS1-depleted cells was examined. HeLa cells were transfected with an siRNA oligo against LATS1 and synchronized at the G1/S boundary using a double-thymidine block. The cells were released and treated with DRB for 1 h at G2 phase (7 h after release), and the G2 checkpoint response or mitotic entry was examined. 1 h after DNA damage in control cells, Chk1 (checkpoint kinase 1) was activated via phosphorylation at S317 (Fig. 7 A, fifth row, lanes 7–10), whereas LATS1-depleted cells showed significantly reduced phosphorylation at that time point (Fig. 7 A, fifth row, lanes 17–20). In addition, the abundance of Wee1 was declined in LATS1-depleted cells (Fig. 7 A, eleventh row, lanes 7–10 and 17–20; and Fig. S4). Concomitantly, inhibitory phosphorylation of Cdk1 (Tyr15) was attenuated in LATS1-depleted cells (Fig. 7 A, eighth row, lanes 7–10 and 17–20; and Fig. S4). However, we could not detect a significant difference of Claspin abundance between control and LATS1-depleted cells (Fig. 7 A, seventh row, lanes 7–10 and 17–20; and Fig. S4), suggesting the LATS1-mediated regulation of Chk1 is less dependent on the Claspin. Overall, mitotic entry after DNA damage was significantly accelerated in LATS1-depleted cells compared with that in control cells (Fig. 7 B), indicating that G2 checkpoint control was compromised when LATS1 was knocked down.
Figure 7.
G2 checkpoint defect in LATS1-depleted cells. (A) LATS1-depleted cells showed a reduced G2 checkpoint response. HeLa cells were synchronized at the G1/S boundary by a double-thymidine block. siRNA transfection was performed at the time of first release from the thymidine block. The transfected cells were pulsed with solvent or 0.5 µM DRB (1 h) 7 h after release from the G1/S boundary, subsequently cultured in complete medium containing Noc, and collected at the indicated times. The cells were then subjected to immunoblot analysis with the indicated antibodies. (B) LATS1-depleted cells show abrogated G2 arrest after DNA damage. HeLa cells stably expressing Histone H2B-GFP were transfected with the indicated siRNA oligos and synchronized at G1/S by a double-thymidine block. Cells were released from block, allowed to progress to G2, and pulsed for 1 h with DRB as described in A. Mitotic entry was monitored using time-lapse microscopy. Mitotic cells in three different fields were counted at the indicated time points (percentage of cells). The graph shows the mean of these counts, and error bars indicate the SDs. Three independent experiments showed the same results. (C) Role of the LATS1-dependent phosphorylation of MYPT1 on G2 checkpoint control. HeLa cells stably expressing mock or MYPT1 (wild type [WT] and 5D) were treated with siRNA oligo specifically targeting the 3′ untranslated region of MYPT1 RNA to replace the endogenous gene with each mutant. The cells were synchronized and pulsed with DRB as in B and subjected to time-lapse microscopy to monitor mitotic entry. Mitotic cells in three different fields were counted at the indicated time points (percentage of cells). The graph shows the mean of these counts, and error bars indicate the SDs. Two independent experiments showed the same results. (D) A proposed model for the G2 DNA damage checkpoint. For simplicity, many downstream targets of ATM and PLK1 and other G2 checkpoint components have been omitted. Arrows indicate stimulatory interactions, and blunt lines indicate inhibitory interactions. LATS1-mediated phosphorylation of MYPT1 (S445) activates MYPT1–PP1C toward PLK1, suppressing PLK1 activity. Because suppression of PLK1 activity is essential for the establishment and maintenance of an efficient G2 checkpoint, LATS1 is positively involved in this process. DSB, double-strand DNA break; Ctrl, control; P, phosphorylated; APC/C, anaphase-promoting complex/cyclosome.
MYPT1-depleted cells also entered mitosis more quickly than control cells (Fig. 7 B), indicating that MYPT1 also contributes to G2 checkpoint. But mitotic entry of MYPT1-depleted cells was not faster than that of LATS1 knockdown cells (Fig. 7 B). We next examined the effect of LATS1-dependent phosphorylation of MYPT1 on G2 checkpoint control. HeLa cells stably expressing mock or MYPT1 (wild type and 5D) were treated with siRNA oligo specifically targeting the 3′ untranslated region of endogenous MYPT1 RNA. Under these conditions, wild-type MYPT1 could rescue the deficient G2 checkpoint of MYPT1-depleted cells (Fig. 7 C, compare mock with MYPT1-WT). Furthermore, 5D MYPT1–expressing cells arrested at G2 more efficiently than wild-type MYPT1–expressing cells (Fig. 7 C), indicating that LATS1-mediated phosphorylation of MYPT1 is involved in G2 checkpoint. These results suggest that the LATS1–MYPT1 pathway contributes to the G2 DNA damage response.
