Abstract
Mutations in the DMD gene, encoding the dystrophin protein, are responsible for the dystrophinopathies Duchenne Muscular Dystrophy (DMD), Becker Muscular Dystrophy (BMD), and X-linked Dilated Cardiomyopathy (XLDC). Mutation analysis has traditionally been challenging, due to the large gene size (79 exons over 2.2 Mb of genomic DNA). We report a very large aggregate data set comprised of DMD mutations detected in samples from patients enrolled in the United Dystrophinopathy Project, a multicenter research consortium, and in referral samples submitted for mutation analysis with a diagnosis of dystrophinopathy. We report 1111 mutations in the DMD gene, including 891 mutations with associated phenotypes. These results encompass 506 point mutations (including 294 nonsense mutations) and significantly expand the number of mutations associated with the dystrophinopathies, highlighting the utility of modern diagnostic techniques. Our data supports the uniform hypermutability of CGA>TGA mutations, establishes the frequency of polymorphic muscle (Dp427m) protein isoforms and reveals unique genomic haplotypes associated with `private' mutations. We note that 60% of these patients would be predicted to benefit from skipping of a single DMD exon using antisense oligonucleotide therapy, and 62% would be predicted to benefit from an inclusive multi-exon skipping approach directed toward exons 45 through 55.
Keywords: dystrophinopathy, Duchenne Muscular Dystrophy, DMD, Becker Muscular Dystrophy, BMD, mutation detection
Introduction
Mutation detection in the DMD gene (MIM# 300377) has historically been challenging, due primarily to the large size of the gene. Early studies found that approximately two-thirds of the mutations causing DMD were due to deletions of one or more exons, and the observation that these exonic deletions cluster in hotspots led to the use of the multiplex polymerase chain reaction (PCR) test as the standard method of diagnosis for nearly fifteen years (Beggs, et al., 1990; Chamberlain, et al., 1990). This multiplex PCR detected up to ninety-eight percent of deletions (Beggs, et al., 1990), but could not detect the approximately 5% of DMD mutations due to exonic duplications. Detections of duplications require gene dosage tests (which also detect deletions); these include Southern blot analysis (Curtis and Haggerty, 2001; Den Dunnen, et al., 1989), dosimetric PCR-based methods (Abbs and Bobrow, 1992; Frisso, et al., 2004), or techniques such as multiplex amplifiable probe hybridization (MAPH) (White, et al., 2002) and multiplex ligation-dependent probe amplification (MLPA) (Schwartz and Duno, 2004). None of these methods detect point mutations, including premature stop codon (nonsense) mutations, subexonic insertions or deletions, splice site mutations, and missense mutations.
The search for economical methods for point mutation detection led to a variety of strategies, including multiplex single strand conformational polymorphism analysis (SSCP) (Mendell, et al., 2001), denaturing high performance liquid phase chromatography (Bennett, et al., 2001), denaturing gradient gel-electrophoresis (DGGE) (Dolinsky, et al., 2002; Hofstra, et al., 2004). We have previously reported the development of a semi-automated direct sequencing technique that provided for the economical and rapid sequencing of all of the coding exons of the DMD gene, along with flanking intronic sequences, and promoters (Flanigan, et al., 2003). This method, called single condition amplification/internal primer (SCAIP) sequencing, detects deletions of all DMD exons, and has several advantages in the detection of point mutations, including a higher sensitivity than screening methods based on conformational (SSCP) or heteroduplex (dHPLC) analysis. It has also been adapted to other large multi-exon genes, including the three genes encoding collagen VI subunits, mutations of which are associated with Ullrich Congenital Muscular Dystrophy and Bethlem Myopathy (Lampe, et al., 2005).
Combined with dosimetric duplication analysis, SCAIP is a powerful tool, and direct sequencing methodologies are now the gold standard for genomic point mutation detection. We have previously reported the results of a survey of unselected clinic patients using these modern diagnostic techniques (Dent, et al., 2005), and others have reported similar surveys in referral patient sets (rather than unselected clinic survey populations) (Prior and Bridgeman, 2005; Yan, et al., 2004).
Herein we report results using the SCAIP technique in combination with either MLPA or MAPH testing in a very large sample population. This population is comprised of (1) research subject samples evaluated through the United Dystrophinopathy Project (UDP), a seven-center consortium consisting of a prospective longitudinal natural history study, a genotype/phenotype database, and self-report patient registry (N = 858 samples tested), and (2) specimens referred for molecular analysis outside of the UDP (N = 428 samples tested). We report the spectrum of mutations detected by these techniques in 1111 subjects, and discuss genotype/phenotype correlations in the subset of male patients (N = 794) with definitive phenotypic information. We provide frequencies of polymorphisms within the exons and flanking intronic regions of the DMD gene.
Using the frequencies of exonic polymorphisms, we have identified an idealized version of coding region of the Dp427m transcript, in which each nucleotide position is represented by the major allele; this idealized transcript is observed at a frequency of 9% in this population and may prove useful as a reference sequence for the DMD gene. We propose the existence of eight common protein variants of the dystrophin 427 kiloDalton (kDa) muscle isoform, based upon amino acid polymorphism haplotypes, and suggest that the identification and further study of these minor variants may shed light on variation in disease phenotype. Lastly, we analyze point mutation spectra, and in contrast to previous analysis of smaller DMD data sets we confirm that CpG codons are uniformly more likely to undergo transitions to stop codons.
