Abstract
Epidermal Growth Factor Receptor (EGFR) is a cell surface protein overexpressed in cancerous cells. It is known to be the most common oncongene. EGFR concentration also increases in the serum of cancer patients. The detection of small changes in the concentration of EGFR can be critical for early diagnosis, resulting in better treatment and improved survival rate of cancer patients. This article reports an RNA aptamer based approach to selectively capture EGFR protein and an electrical scheme for its detection. Pairs of gold electrodes with nanometer separation were made through confluence of focused ion beam scratching and electromigration. The aptamer was hybridized to a single stranded DNA molecule, which in turn was immobilized on SiO2 surface between the gold nanoelectrodes. The selectivity of the aptamer was demonstrated by using control chips with mutated non–selective aptamer and with no aptamer. Surface functionalization was characterized by optical detection and two orders of magnitude increase in direct current (DC) was measured when selective capture of EGFR occurred. This represents an electronic biosensor for the detection of proteins of interest for medical applications.
Keywords: Epidermal Growth Factor Receptor (EGFR), Focused Ion Beam (FIB) milling, electromigration, DNA, Aptamer, Break-junction
1. Introduction
Evolution of proteomics has enabled better understanding of disease progression, and helped in establishing new biomarkers for early diagnosis, appropriate treatment and faster drug discovery. Overexpression of Epidermal Growth Factor Receptor (EGFR) is considered to be involved in several types of cancers. Its overexpression has been reported as an indication for many cancers like breast, lung, cervical, bladder, esophageal and ovarian cancer [1–6]. The presence of elevated or reduced levels of a biomarker can point out the presence or onset of a disease, specify the phase of that disease and enable monitoring of changes in homeostasis [7]. Elevated levels of EGFR in the serum of lung cancer patients has been reported before. The normal EGFR concentration can be in the range of 45–78 ng/ml, whereas it may be as much as an order higher in the lymph node metastasis of lung cancer patients (850 ng/ml) [8, 9]. The early detection of a few tens to hundreds of ng/ml EGFR can be important to improve the prognosis of the patients.
A number of approaches have been reported for the detection of EGFR and its interactions. These techniques incorporate immunohistochemical, flow cytometric, amperometric, mechanical and optical detection modalities [10–15]. Optical waveguides have also been used for label-free sensing to study cell biology and for biomolecular detection [16–19]. These methods of detecting proteins are generally categorized as labeled detection and label-free detection methods. In the case of labeled detection, presence of proteins is confirmed by some secondary molecule (organic, inorganic or radioactive) that binds to the proteins. Optical detection utilizes a fluorescent tag attached to the proteins and the change in fluorescent intensity after binding signals the presence of proteins. Whereas electrical detection directly interrogates the presence of protein molecules. It entails efficient device fabrication in order to define contact structures with a molecular dimension separation. Charge transport properties of a single molecule have also been studied with such structures. Such works have been primarily to identify molecules that may be used as functional electronics components [20–22]. It was recently reported that a single molecule was captured by a probing aptamer between nanogapped single-walled carbon nanotubes (SWCNTs) and charge-transport properties were characterized [23].
There have been reports of many types of metal-molecule-metal structures to measure electrical behavior of molecules [22]. Such structures can be broadly classified as vertical device structures (VDS) and lateral device structures (LDS). VDS utilize a self-assembled monolayer of molecules grown on a metal surface which acts as one contact. The second contact is made through conductive probes of atomic force microscopes, scanning tunneling microscopes or crosswire arrangements [24–26]. LDS involves a pair of metal contacts with nanometer sized separation and the probes selective to protein molecules are immobilized between the contact structures. Such devices that contain a thin metal strip, in that a gap is created by some technique, are known as break-junctions. Such gaps can be formed through e-beam lithography, electrodeposition of metals or electromigration induced “breaking” of metal lines [27–31]. These techniques are either slow and costly, mainly because of e-beam writing of the whole structures/lines or have very low yield of functional devices. This article reports electrical detection of EGFR molecules by making use of a new class of break-junctions where a focused ion beam (FIB) is used to scratch a thin metal strip (made with optical lithography and lift-off), followed by conventional microscale probing and electromigration induced breaking to produce break-junctions with high throughput, greater yield, low cost and at exact spot, as recently reported [32].
Antibodies are mostly used to functionalize surfaces for protein binding and detection. The antibody-based devices are not well-suited for field-deployable or point-of-care modalities because these require specific ranges of temperature, humidity and ionic strengths of the buffer solution in order to retain their function [33]. Low ionic strength is necessitated to prevail over surface Debye screening but it causes poor interaction between target and surface probes [33]. Recent works have shown significant advancements introducing aptamers for several types of target molecules [34–41]. Aptamers are reported to be as selective and sensitive as antibodies but exceedingly stable over changing temperatures, humidity and ionic concentrations [42].