Discussion
Phosphoproteomic screening identified MYPT1 as a new target for LATS1. MYPT1 is a part of MP, a holoenzyme comprising three subunits: a catalytic subunit, PP1C; a targeting subunit, MYPT1; and a smaller subunit of 20 kD, M20. MP reduces the phosphorylation of the myosin light chain (S19) to inhibit actomyosin contraction (Matsumura and Hartshorne, 2008). MYPT1 modulates the targeting of a substrate, phosphorylated myosin light chain, to MP (Cohen, 2002). However, a recent study indicates that MYPT1 has another target, PLK1 (Yamashiro et al., 2008), which is an important player in mitosis (Glover, 2005). MYPT1 is specifically phosphorylated by Cdk1 at S473 during mitosis, the site selectively bound by PLK1 via its polo box domain. MYPT1–PP1C subsequently antagonizes PLK1 by removing its activation phosphorylation site (T210; Yamashiro et al., 2008). The present study identified S473 as a one of the significant LATS1 phosphorylation sites within MYPT1 (Fig. 3, B and E). Of the five identified phosphorylation sites, alanine substitution at S445 showed the strongest effects in the in vitro phosphorylation assay (87.8% reduction in phosphorylation), whereas S473 showed the weakest (26.6% reduction; Fig. 3 E). Consistent with these results, S445 is highly conserved from flies to humans and is critical for MYPT1–PP1C activity toward PLK1 (Fig. 3 B, residue shown in blue; and Fig. 5, C and F). We observed that full-length PLK1 was coprecipitated with the 5A MYPT1 mutant (containing S473A; Fig. S3 B). Pull-down analysis further revealed that PLK1 formed a complex with the C terminus of MYPT1 (CD, amino acids 654–1,030), indicating that more than two binding sites for PLK1 exist within MYPT1 (Fig. S3 C). These results suggest that binding to MYPT1 might not be a major determinant governing the dephosphorylation of PLK1 by MYPT1–PP1C. Indeed, no significant change in the interaction between the S445A mutant form of MYPT1 and PLK1 was observed (Fig. S3 B). Furthermore, centrosomal localization of MYPT1 was not influenced by LATS1 depletion even after IR (Fig. S5). We also found the 5A mutation of MYPT1 had no significant effect on the interaction with PP1C (Fig. S3 D). These results suggest that LATS1-mediated phosphorylation (S445) does not affect the affinity of MYPT1 to PLK1 or to PP1C but rather modulates MYPT1–PP1C activity toward PLK1. Additionally, it is interesting to note that abundance of both PLK1 and MYPT1 can be modulated by LATS1 or its phosphorylation signal (Fig. 4 A, Fig. 5 D, Fig. 7 A, and Fig. S3 B). Because both protein levels are regulated by the ubiquitin–proteasome system (Bassermann et al., 2008; Twomey et al., 2010), LATS1-mediated phosphorylation might also be involved in the posttranslational control of these proteins.
The MEN counteracts Cdk activity at mitotic exit in budding yeast (Stegmeier and Amon, 2004). Interestingly, the septation initiation network, the equivalent to MEN in fission yeast, counteracts Cdk activity not at mitotic exit but rather at mitotic entry (Trautmann et al., 2001). When the fission yeast Cdc14 homologue Clp1 is deleted, cells enter mitosis precociously, whereas mitotic entry is delayed in Clp1-overexpressing cells (Trautmann et al., 2001). Similar to Clp1 in fission yeast, human Cdc14B is also involved in the regulation of mitotic entry, particularly during G2 DNA damage response (Bassermann et al., 2008). In response to genotoxic stress in G2, Cdc14B translocates from the nucleolus to the nucleoplasm and induces the activation of the E3 ubiquitin ligase complex, anaphase-promoting complex/cyclosomeCdh1, which targets PLK1 for degradation (Bassermann et al., 2008). PLK1 suppression stabilizes Wee1, or Claspin to promote ATR (ataxia telangiectasia and rad3 related)-mediated phosphorylation and activation of Chk1. These processes maintain Cdk1 suppression (Bartek and Lukas, 2007; Harper and Elledge, 2007). Thus, Cdc14B mediates the G2 DNA damage response via anaphase-promoting complex/cyclosomeCdh1–induced PLK1 degradation (Fig. 7 D; Bassermann et al., 2008). As reported by Hamilton et al. (2009), the ATM–RASSF1A–MST2 pathway activates LATS1 to induce stabilization of the YAP1–p73 transcriptional complex after genotoxic stress. The present study provides evidence for a new pathway linking DNA damage to PLK1 suppression via LATS1–MYPT1–PP1C. In response to genotoxic stress, ATM activates LATS1 to inhibit mitotic entry via MYPT1–PP1C-mediated PLK1 suppression (Fig. 7 D). These results, and those of Bassermann et al. (2008), indicate that components of the mammalian MEN signaling pathway (LATS1, Cdc14B, and Cdh1) play an important role for the G2 DNA damage checkpoint response. However, a recent study (Mocciaro et al., 2010) contradicts the roles of Cdc14B and Cdh1 in this checkpoint (Sudo et al., 2001; Bassermann et al., 2008). Further study is required to clarify this issue.
A recent cell cycle study has highlighted the link between kinases and phosphatases (Barr et al., 2011). For example, mitotic entry cannot only be explained by a canonical mechanism that regulates Cdk1 via synthesis and stability of Cyclin B or via the activation of Cdc25 phosphatases. To enable mitotic entry, Greatwall (Gwl) kinase inhibits PP2A and sensitizes cells to Cdk1 activation. Gwl phosphorylates two small, closely related heat-stable proteins called endosulfine-α and ARPP-19 (cAMP-regulated phosphoprotein-19; Gharbi-Ayachi et al., 2010; Mochida et al., 2010). Phosphorylated endosulfine-α and ARPP-19 specifically bind to PP2A-Bδ and almost completely inhibit its activity (Gharbi-Ayachi et al., 2010; Mochida et al., 2010). Interestingly, both LATS1 and Gwl (also known as MASTL) belong to the AGC (PKA, PKG, and PKC) kinase family (Pearce et al., 2010) and have similar catalytic domain structures. Furthermore, some kinases in the AGC kinase family, such as Rho-associated kinase, myotonic dystrophy kinase-related Cdc42-binding kinase, and myotonic dystrophy protein kinase, modulate PP1C activity (Kimura et al., 1996; Murányi et al., 2001; Wilkinson et al., 2005). Therefore, together with the present study, the property of phosphatase regulation is likely to be one of the characteristics in this kinase family.