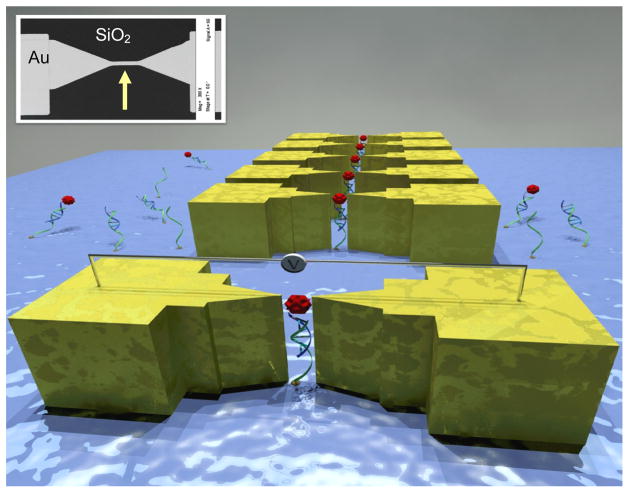
This article reports cheap and rapid nano-electrode manufacturing and direct current detection of low concentration of EGFR (50 μg/ml) binding to RNA aptamers. The effective attachment of RNA aptamers on a chemically modified silicon dioxide surface and EGFR binding to aptamer was detected from direct current electronic measurements. Fig. 1 shows a sketch of the sensor that describes the mechanism of the transduction. The inset shows a scanning electron microscope (SEM) micrograph of the actual device layout. Attachment of RNA aptamer to SiO2 surface and RNA-protein complex interactions were verified with optical measurements as well.
Figure 1.
A 3D sketch of the device that demonstrates the mechanism of the electronic detection (not to scale). Inset shows an SEM micrograph of the thin Au line on SiO2 chip.
2. Materials and methods
2.1. Materials
The chemicals used for this work were 3′-Aminopropyltriethoxysilane (APTES); 1,4-Phenylene diisothiocyanate (PDITC); Ethanol; Isopropyl alcohol (IPA); Acetone; Dimethyl sulfoxide (DMSO); Pyridine; 6–amino–1–hexanol; N,N-dimethylformamide (DMF); N,N-diisopropylethylamine (DIPEA); Phosphate buffered Saline (PBS); Magnesium chloride; Hybridization buffer solution and Diethylpyrocarbonate (DEPC) treated de-ionized (DI) water. The chemicals were obtained from Sigma-Aldrich (St. Louis, MO, USA). The 5′-amine modified single-strand DNA (ssDNA) were purchased from Alpha DNA (Montreal, Quebec, Canada).
The target molecule was recombinant human EGFR/ErbB1 Fc Chimera (R&D Systems, Minneapolis, MN). The receptor, as per the supplier, was able to bind recombinant human EGF in a functional ELISA at Kd<8nM.
2.2. Methods
The reported work included three distinct aims that were accomplished towards the final goal of a device that can detect EGFR in real-time. The first section involves the fabrication of nanogap break-junctions. The second one elucidates the immobilization of selective probes (aptamers) on the SiO2 substrates. The third and most important part demonstrates the integration of protein binding and its optical and electrical detection.
2.2.1. Controlled fabrication of nanogap break-junctions
A two-step photolithography process was carried out to define metallic structures on SiO2 chips. The first layer of pattern was used to create 3 μm wide lines of titanium/gold (thickness 50 Å/150 Å) using metal lift-off. The second step of photolithography defined 200×200 μm2 probing pads that were made of Ti/Au (thickness 100 Å/500 Å) and aligned to the far edges of lines made in first lithography/metallization/lift-off. Inset to Fig. 1 shows the device at this step. The metal lines were partially scratched by FIB milling process (at spot highlighted with arrow in inset to Fig. 1). These scratched parts were where the nanogap break-junctions were ultimately made. FIB scratching with gallium ions was done at 30 kV acceleration voltage, 1 pA milling current and 2 sec per micron scan rate of the beam to achieve optimum scratch depth. The current-voltage (I–V) characteristics across the metal lines were determined using Agilent 4155C semiconductor parameter analyzer on a probe station. The I–V measurements before and after the nanomanufacturing of metallic break-junctions were recorded to demonstrate the electromigration effect. The essential advantage of FIB scratching followed by electromigration is to increase the yield of useful devices with desired gap between electrodes, make nanogap breaks at precise location on the line, and narrow the distribution of the nanogap sizes. These are not possible with approaches involving only electromigration [21, 43–45].