PLK1 is overexpressed in many cancers, and its expression levels often correlate with a poor prognosis (Strebhardt and Ullrich, 2006). Conversely, LATS1 expression is reduced in breast cancer patients because of hypermethylation of its promoter region, and the decreased expression of LATS1 is significantly associated with a poor prognosis (Takahashi et al., 2005). In addition, LATS1 KO mice develop soft tissue sarcomas and ovarian tumors (St John et al., 1999), indicating that LATS1 functions as a tumor suppressor. In the present study, LATS1 inhibited PLK1 activity via the MYPT1–PP1C. Thus, the oncogenic properties of PLK1, such as inducing genomic instability, would be enhanced under the condition of reduced LATS1 expression, consequently resulting in tumor formation and/or a poor prognosis. Thus, our data raise the intriguing possibility that cancer patients showing reduced LATS1 expression are good candidates for treatment using PLK1 inhibitors. Further studies will be required to assess the potential role of LATS1 for the PLK1 inhibitor treatment.
Materials and methods
Sample preparation for phosphoproteomic screening
HeLa cells were sonicated in chilled extraction buffer (20 mM Hepes/NaOH, pH 7.9, 0.25 M sucrose, 1.5 mM MgCl2, 10 mM KCl, 0.5% NP-40, and protease inhibitors) for 5 min. The lysates were centrifuged at 1,500 g for 10 min, and the supernatant was incubated with thermosensitive alkaline phosphatase (10 U/mg protein; Promega) at 37°C for 60 min followed by heat inactivation (75°C for 30 min). An in vitro kinase assay was performed by using recombinant active LATS1 (see In vitro kinase assays for details). Tris-HCl buffer, pH 9.0, urea, and octylglycoside were added to the reaction mixture at final concentrations of 0.1 M, 8 M, and 0.4%. The solution was reduced with DTT, alkylated with iodoacetamide, and digested with Lys-C followed by dilution (Saito et al., 2006). The digested samples were desalted using StageTips (Thermo Fisher Scientific) with C18 disk membranes (3M; Empore; Rappsilber et al., 2003). Dimethyl labeling of the tryptic digest with formaldehyde was performed as follows. The digested peptides were dried up with a centrifugal evaporator (CC-105; TOMY) and then dissolved in 100 mM triethylammonium bicarbonate. Normal and [2H2, 13C]-labeled formaldehyde solution was added to the control and the LATS1-treated sample, respectively. After addition of sodium cyanoborohydride, the mixture was incubated at RT for 1 h. Reaction was terminated by adding aqueous ammonia, and then, the samples were desalted using StageTips. Phosphopeptides were enriched by metal oxide chromatography using a lactic acid–modified titania tip as previously described (Sugiyama et al., 2007) with slight modifications. In brief, custom-made metal oxide chromatography tips were prepared by packing methanolic slurry of titania bulk beads (0.5 mg beads per 10-µl pipette tip) into pipette tips inserted with a C8 disk membrane (Rappsilber et al., 2007). The samples were diluted with an equal volume of solution A (300 mg/ml lactic acid and 0.1% trifluoroacetic acid in 80% acetonitrile) and loaded onto the tips. After washing, phosphopeptides were eluted with 0.5% piperidine.
Nano–liquid chromatography/MS/MS analysis and database searches
Nano–liquid chromatography/MS/MS analyses were performed by using an Orbitrap system (LTQ-Orbitrap XL; Thermo Fisher Scientific), a pump (UltiMate 3000; Dionex) with a flow manager (FLM-3000; Germering), and an autosampler (HTC PAL; CTC Analytics). 3-µm C18-AQ materials (ReproSil-Pur 120; Dr. Maisch, GmbH) were packed into a self-pulled needle (150-mm length × 100-µm inside diameter, 6-µm opening) with a nitrogen-pressurized column loader cell (Nikkyo Technos, Co., Ltd.) to prepare an analytical column. The mobile phases consisted of 0.5% acetic acid (A) and 0.5% acetic acid and 80% acetonitrile (B). A three-step linear gradient of 5–10% B in 5 min, 10–40% B in 60 min, 40–100% B in 5 min, and 100% B for 10 min was used at a flow rate of 500 nl/min. A spray voltage of 2,400 V was applied via the metal connector. The MS scan range was mass per charge of 300–1,500. The top 10 precursor ions were selected in MS scan by Orbitrap with R = 60,000 for MS/MS scans by ion trap in the automated gain control mode in which automated gain control values of 5.00 × 105 and 104 were set for full MS and MS/MS, respectively. To minimize repetitive MS/MS scanning, a dynamic exclusion time was set as 20 s with a repeat count of 1 and an exclusion list size of 500. The normalized collision-induced dissociation was set to be 35.0. A lock mass function was used for the LTQ Orbitrap to obtain constant mass accuracy during analyses.
Peptides and proteins were identified via automated database searching using Mascot version 2.3 (Matrix Science) against UniProt/SwissProt release 2010_11 (release date: November 2, 2010) with a precursor mass tolerance of 3 ppm, a fragment ion mass tolerance of 0.8 D, and strict trypsin specificity allowing for up to two missed cleavages. Cysteine carbamidomethylation was set as a fixed modification, and methionine oxidation, phosphorylation of serine, threonine, and tyrosine, and [1H4, 12C2/2H4, 13C2] dimethylation of N-terminal α-amino groups and lysine ε-amino groups in each peptide were allowed as variable modifications. Peptides were considered identified if the Mascot score was over the 95% confidence limit based on the identity score of each peptide, and at least three successive y or b ions with two and more y, b, and/or precursor origin neutral loss ions were observed. Phosphorylation sites were unambiguously determined when b or y ions, which occurred between the existing phosphorylated residues, were observed in the peak list of fragment ions (Sugiyama et al., 2008). False-positive rates were estimated to be 1.65% by searching against a randomized decoy database created by the Mascot Perl program supplied by Matrix Science. After identification, the peptide peak area of each peptide was integrated using Mass Navigator v1.2 (Mitsui Knowledge Industry), and the peak area ratio of LATS1-treated to control sample was calculated.
Plasmids
Human Gateway entry clones (Human Gene and Protein database clone numbers: MYPT1, FLJ94919AAAF; PLK1, FLJ80030AAAN; and PP1C-β, FLJ92643AAAF) were obtained from the National Institute of Technology and Evaluation Biological Resource Center. The cDNAs in these entry clones were subcloned into the destination expression vectors according to manufacturer’s instruction (Invitrogen). Mutations in MYPT1 cDNAs were introduced in vitro using the lightning multisite-directed mutagenesis kit (QuikChange; Agilent Technologies) and confirmed by DNA sequencing.
Cell culture, synchronization, and transfections
MEFs were prepared from a 13.5 d postcoitum embryo as follows: the viscera of each embryo were removed under a dissecting microscope (S6 E; Leica), and the embryo was washed in PBS and incubated with TrypLE Express (Invitrogen) for 20 min at 37°C. The cell suspension was then passed several times through a pipette and filtered through a 100-µm sterile cell strainer to remove cell clumps. The cell suspension prepared from each embryo was washed with medium containing 10% FBS and plated onto 100-mm culture dishes (Naoe et al., 2010). MEFs, HeLa cells, 293T cells, and U2OS cells were maintained in DME/F12 (Sigma-Aldrich) supplemented with 10% FBS under an atmosphere of 5% CO2 at 37°C. HeLa cells were synchronized at the G1/S boundary using a double-thymidine block and release protocol (24-h incubation with 2 mM thymidine, thymidine-free incubation for 8 h, and thymidine incubation for 14 h). Mitotic cells were collected by mechanical shake-off from the culture plate after 12–15 h of treatment with 60 ng/ml Noc. Cells were subjected to transient transfection using the FuGENE HD reagent (Roche) or the X-tremeGENE 9 DNA transfection reagent (Roche). Transfection of the RNAi oligo was performed using RNAiMAX reagent (Lipofectamine; Invitrogen). The target sequences of siRNA used in this study were as follows: firefly luciferase (GL2 as a control), 5′-CGTACGCGGAATACTTCGAdTdT-3′; LATS1 siRNA-1, 5′-ATTCGGGAATCCCTTAGGAdTdT-3′, and siRNA-2, 5′-GCAATTGAATTCATTAGTAdTdT-3′ (Kuninaka et al., 2005); and MYPT1 siRNA-1, 5′-AGTACTCAACCATAATTAAdTdT-3′, and siRNA-2, 5′-GCGCCAGAAGACCAAGGTGdTdT-3′ (Yamashiro et al., 2008). In each RNAi experiment, essentially the same results were observed using two independent RNAi sequences. For the production of stable cell lines, HeLa cells were cotransfected with pMX Myc-LATS1-wild type or pMX Myc-LATS1-KD using Plat-A packaging cells (Kitamura et al., 2003), and transfectants were selected in the presence of 2 µg/ml puromycin for 7 d. IR was performed using an x-ray apparatus (MBR-1520R-3; Hitachi) calibrated to deliver 0.52 Gy γ irradiation per minute.