2.2.2. Aptamer preparation
Aptamer was isolated with a well-known process called “Systematic Evolution of Ligands by Exponential Enrichment” (SELEX) as reported before [46, 47]. An iterative selection of binding species against purified human EGFR (R&D Systems, Minneapolis, MN) was done to isolate anti-EGFR RNA aptamer [48, 49]. The high affinity (Kd = 2.4 nM) anti-EGFR RNA aptamer and a non-functional mutated aptamer were extended with a capture sequence. The extended anti-EGFR aptamer (5′-GGC GCU CCG ACC UUA GUC UCU GUG CCG CUA UAA UGC ACG GAU UUA AUC GCC GUA GAA AAG CAU GUC AAA GCC GGA ACC GUG UAG CAC AGC AGA GAA UUA AAU GCC CGC CAU GAC CAG-3′); extended mutant aptamer (5′-GGC GCU CCG ACC UUA GUC UCU GUU CCC ACA UCA UGC ACA AGG ACA AUU CUG UGC AUC CAA GGA GGA GUU CUC GGA ACC GUG UAG CAC AGC AGA GAA UUA AAU GCC CGC CAU GAC CAG-3′) and amino modified substrate-anchored probe ssDNA (5′-amine-CTG GTC ATG GCG GGC ATT TAA TTC-3′) were used. The capture sequence has been highlighted in bold font. The capture sequence was the modification of isolated anti-EGFR aptamer and the mutant aptamer. This was made by extending the DNA template at its 3′ end and had 24 nucleotides. The extended capture sequence didn’t disrupt the aptamer structure but was used to immobilize aptamers on the surface through duplex-formation with the surface-bound ssDNA probe molecules.
2.2.3. Immobilization of ssDNA and probing aptamers on the SiO2 chip surface
A thermally oxidized silicon wafer was diced into 4×4 mm2 small chips. The chips were initially cleaned in oxygen plasma for 15 minutes using Ar+O2 at 200 W. This also resulted in a hydrophilic SiO2 surface. The chips were then immediately immersed in a solution of 2% APTES in ethanol for 1 hour at room temperature to get the surfaces silanized. The chips were then sequentially rinsed with IPA and DI water and dried in nitrogen flow, followed by curing at 115 °C for 30 minutes. The chips were then submerged in a solution of 1 mM PDITC in DMSO containing 10% pyridine for 5 to 7 hours at 45 °C in order to get PDITC homobifunctional linker molecules attached onto the silanized surface. The chips were then washed sequentially with ethanol and DI water and dried in nitrogen flow. A volume of 8 μl of amine modified ssDNA solution (5 μM concentration of ssDNA in DI water with 50% DMSO, 1% Pyridine) was placed on each substrate and allowed to incubate in a humidity chamber for 16 to 18 hours at 45 °C to attach amino modified ssDNA to PDITC. After ssDNA attachment, each chip was rinsed sequentially with ethanol and DEPC treated DI water and dried with nitrogen. The functionalized surface was then deactivated and unbound reactive groups of PDITC capped by submerging the chips in a blocking buffer (BB) solution that contained 50 mM 6-amino-1-hexanol with 150 mM DIPEA in DMF for 1 hour. The chips were then rinsed with DMF, ethanol, DEPC treated DI water and dried in nitrogen stream. After that, a volume of 8 μl of anti-EGFR RNA aptamer solution was placed on every substrate for hybridization by incubating the chips in the buffer solution (100 nM of anti-EGFR aptamer with 5:1 of DI water and hybridization buffer) at 42 °C for 1 hour. The chips were then washed thoroughly with DEPC treated DI water and dried with nitrogen gas. The capture extension of the anti-EGFR RNA aptamer hybridized with the surface bound complementary ssDNA. In one set of controls, the mutant aptamer was hybridized to surface-bound ssDNA on separate chips using identical protocol. The presence of ssDNA and RNA aptamers immobilized on the SiO2 surface was determined by fluorescence measurements of Acridine Orange (AO) stain at an excitation wavelength of 460 nm and the emission wavelength of 650 nm using a confocal microscope. Another set of controlled chips had no aptamer but underwent all other steps like the aptamer chips.
2.2.4. Protein binding to the probing aptamer
Chips with the attached RNA aptamer were then incubated in 50 ng/μl of EGFR protein prepared in PBS with 5 mM Mg2+ at 37 °C for 45 minutes by putting 8 μl of solution on each substrate. The chips were washed thoroughly with PBS and DEPC treated DI water and dried in the nitrogen flow. The chips were analyzed for the captured EGFR protein by optical detection of fluorescent stain Sypro Ruby Protein Blot at 488 nm wavelength. The fluorescence intensity analysis was done with ImageJ software [50]. Essentially, the surface bound ssDNA was used to immobilize an anti-EGFR aptamer and subsequently the aptamer was used to capture the EGFR protein.
After confirming the attachment chemistry for RNA immobilization and selectivity of anti-EGFR aptamer on plain chips, electrical detection of EGFR protein was performed using nanomanufactured metallic break-junction devices on oxidized silicon chips. An exactly identical attachment chemistry as for plain chips was employed for these devices. Although the concentration of EGFR used was high but the theoretical limit of detection (LOD) of the device is much smaller. As a first order estimate, the protein footstep is about 11 nm and in a nanogap 50 nm long and 3 μm wide, we would need about 400 EGFR copies to fill the whole area of nanogap [51]. However, surface charges at nanoscale, time of detection i.e. time it would take for EGFR to reach the capture region just by diffusion [52], steric hindrance, conformation of molecules will all contribute to LOD of this framework.
Semiconductor parameter analyzer recorded the direct current tunneling through the nanometer scale electrode separation before and after the protein binding to the probing aptamers in between these nano-electrodes.
3. Results and discussion
3.1. Fabrication and characterization of nanometer-sized break-junctions
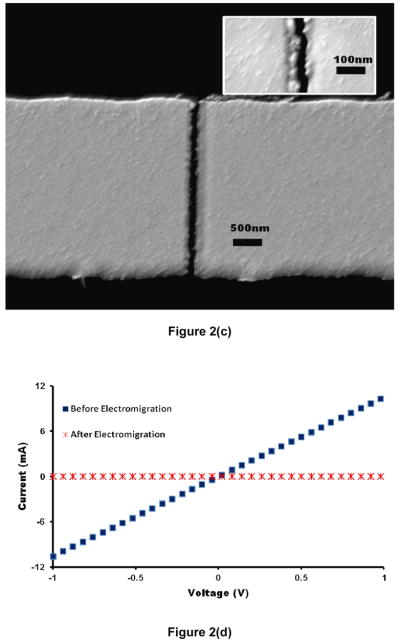
Optimum FIB scratches were obtained by utilizing our previously reported protocol [32]. The I–V data recorded after FIB scratching of the metal lines showed a linear ohmic behavior that confirmed electrical and physical continuity of the metal lines (Fig. 2(a)). A voltage sweep from 0 to 4 V broke the metal lines due to electromigration. Fig. 2(b) shows a sudden drop in current that depicts an absolute break in the metal line. The SEM micrographs also showed a complete break in the metal lines with nano-scale separation (shown clearly in the inset of Fig. 2(c)). Figure 2(d) demonstrates the comparison of I–V data recorded for the same voltage range (−1 to +1 V) before and after the application of electromigration-inducing bias. All data and micrographs of Fig. 2 are representative for the rest of the devices on the chips.
Figure 2.
Characterization of the device (a) Representative I–V data after FIB scratching of metal line shows linear ohmic behavior. (b) A sudden drop in current confirms the complete breaking of metal line due to electromigration and results in nanogap break-junctions. (c) SEM micrograph showing a clean break between nanoelectrodes. Inset shows magnified view of a break-junction with a gap smaller than 30 nm. (d) Comparison of I–V data for a representative break-junction before and after the electromigration.
A very small and random current flow was observed after the electrode separation that showed tunneling current characteristics. The tunneling current in an arrangement of the nanogap electrodes with vacuum as the insulator between them can be approximated as a function of the average barrier height relative to the Fermi level of the negative electrode, the barrier width and the applied voltage across the nanoelectrodes [53].
3.2. Optical detection of immobilized aptamer and captured protein
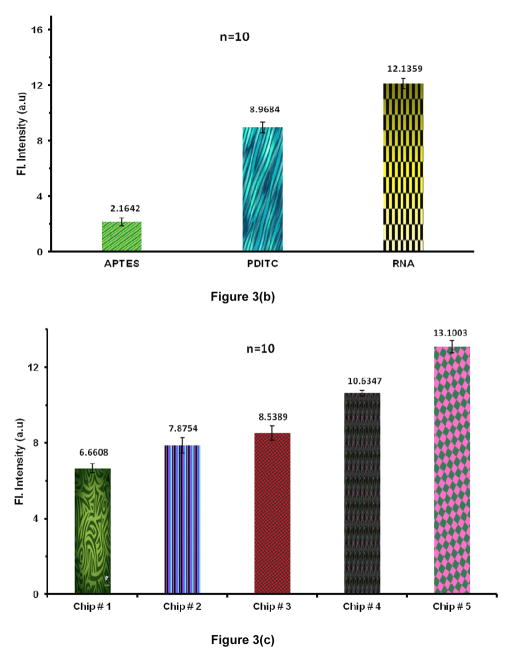
The surface binding and the presence of ssDNA and RNA aptamer immobilized on the plain SiO2 chips was confirmed by AO stain and the fluorescence measurements. Data of Fig. 3(a) confirms the binding of ssDNA to the functionalized plain SiO2 surface while Figure 3(b) depicts the hybridization of RNA aptamer to the surface-bound ssDNA. AO stain gives green fluorescence when it interacts with ssDNA whereas a flame-red fluorescence is observed when it interacts with double stranded nucleic acids [54]. The AO stain binds electrostatically to the nucleic acids as it carries positive charge. Electrostatic communication with non-specific polyanions was avoided by using a very low concentration of AO stain (0.2% v/v) and providing a buffer solution with Mg+2 and Na+ cations that competed binding to the nucleic acids [55].
Figure 3.