Immunoprecipitation, GST pull-down, Western blotting, and antibodies
For immunoprecipitation or GST pull-down assay, HeLa or 293T cells transfected with the indicated vectors were washed with PBS and lysed by incubation for 15 min on ice with lysis buffer (0.5% NP-40, 25 mM Tris-Cl, pH 7.5, 137 mM NaCl, 1 mM EDTA, 5% glycerol, 1 mM DTT, 20 mM β-glycerophosphate, 1 mM sodium vanadate, and protease inhibitor [Complete Mini; Roche]). After centrifugation of the lysate at 14,000 g for 20 min, portions of the supernatant were incubated for 1 h at 4°C with specific antibodies immobilized on magnetic Dynabeads (Invitrogen) or indicated GST fusion proteins immobilized on glutathione–Sepharose beads (GE Healthcare). The beads were then separated by centrifugation and washed with lysis buffer, and the bound proteins were eluted by boiling for 5 min in SDS sample buffer and subjected to immunoblot analysis. For Western blotting, proteins fractionated by SDS-PAGE were electroblotted onto nitrocellulose membrane (Hybond ECL; GE Healthcare) using a semidry transfer apparatus. The membranes were blocked with 5% nonfat dried milk in PBS for 30 min at RT and incubated overnight at 4°C with primary antibodies diluted in Can Get Signal solution 1 (TOYOBO Co., Ltd.). Membranes were washed three times with PBST (PBS and 0.1% Tween 20) and incubated for 40 min with HRP-conjugated secondary antibodies (GE Healthcare), which were diluted in Can Get Signal solution 2. After three washes with PBST, the proteins were detected using ECL Plus (GE Healthcare) and a chemiluminescence detector (LAS-3000; Fujifilm). The antibodies used for the immunoprecipitation and Western blotting experiments were as follows: monoclonal anti–c-Myc (clone 9E10; Santa Cruz Biotechnology, Inc.), monoclonal anti-HA (3F10; Roche), rabbit anti-LATS1 (Bethyl Laboratories, Inc.), monoclonal anti-Plk1 (Invitrogen), rabbit anti-PP1C (Bethyl Laboratories, Inc.), rabbit anti-MYPT1 (Santa Cruz Biotechnology, Inc.), monoclonal anti-PLK1 (pT210; BD), rabbit anti-pCdk1(pThr14pTyr15; Abcam), monoclonal anti-Cdk1 (A17; Abcam), monoclonal anti-Wee1 (Santa Cruz Biotechnology, Inc.), rabbit anti-Claspin (Bethyl Laboratories, Inc.), monoclonal anti–α-tubulin (clone B-5-1-2; Sigma-Aldrich), monoclonal anti-Cdc25A (Santa Cruz Biotechnology, Inc.), monoclonal anti-MPM2 (Millipore), sheep anti-MYPT1 (pS445; provided by D. Alessi and J. Hastie, University of Dundee, Dundee, Scotland, UK), rabbit anti-pChk1 (Cell Signaling Techonology), monoclonal anti-Chk1 (Santa Cruz Biotechnology, Inc.), monoclonal anti-FLAG (M2; Sigma-Aldrich), and monoclonal anti-GST (Nacalai Tesque).
LATS1 KO mice
LATS1 KO mice were generated by homologous recombination using standard techniques (Fig. S2). Mice were backcrossed for more than five generations onto a C57BL/6 background. All animal experiments were approved by the Animal Ethics Committees of Keio University. Genotyping of mutant mice was performed using a PCR protocol based and the primers NeoR-S2, 5′-AGACAATCGGCTGCTCTGAT-3′; WTSex4S, 5′-CAAGATGCCCAGGATCAAAT-3′; and WTSin4AS2, 5′-AACAAAAATGGGCTGGTGAG-3′.
Immunofluorescence microscopy
MEFs or HeLa cells were seeded onto glass-bottom, 35-mm dishes to ∼70% confluence. A preextraction protocol was performed using 7.5 µg/ml digitonin in KHM buffer (25 mM Hepes-KOH, pH 7.2, 125 mM potassium acetate, and 2.5 mM magnesium acetate) for 5 min at RT before fixing with chilled absolute methanol for 10 min at −20°C (Hirota et al., 2000). Cells were then incubated and stained with the following antibodies: rabbit anti-LATS1 antibody (Bethyl Laboratories, Inc.), rabbit anti-MYPT1 antibody (Covance), rabbit anti-PLK1 antibody (Bethyl Laboratories, Inc.), monoclonal mouse anti–γ-tubulin (clone GTU-88; Sigma-Aldrich), rabbit anti–γ-tubulin (Sigma-Aldrich), and monoclonal mouse anti–c-Myc antibody (clone 9E10). This was followed by incubation with Alexa Fluor 488 goat anti–mouse IgG and Alexa Fluor 555 donkey anti–rabbit IgG (Invitrogen). Stained cells were mounted with DAPI Fluoromount-G (SouthernBiotech). Images were collected at RT (22°C) using 63×, 1.4 NA oil immersion lenses (Leica) on an upright microscope (DM5000; Leica) coupled to a confocal laser scanner (TCS SP5; Leica). LAS AF SP5 software (Leica) was used for data acquisition. All digital images were imported into Photoshop (Adobe) and adjusted for gain, contrast, and γ settings.
In vitro kinase assays
GST-LATS1 (catalytic domain), GST-Cdk1–Cyclin B, or GST-PLK1 (full length) were obtained from Carna Biosciences, Inc. Kinase reactions were conducted at 30°C for 30 min in a final volume of 20 µl containing 20 mM Tris, pH 7.4, 10 mM MgCl2, 10 µCi γ-[32P]ATP (3,000 Ci/mmol; Institute of Isotopes Co., Ltd.), and 1 mM DTT. Reactions were stopped by adding 4× SDS sample buffer and boiled at 95°C for 5 min. Samples were resolved by SDS-PAGE on 5–20% gradient gels and visualized by autoradiography. The intensity of the bands was quantified using BAS-5000 (Fujifilm).
In vitro phosphatase assays
Myc-MYPT1–transfected 293T cell lysates were subjected to immunoprecipitation using an anti-Myc antibody (Santa Cruz Biotechnology, Inc.). Immunoprecipitates were washed three times in lysis buffer and preincubated at 4°C for 10 min with PP1 assay buffer (20 mM Tris-HCl, pH 7.4, 1% Triton X-100, 250 mM sucrose, 1 mM MnCl2, and 0.1% β-mercaptoethanol) and 1.6 µg GST-PP1C. Next, 0.2 µg of active PLK1 (Carna Biosciences, Inc.) was added to the reaction solution to a final volume of 20 µl. Phosphatase reactions were performed at 30°C for 30 min and stopped by adding 4× SDS sample buffer. The samples were boiled at 95°C for 5 min and resolved by SDS-PAGE on 5–20% gradient gels. Phosphatase activity was quantified by subtracting the anti-PLK1 (pT210) band intensity of each MYPT1-overexpressing cell from that of mock-transfected cells and shown as the relative intensity to mock control (percentage of control).