(a) Acridine Orange (AO) stain intensity measurements (background subtracted) depicts the presence of surface-bound DNA by comparing it with control chips. (n=10). (b) Fluorescence intensity measurements (background subtracted) using AO confirms the RNA hybridization to surface bound ssDNA when compared to control chips (n=10). (c) Sypro staining shows the selective capture of EGFR on functionalized SiO2. From L to R: Chip # 1: Control chip treated with blocking buffer (BB) but no aptamer; Chip # 2: Control chip with no BB treatment and no aptamer; Chip # 3: Control chip functionalized with mutant aptamer and treated with BB; Chip # 4: Functionalized chip with EGFR aptamer and captured protein after BB treatment; Chip # 5: Functionalized chip with EGFR aptamer and captured protein without using BB. (Error bars represent the standard deviation for n = 10)
EGFR binding to the probing aptamer onto the plain chip surface was confirmed with the Sypro Ruby protein gel stain. Sypro Ruby is a ruthenium based stain used to detect the amino acids lysine, arginine & histidine [56]. Figure 3(c) shows a clear enhancement of fluorescent intensity measured from the chips with anti-EGFR RNA aptamer probes and EGFR protein as compared to that for negative control chips that included probe-functionalized chips not exposed to the target proteins and the chips exposed to the target protein but without probe aptamer immobilized on the surface. The fluorescent intensity was at a maximum when use of a blocking buffer (BB) was omitted and therefore, unreacted PDITC moieties also contributed towards protein capturing. On the other hand, probe-functionalized chips with BB treatment selectively captured the EGFR protein as revealed by a marked increase in fluorescence intensity. The BB was thus used in subsequent experiments to avoid non-specific adsorption/binding on gold or oxide surfaces,
3.3. Electrical detection of EGFR binding
Fig. 4(a) shows representative data from one device that shows the electronic detection of selective EGFR binding. The control chips with exactly same devices but with mutant aptamer or no aptamer at all showed no change in conductivity (Figure 4(b)). The scrambled sequence of a mutant aptamer prevented it from recognizing the EGFR whereas anti-EGFR RNA aptamer selectively captured EGFR. The I–V measurements recorded from -1 to +1 V across the metal break-junctions, showed a significant increase in current after the capture of EGFR by surface immobilized aptamers. There were two orders of reduction in the resistance through the the nanoelectrodes indicating the conducting behavior of proteins that bridged the nanogap between the electrodes. The yield of devices was 60% that can be possibly increased by tagging the protein molecules with conducting nanoparticles, e.g. of gold. Serum EGFR levels elevate in many cancers and aptamers isolated through SELEX show high-affinity binding to these. The non-specific binding of the aptamer with non-target serum proteins has already been studied and found to be very atypical [8, 57–59]. Therefore the presented data shows the power of this electronic biosensor for direct detection of EGFR present in human serum or plasma.
Figure 4.
Comparison of I–V data for a representative break-junction before and after the surface modification and exposure to EGFR: (a) Anti-EGFR aptamer chip shows current increase due to the capture of EGFR bridging the nanogap; (b) Mutant aptamer functionalized control chip shows no change in conductivity, inset shows data for control chip with no aptamer and thus no change in conductivity. Both controls show no capture of EGFR.
4. Conclusion
A metallic break-junction framework with precise nanometer-sized separation is presented that utilizes aptamers for selective EGFR capture. The aptamer specifically binds, recognizes and captures EGFR in break-junctions. The capture is electrically detected. The integration of FIB scratching with electromigration provides rapid, cheap and controlled manufacturing of nanoscale break-junctions with high yield. The presence of protein between the electrodes shows a robust increase in conductivity between otherwise insulated nanoelectrodes. The detection of a disease linked protein can help in early diagnosis and improved therapy. Such proteonic chips can find applications in many electrical sensors for detection of biologically relevant molecules and species.
Acknowledgments
The authors would like to thank Mohammed A.I. Mahmood and Syed E.A. Quadri for help with photolithography; F. Z. Amir for her help with electron microscopy and Yuan Wan for his guidance through surface functionalization. This work was supported by National Science Foundation CAREER grant to S.M.I. (ECCS-0845669). Azhar Ilyas and Waseem Asghar were partially supported by a fellowship from the Consortium for Nanomaterials for Aerospace Commerce and Technology (CONTACT) program, Rice University, Houston, TX, USA. Partial chip fabrication was carried out at Characterization Center for Materials and Biology (C2MB), and NanoFab, University of Texas at Arlington and Cleanroom Research Laboratory, University of Texas at Dallas.