Time-lapse imaging
HeLa cells stably expressing the GFP-tagged Histone H2B were grown onto a glass-bottomed dish (IWAKI) in DME/F12 supplemented with 10% FBS. During recording, cell cultures were kept at 37°C with 5% CO2. Time-lapse fluorescence and differential interference contrast video microscopy were performed using a microscope (LCV110; Olympus); images were acquired using a charge-coupled device camera (Retiga EXi; QImaging) at U Plan Super Apochromatic 40×, 0.95 NA objective every 10 min and analyzed using MetaMorph (Molecular Devices).
Statistical analysis
Two-tailed Student’s t test was used to determine comparative significance. The result was considered significant at P < 0.05.
Online supplemental material
Fig. S1 shows the mapping of LATS1 phosphorylation sites within MYPT1 by using single amino acid substitution mutants. Fig. S2 demonstrates the characterization of LATS1 KO mice. Fig. S3 shows the interaction between LATS1 and MYPT1. Fig. S4 demonstrates a single gel analysis of the samples shown in Fig. 7 A. Fig. S5 depicts that LATS1 depletion had no effect on the localization of endogenous MYPT1 and PLK1 after IR. Table S1 demonstrates mass spectrometric analysis of immunopurified MYPT1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201110110/DC1.
Supplementary Material
Acknowledgments
We would like to thank Dr. H. Yoshihara for help during the initial phase of the project; Y. Igarashi, S. Ohnuma, and M. Tsukahara for technical assistance; and all members of the Saya laboratory for helpful comments, discussion, and support.
This study was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and Keio Gijuku Academic Development Funds.
Footnotes
Abbreviations used in this paper:
- ANK
- ankyrin repeats domain
- ATM
- ataxia telangiectasia mutated
- CD
- C-terminal domain
- DRB
- doxorubicin
- Gwl
- Greatwall
- Hpo
- Hippo
- IR
- ionizing radiation
- KD
- kinase dead
- KO
- knockout
- MD
- middle domain
- MEF
- mouse embryonic fibroblast
- MEN
- mitotic exit network
- MP
- myosin phosphatase
- MS
- mass spectrometry
- Noc
- nocodazole
References
- Barr F.A., Elliott P.R., Gruneberg U. 2011. Protein phosphatases and the regulation of mitosis. J. Cell Sci. 124:2323–2334 10.1242/jcs.087106 [DOI] [PubMed] [Google Scholar]
- Bartek J., Lukas J. 2007. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19:238–245 10.1016/j.ceb.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Bassermann F., Frescas D., Guardavaccaro D., Busino L., Peschiaroli A., Pagano M. 2008. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 134:256–267 10.1016/j.cell.2008.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothos J., Tuttle R.L., Ottey M., Luca F.C., Halazonetis T.D. 2005. Human LATS1 is a mitotic exit network kinase. Cancer Res. 65:6568–6575 10.1158/0008-5472.CAN-05-0862 [DOI] [PubMed] [Google Scholar]
- Chan E.H., Nousiainen M., Chalamalasetty R.B., Schäfer A., Nigg E.A., Silljé H.H. 2005. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 24:2076–2086 10.1038/sj.onc.1208445 [DOI] [PubMed] [Google Scholar]
- Cohen P.T.W. 2002. Protein phosphatase 1—targeted in many directions. J. Cell Sci. 115:241–256 http://jcs.biologists.org/content/115/2/241 [DOI] [PubMed] [Google Scholar]
- Gharbi-Ayachi A., Labbé J.-C., Burgess A., Vigneron S., Strub J.-M., Brioudes E., Van-Dorsselaer A., Castro A., Lorca T. 2010. The substrate of Greatwall kinase, Arpp19, controls mitosis by inhibiting protein phosphatase 2A. Science. 330:1673–1677 10.1126/science.1197048 [DOI] [PubMed] [Google Scholar]
- Glover D.M. 2005. Polo kinase and progression through M phase in Drosophila: a perspective from the spindle poles. Oncogene. 24:230–237 10.1038/sj.onc.1208279 [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E., Tao C., Jafar-Nejad H., Halder G. 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8:27–36 10.1038/ncb1339 [DOI] [PubMed] [Google Scholar]
- Hamilton G., Yee K.S., Scrace S., O’Neill E. 2009. ATM regulates a RASSF1A-dependent DNA damage response. Curr. Biol. 19:2020–2025 10.1016/j.cub.2009.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., Elledge S.J. 2007. The DNA damage response: ten years after. Mol. Cell. 28:739–745 10.1016/j.molcel.2007.11.015 [DOI] [PubMed] [Google Scholar]
- Hirota T., Morisaki T., Nishiyama Y., Marumoto T., Tada K., Hara T., Masuko N., Inagaki M., Hatakeyama K., Saya H. 2000. Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J. Cell Biol. 149:1073–1086 10.1083/jcb.149.5.1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K., Pan D. 2005. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 122:421–434 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Iida S., Hirota T., Morisaki T., Marumoto T., Hara T., Kuninaka S., Honda S., Kosai K., Kawasuji M., Pallas D.C., Saya H. 2004. Tumor suppressor WARTS ensures genomic integrity by regulating both mitotic progression and G1 tetraploidy checkpoint function. Oncogene. 23:5266–5274 10.1038/sj.onc.1207623 [DOI] [PubMed] [Google Scholar]