References
- 1.Klijn JGM, Berns P, Schmitz PIM, Foekens JA. The clinical significance of epidermal growth factor receptor (EGF-R) in human breast cancer: a review on 5232 patients. Endocrine reviews. 1992;13:3. doi: 10.1210/edrv-13-1-3. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2004;350:2129. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Kersemaekers AMF, Fleuren GJ, Kenter GG, Van den Broek L, Uljee SM, Hermans J, Van de Vijver MJ. Oncogene alterations in carcinomas of the uterine cervix: overexpression of the epidermal growth factor receptor is associated with poor prognosis. Clinical Cancer Research. 1999;5:577. [PubMed] [Google Scholar]
- 4.Mellon K, Wright C, Kelly P, Horne CH, Neal DE. Original Articles: Bladder Cancer: Long-Term Outcome Related to Epidermal Growth Factor Receptor Status in Bladder Cancer. The Journal of urology. 1995;153:919–25. [PubMed] [Google Scholar]
- 5.Inada S, Koto T, Futami K, Arima S, Iwashita A. Evaluation of malignancy and the prognosis of esophageal cancer based on an immunohistochemical study (p53, E-cadherin, epidermal growth factor receptor) Surgery today. 1999;29:493–503. doi: 10.1007/BF02482343. [DOI] [PubMed] [Google Scholar]
- 6.Fischer-Colbrie J, Witt A, Heinzl H, Speiser P, Czerwenka K, Sevelda P, Zeillinger R. EGFR and steroid receptors in ovarian carcinoma: comparison with prognostic parameters and outcome of patients. Anticancer research. 1997;17:613–9. [PubMed] [Google Scholar]
- 7.Dalton WS, Friend SH. Cancer biomarkers--an invitation to the table. Science. 2006;312:1165–8. doi: 10.1126/science.1125948. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki H, Yukiue H, Mizuno K, Sekimura A, Konishi A, Yano M, Kaji M, Kiriyama M, Fukai I, Yamakawa Y. Elevated serum epidermal growth factor receptor level is correlated with lymph node metastasis in lung cancer. International Journal of Clinical Oncology. 2003;8:79–82. doi: 10.1007/s101470300014. [DOI] [PubMed] [Google Scholar]
- 9.Carney WP, Burrell M, Morris LD, Hamer PJ. Normal levels of serum EGFr and decreases in several cancers. Proceddings AACR. 2002;43 [Google Scholar]
- 10.Atkins D, Reiffen KA, Tegtmeier CL, Winther H, Bonato MS, Storkel S. Immunohistochemical detection of EGFR in paraffin-embedded tumor tissues: variation in staining intensity due to choice of fixative and storage time of tissue sections. Journal of Histochemistry and Cytochemistry. 2004;52:893. doi: 10.1369/jhc.3A6195.2004. [DOI] [PubMed] [Google Scholar]
- 11.Brockhoff G, Hofstaedter F, Knuechel R. Flow cytometric detection and quantitation of the epidermal growth factor receptor in comparison to Scatchard analysis in human bladder carcinoma cell lines. Cytometry Part A. 1994;17:75–83. doi: 10.1002/cyto.990170110. [DOI] [PubMed] [Google Scholar]
- 12.Lee KW, Hwang KH, Kim CS, Han K, Chung YB, Park JS, Lee YM, Moon DC. Determination of Recombinant human epidermal growth factor (rhEGF) in a pharmaceutical formulation by high performance liquid chromatography with electrochemical detection. Archives of Pharmacal Research. 2001;24:355–9. doi: 10.1007/BF02975106. [DOI] [PubMed] [Google Scholar]
- 13.Kippenberger S, Loitsch S, Guschel M, Müller J, Knies Y, Kaufmann R, Bernd A. Mechanical stretch stimulates protein kinase B/Akt phosphorylation in epidermal cells via angiotensin II type 1 receptor and epidermal growth factor receptor. Journal of Biological Chemistry. 2005;280:3060. doi: 10.1074/jbc.M409590200. [DOI] [PubMed] [Google Scholar]
- 14.Sako Y, Minoghchi S, Yanagida T. Single-molecule imaging of EGFR signalling on the surface of living cells. Nature cell biology. 2000;2:168–72. doi: 10.1038/35004044. [DOI] [PubMed] [Google Scholar]
- 15.Sorkin A, McClure M, Huang F, Carter R. Interaction of EGF receptor and grb2 in living cells visualized by fluorescence resonance energy transfer (FRET) microscopy. Current Biology. 2000;10:1395–8. doi: 10.1016/s0960-9822(00)00785-5. [DOI] [PubMed] [Google Scholar]
- 16.Fang Y, Ferrie A. Optical biosensor differentiates signaling of endogenous PAR1 and PAR2 in A431 cells. BMC cell biology. 2007;8:24. doi: 10.1186/1471-2121-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramsden JJ, Horvath R. Optical biosensors for cell adhesion. Journal of Receptors and Signal Transduction. 2009;29:211–23. doi: 10.1080/10799890903064119. [DOI] [PubMed] [Google Scholar]
- 18.Fang Y. Label-Free Biosensors for Cell Biology. International Journal of Electrochemistry 2011 [Google Scholar]
- 19.Kozma P, Hamori A, Kurunczi S, Cottier K, Horvath R. Grating coupled optical waveguide interferometer for label-free biosensing. Sensors and Actuators B: Chemical 2011 [Google Scholar]
- 20.Aviram A, Ratner MA. Molecular rectifiers. Chemical Physics Letters. 1974;29:277–83. [Google Scholar]
- 21.Iqbal SM, Balasundaram G, Ghosh S, Bergstrom DE, Bashir R. Direct current electrical characterization of ds-DNA in nanogap junctions. Applied Physics Letters. 2005;86:153901. [Google Scholar]
- 22.Reed MA, Zhou C, Muller CJ, Burgin TP, Tour JM. Conductance of a molecular junction. Science. 1997;278:252. [Google Scholar]
- 23.Liu S, Zhang X, Luo W, Wang Z, Guo X, Steigerwald ML, Fang X. Single-Molecule Detection of Proteins Using Aptamer-Functionalized Molecular Electronic Devices. Angewandte Chemie International Edition. 2011;50:2496–502. doi: 10.1002/anie.201006469. [DOI] [PubMed] [Google Scholar]
- 24.Wold DJ, Frisbie CD. Fabrication and Characterization of Metal-Molecule- Metal Junctions by Conducting Probe Atomic Force Microscopy. J Am Chem Soc. 2001;123:5549–56. doi: 10.1021/ja0101532. [DOI] [PubMed] [Google Scholar]
- 25.Kushmerick JG, Holt DB, Yang JC, Naciri J, Moore MH, Shashidhar R. Metal-molecule contacts and charge transport across monomolecular layers: Measurement and theory. Physical review letters. 2002;89:86802. doi: 10.1103/PhysRevLett.89.086802. [DOI] [PubMed] [Google Scholar]
- 26.Datta S, Tian W, Hong S, Reifenberger R, Henderson JI, Kubiak CP. Current-voltage characteristics of self-assembled monolayers by scanning tunneling microscopy. Physical Review Letters. 1997;79:2530–3. [Google Scholar]
- 27.Liu K, Avouris P, Bucchignano J, Martel R, Sun S, Michl J. Simple fabrication scheme for sub-10 nm electrode gaps using electron-beam lithography. Applied Physics Letters. 2002;80:865. [Google Scholar]
- 28.Zhou C, Muller CJ, Deshpande MR, Sleight JW, Reed MA. Microfabrication of a mechanically controllable break junction in silicon. Applied Physics Letters. 1995;67:1160. [Google Scholar]
- 29.Li CZ, He HX, Tao NJ. Quantized tunneling current in the metallic nanogaps formed by electrodeposition and etching. Applied Physics Letters. 2000;77:3995. [Google Scholar]
- 30.Park H, Lim AKL, Alivisatos AP, Park J, McEuen PL. Fabrication of metallic electrodes with nanometer separation by electromigration. Applied Physics Letters. 1999;75:301. [Google Scholar]
- 31.Strachan DR, Smith DE, Johnston DE, Park TH, Therien MJ, Bonnell DA, Johnson AT. Controlled fabrication of nanogaps in ambient environment for molecular electronics. Applied Physics Letters. 2005;86:043109. [Google Scholar]
- 32.Asghar W, Ramachandran PP, Adewumi A, Noor MR, Iqbal SM. Rapid Nanomanufacturing of Metallic Break Junctions Using Focused Ion Beam Scratching and Electromigration. Journal of Manufacturing Science and Engineering. 2010;132:030911. [Google Scholar]
- 33.Christensen SM, Ramachandran PP, Iqbal SM. Electronic Detection of Selective Proteins using Non Antibody-Based CMOS Chip. DNA. 10:391.4. [Google Scholar]
- 34.Song S, Wang L, Li J, Fan C, Zhao J. Aptamer-based biosensors. TrAC Trends in Analytical Chemistry. 2008;27:108–17. [Google Scholar]
- 35.Zhang H, Wang Z, Li XF, Le XC. Ultrasensitive detection of proteins by amplification of affinity aptamers. Angewandte Chemie. 2006;118:1606–10. doi: 10.1002/anie.200503345. [DOI] [PubMed] [Google Scholar]
- 36.Hamaguchi N, Ellington A, Stanton M. Aptamer Beacons for the Direct Detection of Proteins. Analytical Biochemistry. 2001;294:126–31. doi: 10.1006/abio.2001.5169. [DOI] [PubMed] [Google Scholar]
- 37.Maehashi K, Katsura T, Kerman K, Takamura Y, Matsumoto K, Tamiya E. Label-Free Protein Biosensor Based on Aptamer-Modified Carbon Nanotube Field-Effect Transistors. Anal Chem. 2007;79:782–7. doi: 10.1021/ac060830g. [DOI] [PubMed] [Google Scholar]
- 38.Ylera F, Lurz R, Erdmann VA, Fürste JP. Selection of RNA Aptamers to the Alzheimer’s Disease Amyloid Peptide. Biochemical and Biophysical Research Communications. 2002;290:1583–8. doi: 10.1006/bbrc.2002.6354. [DOI] [PubMed] [Google Scholar]