- Ito M., Nakano T., Erdődi F., Hartshorne D.J. 2004. Myosin phosphatase: structure, regulation and function. Mol. Cell. Biochem. 259:197–209 10.1023/B:MCBI.0000021373.14288.00 [DOI] [PubMed] [Google Scholar]
- Jang Y.J., Ma S., Terada Y., Erikson R.L. 2002. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J. Biol. Chem. 277:44115–44120 10.1074/jbc.M202172200 [DOI] [PubMed] [Google Scholar]
- Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., Yamamori B., Feng J., Nakano T., Okawa K., et al. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science. 273:245–248 10.1126/science.273.5272.245 [DOI] [PubMed] [Google Scholar]
- Kitamura T., Koshino Y., Shibata F., Oki T., Nakajima H., Nosaka T., Kumagai H. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31:1007–1014 10.1016/S0301-472X(03)00260-1 [DOI] [PubMed] [Google Scholar]
- Kumagai A., Dunphy W.G. 1996. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science. 273:1377–1380 10.1126/science.273.5280.1377 [DOI] [PubMed] [Google Scholar]
- Kuninaka S., Nomura M., Hirota T., Iida S., Hara T., Honda S., Kunitoku N., Sasayama T., Arima Y., Marumoto T., et al. 2005. The tumor suppressor WARTS activates the Omi/HtrA2-dependent pathway of cell death. Oncogene. 24:5287–5298 10.1038/sj.onc.1208682 [DOI] [PubMed] [Google Scholar]
- Lindqvist A., Rodríguez-Bravo V., Medema R.H. 2009. The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J. Cell Biol. 185:193–202 10.1083/jcb.200812045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E., Guillamot M., de Cárcer G., Eguren M., Trickey M., García-Higuera I., Moreno S., Yamano H., Cañamero M., Malumbres M. 2010. Targeting mitotic exit leads to tumor regression in vivo: Modulation by Cdk1, Mastl, and the PP2A/B55α,δ phosphatase. Cancer Cell. 18:641–654 10.1016/j.ccr.2010.10.028 [DOI] [PubMed] [Google Scholar]
- Matsumura F., Hartshorne D.J. 2008. Myosin phosphatase target subunit: Many roles in cell function. Biochem. Biophys. Res. Commun. 369:149–156 10.1016/j.bbrc.2007.12.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocciaro A., Berdougo E., Zeng K., Black E., Vagnarelli P., Earnshaw W., Gillespie D., Jallepalli P., Schiebel E. 2010. Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J. Cell Biol. 189:631–639 10.1083/jcb.200910057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida S., Maslen S.L., Skehel M., Hunt T. 2010. Greatwall phosphorylates an inhibitor of protein phosphatase 2A that is essential for mitosis. Science. 330:1670–1673 10.1126/science.1195689 [DOI] [PubMed] [Google Scholar]
- Mohl D.A., Huddleston M.J., Collingwood T.S., Annan R.S., Deshaies R.J. 2009. Dbf2–Mob1 drives relocalization of protein phosphatase Cdc14 to the cytoplasm during exit from mitosis. J. Cell Biol. 184:527–539 10.1083/jcb.200812022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murányi A., Zhang R., Liu F., Hirano K., Ito M., Epstein H.F., Hartshorne D.J. 2001. Myotonic dystrophy protein kinase phosphorylates the myosin phosphatase targeting subunit and inhibits myosin phosphatase activity. FEBS Lett. 493:80–84 10.1016/S0014-5793(01)02283-9 [DOI] [PubMed] [Google Scholar]
- Nakai R., Iida S., Takahashi T., Tsujita T., Okamoto S., Takada C., Akasaka K., Ichikawa S., Ishida H., Kusaka H., et al. 2009. K858, a novel inhibitor of mitotic kinesin Eg5 and antitumor agent, induces cell death in cancer cells. Cancer Res. 69:3901–3909 10.1158/0008-5472.CAN-08-4373 [DOI] [PubMed] [Google Scholar]
- Naoe H., Araki K., Nagano O., Kobayashi Y., Ishizawa J., Chiyoda T., Shimizu T., Yamamura K., Sasaki Y., Saya H., Kuninaka S. 2010. The anaphase-promoting complex/cyclosome activator Cdh1 modulates Rho GTPase by targeting p190 RhoGAP for degradation. Mol. Cell. Biol. 30:3994–4005 10.1128/MCB.01358-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2:21–32 10.1038/35048096 [DOI] [PubMed] [Google Scholar]
- Olsen J.V., Vermeulen M., Santamaria A., Kumar C., Miller M.L., Jensen L.J., Gnad F., Cox J., Jensen T.S., Nigg E.A., et al. 2010. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3:ra3 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- Pan D. 2010. The hippo signaling pathway in development and cancer. Dev. Cell. 19:491–505 10.1016/j.devcel.2010.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce L.R., Komander D., Alessi D.R. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11:9–22 10.1038/nrm2822 [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M. 2003. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75:663–670 10.1021/ac026117i [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Saito H., Oda Y., Sato T., Kuromitsu J., Ishihama Y. 2006. Multiplexed two-dimensional liquid chromatography for MALDI and nanoelectrospray ionization mass spectrometry in proteomics. J. Proteome Res. 5:1803–1807 10.1021/pr0601178 [DOI] [PubMed] [Google Scholar]