- 39.Fischer NO, Tarasow TM, Tok JBH. Aptasensors for biosecurity applications. Current Opinion in Chemical Biology. 2007;11:316–28. doi: 10.1016/j.cbpa.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Wang Z, Li XF, Le XC. Ultrasensitive detection of proteins by amplification of affinity aptamers. Angewandte Chemie (International ed in English) 2006;45:1576–80. doi: 10.1002/anie.200503345. [DOI] [PubMed] [Google Scholar]
- 41.Lion N, Rohner TC, Dayon L, Arnaud IL, Damoc E, Youhnovski N, Wu ZY, Roussel C, Josserand J, Jensen H. Microfluidic systems in proteomics. Electrophoresis. 1921;2003:3533–62. doi: 10.1002/elps.200305629. [DOI] [PubMed] [Google Scholar]
- 42.Bunka DHJ, Stockley PG. Aptamers come of age–at last. Nature Reviews Microbiology. 2006;4:588–96. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- 43.Ghosh S, Halimun H, Mahapatro AK, Choi J, Lodha S, Janes D. Device structure for electronic transport through individual molecules using nanoelectrodes. Applied Physics Letters. 2005;87 [Google Scholar]
- 44.Mahapatro AK, Ghosh S, Janes DB. Nanometer scale electrode separation (nanogap) using electromigration at room temperature. Nanotechnology, IEEE Transactions on. 2006;5:232–6. [Google Scholar]
- 45.Mahapatro AK, Jeong KJ, Lee GU, Janes DB. Sequence specific electronic conduction through polyion-stabilized double-stranded DNA in nanoscale break junctions. Nanotechnology. 2007;18 [Google Scholar]
- 46.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 47.Stoltenburg R, Reinemann C, Strehlitz B. SELEX--a (r) evolutionary method to generate high-affinity nucleic acid ligands. Biomolecular engineering. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Li N, Ebright JN, Stovall GM, Chen X, Nguyen HH, Singh A, Syrett A, Ellington AD. Technical and biological issues relevant to cell typing with aptamers. Journal of proteome research. 2009;8:2438–48. doi: 10.1021/pr801048z. [DOI] [PubMed] [Google Scholar]
- 49.Li N, Nguyen HH, Byrom M, Ellington AD. Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS One. 2011;6:e20299. doi: 10.1371/journal.pone.0020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.NIH ImageJ.
- 51.Ilyas A, Asghar W, Billo JA, Syed EAQ, Iqbal SM. IEEE/NIH Life Science Systems and Applications Workshop (LiSSA) 2011. Bethesda, MD: IEEE; 2011. From Molecular Electronics to Proteonics: Break Junctions for Biomarker Detection; p. 4. [Google Scholar]
- 52.Nair PR, Alam MA. Performance limits of nanobiosensors. Appl Phys Lett. 2006;88:233120–3. [Google Scholar]
- 53.Wang W, Lee T, Reed MA. Mechanism of electron conduction in self-assembled alkanethiol monolayer devices. Physical Review B. 2003;68:35416. doi: 10.1196/annals.1292.001. [DOI] [PubMed] [Google Scholar]
- 54.McMaster GK, Carmichael GG. Analysis of single-and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proceedings of the National Academy of Sciences of the United States of America. 1977;74:4835. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Traganos F, Darzynkiewicz Z, Sharpless T, Melamed MR. Simultaneous staining of ribonucleic and deoxyribonucleic acids in unfixed cells using acridine orange in a flow cytofluorometric system. Journal of Histochemistry and Cytochemistry. 1977;25:46. doi: 10.1177/25.1.64567. [DOI] [PubMed] [Google Scholar]
- 56.Lopez MF, Berggren K, Chernokalskaya E, Lazarev A, Robinson M, Patton WF. A comparison of silver stain and SYPRO Ruby Protein Gel Stain with respect to protein detection in two-dimensional gels and identification by peptide mass profiling. Electrophoresis. 2000;21:3673–83. doi: 10.1002/1522-2683(200011)21:17<3673::AID-ELPS3673>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 57.Hicke BJ, Stephens AW. Escort aptamers: a delivery service for diagnosis and therapy. Journal of Clinical Investigation. 2000;106:923–8. doi: 10.1172/JCI11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wan Y, Tan J, Asghar W, Kim Y-t, Liu Y, Iqbal SM. Velocity Effect on Aptamer-Based Circulating Tumor Cell Isolation in Microfluidic Devices. The Journal of Physical Chemistry B. 2011;115:13891–6. doi: 10.1021/jp205511m. [DOI] [PubMed] [Google Scholar]
- 59.Wan Y, Kim Y-t, Li N, Cho SK, Bachoo R, Ellington AD, Iqbal SM. Surface Immobilized Aptamers for Cancer Cell Isolation and Microscopic Cytology. Cancer Research. 2010;70:11. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]