- Schmitz M.H., Held M., Janssens V., Hutchins J.R., Hudecz O., Ivanova E., Goris J., Trinkle-Mulcahy L., Lamond A.I., Poser I., et al. 2010. Live-cell imaging RNAi screen identifies PP2A-B55alpha and importin-beta1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12:886–893 10.1038/ncb2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits V.A.J., Klompmaker R., Arnaud L., Rijksen G., Nigg E.A., Medema R.H. 2000. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2:672–676 10.1038/35023629 [DOI] [PubMed] [Google Scholar]
- St John M.A., Tao W., Fei X., Fukumoto R., Carcangiu M.L., Brownstein D.G., Parlow A.F., McGrath J., Xu T. 1999. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat. Genet. 21:182–186 10.1038/5965 [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Strebhardt K., Ullrich A. 2006. Targeting polo-like kinase 1 for cancer therapy. Nat. Rev. Cancer. 6:321–330 10.1038/nrc1841 [DOI] [PubMed] [Google Scholar]
- Sudo T., Ota Y., Kotani S., Nakao M., Takami Y., Takeda S., Saya H. 2001. Activation of Cdh1-dependent APC is required for G1 cell cycle arrest and DNA damage-induced G2 checkpoint in vertebrate cells. EMBO J. 20:6499–6508 10.1093/emboj/20.22.6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N., Masuda T., Shinoda K., Nakamura A., Tomita M., Ishihama Y. 2007. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol. Cell. Proteomics. 6:1103–1109 10.1074/mcp.T600060-MCP200 [DOI] [PubMed] [Google Scholar]
- Sugiyama N., Nakagami H., Mochida K., Daudi A., Tomita M., Shirasu K., Ishihama Y. 2008. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 4:193 10.1038/msb.2008.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M., Morgan D.O. 2007. Finishing mitosis, one step at a time. Nat. Rev. Mol. Cell Biol. 8:894–903 10.1038/nrm2276 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Miyoshi Y., Takahata C., Irahara N., Taguchi T., Tamaki Y., Noguchi S. 2005. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin. Cancer Res. 11:1380–1385 10.1158/1078-0432.CCR-04-1773 [DOI] [PubMed] [Google Scholar]
- Totsukawa G., Yamakita Y., Yamashiro S., Hosoya H., Hartshorne D.J., Matsumura F. 1999. Activation of myosin phosphatase targeting subunit by mitosis-specific phosphorylation. J. Cell Biol. 144:735–744 10.1083/jcb.144.4.735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautmann S., Wolfe B.A., Jorgensen P., Tyers M., Gould K.L., McCollum D. 2001. Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol. 11:931–940 10.1016/S0960-9822(01)00268-8 [DOI] [PubMed] [Google Scholar]
- Tsvetkov L., Stern D.F. 2005. Phosphorylation of Plk1 at S137 and T210 is inhibited in response to DNA damage. Cell Cycle. 4:166–171 10.4161/cc.4.1.1348 [DOI] [PubMed] [Google Scholar]
- Twomey E., Li Y., Lei J., Sodja C., Ribecco-Lutkiewicz M., Smith B., Fang H., Bani-Yaghoub M., McKinnell I., Sikorska M. 2010. Regulation of MYPT1 stability by the E3 ubiquitin ligase SIAH2. Exp. Cell Res. 316:68–77 10.1016/j.yexcr.2009.09.001 [DOI] [PubMed] [Google Scholar]
- Watanabe N., Arai H., Nishihara Y., Taniguchi M., Watanabe N., Hunter T., Osada H. 2004. M-phase kinases induce phospho-dependent ubiquitination of somatic Wee1 by SCFbeta-TrCP. Proc. Natl. Acad. Sci. USA. 101:4419–4424 10.1073/pnas.0307700101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S., Paterson H.F., Marshall C.J. 2005. Cdc42-MRCK and Rho-ROCK signalling cooperate in myosin phosphorylation and cell invasion. Nat. Cell Biol. 7:255–261 10.1038/ncb1230 [DOI] [PubMed] [Google Scholar]
- Wu Y., Murányi A., Erdődi F., Hartshorne D.J. 2005. Localization of myosin phosphatase target subunit and its mutants. J. Muscle Res. Cell Motil. 26:123–134 10.1007/s10974-005-2579-5 [DOI] [PubMed] [Google Scholar]
- Yamashiro S., Yamakita Y., Totsukawa G., Goto H., Kaibuchi K., Ito M., Hartshorne D.J., Matsumura F. 2008. Myosin phosphatase-targeting subunit 1 regulates mitosis by antagonizing polo-like kinase 1. Dev. Cell. 14:787–797 10.1016/j.devcel.2008.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Li D.M., Chen W., Xu T. 2001. Human homologue of Drosophila lats, LATS1, negatively regulate growth by inducing G(2)/M arrest or apoptosis. Oncogene. 20:6516–6523 10.1038/sj.onc.1204817 [DOI] [PubMed] [Google Scholar]
- Zagórska A., Deak M., Campbell D.G., Banerjee S., Hirano M., Aizawa S., Prescott A.R., Alessi D.R. 2010. New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Sci. Signal. 3:ra25 10.1126/scisignal.2000616 [DOI] [PubMed] [Google Scholar]
- Zhao B., Li L., Lei Q., Guan K.L. 2010. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24:862–874 10.1101/gad.1909210 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.