Methods
Patient ascertainment
Mutation results were derived from two cohorts. Patients in the UDP are selected by strict diagnostic criteria that include either (1) clinical features consistent with DMD or BMD and an X-linked family history; or (2) muscle biopsy showing alteration in dystrophin expression by immunofluorescence, immunohistochemistry, or immunoblot; or (3) a mutation in the DMD gene previously detected by clinical testing. After informed consent is obtained (under IRB-approved protocols), blood samples are obtained for DNA analysis; patients are examined; and data is extracted from clinical records for inclusion in the UDP database.
Phenotypes in the UDP data set are determined using the directive of “best clinical diagnosis”. Neither mutation class nor protein expression alone can distinguish phenotype, and in at least 1/3 of dystrophinopathy cases no family history is available to guide prognosis (i.e., based on the clinical course of an affected maternal relative). Furthermore, treatment with steroids may delay progression to a degree that makes a clinical distinction between a steroid-responsive DMD and an untreated BMD patient difficult. Therefore, we defined phenotypes based on the expert clinical opinion of trained neuromuscular physicians at each tertiary center, combining available information regarding clinical presentation features, family history, protein expression, and mutation class. Under the UDP, we utilize three diagnostic classes for male subjects, originally defined by age at loss of ambulation: DMD (loss of ambulation at < age 12); intermediate muscular dystrophy (IMD; loss of ambulation between age 12 and age 15); and BMD (loss of ambulation at > age 15). For patients still walking, the UDP physician makes a “best clinical diagnosis” based on all available data. As part of the UDP protocol, this diagnosis is reported to the central coordinating center at the University of Utah. Patients who are enrolled in the UDP are identified as such in Supp. Table S1 by the presence of a unique UDP identifier number in the column headed “UDP ID”.
Samples sent for analysis in the clinical testing lab are from patients selected for testing by referring clinical physicians, and diagnostic criteria are therefore outside of the control of the authors. For clinical samples, we accepted the referring physician's clinical diagnosis, and sought confirmatory diagnostic information from family history or muscle biopsy criteria wherever possible. Wherever information regarding clinical phenotype was not provided, we use the convention of “B/DMD” to signify phenotype (see Supp. Table S1, in which patients identified via clinical referral have no associated UDP ID number). We have excluded all B/DMD patients in the analysis of genotype/phenotype correlations presented below.
Mutation Analysis
SCAIP testing was performed as previously described (Flanigan, et al., 2003). The presence of single exon deletions, and the extent of multi-exon deletions, are confirmed using an independent set of primers; and in non-deleted samples, SCAIP-generated nucleotide sequence traces were analyzed using the base-calling sequence software described previously (Flanigan, et al., 2003). In selected patients, mRNA was isolated from archived muscle biopsy tissue, and reverse transcription PCR prior to cDNA sequencing, using conditions and primers as published elsewhere (Roberts, et al., 1991). Nucleotide positions were determined according to the standard reference DMD sequence used for mutation analysis (GenBank accession number NM_004006.1). Nucleotide numbering reflects cDNA numbering with +1 corresponding to the A of the ATG translation initiation codon in the reference sequence NM_004006.1, according to journal guidelines (www.hgvs.org/mutnomen). The initiation codon is codon 1.
Duplication analysis was performed using Multiplex Ligation-dependent Probe Amplification (Salsa MLPA kit P034/P035 DMD/Becker MLPA; MRC Holland, Inc.). Some duplication mutations were previously reported as detected in a referral laboratory, using methods published elsewhere (White, et al., 2006). Previously unreported duplications were detected by use of the Salsa MLPA kit according to the manufacturer's instructions with the following modifications: PCR volumes are decreased to one half, and injections are made at twice the manufacturer's recommended injection duration during injection into the ABI 3730 capillary sequencing machine.
Calculation of per nucleotide per generation rate of mutations of type x was estimated as: μx = mnx/Ntx, where m is the disease incidence, n if the number of patients with mutation type x, N is the total number of unrelated mutations, and t is the target size of mutation type x. The disease incidence m (defined as the per generation rate) was assumed to be 1/3 * 1/3500 reflecting the estimated population incidence of DMD as 1:3,500 live male births. The number of patients carrying a mutation of type x defined nx, and the target size for mutations of type x defined tx (in nucleotides) based on muscle transcript isoform Dp427m (NM_004006.1). In this study, N was assumed to be 967 unrelated patients, although unknown patient ascertainment bias between the point mutation classes and deletion/duplication classes will affect the relative magnitude these measures of μx.
Single Nucleotide Polymorphism (SNP) frequency determination
SCAIP analysis results in sequencing of ~84.5 kilobases (kb) of sequence in each individual, including exonic sequence (14.5 kb) and flanking intronic sequence (70 kb). SNP positions were identified by high quality discrepancy analysis (Phrap score > 20) and confirmed by manual inspection. Allele frequencies at each position were calculated using the number of alleles from unrelated individuals with the high quality coverage depth in the resequenced patient population as the denominator. Alignment to the March 2006 human reference sequence (Hg18, NCBI Build 36.1) and the chimpanzee Mar. 2006 (panTro2) assembly was used to identify known SNP positions and to infer the ancestral allele, respectively. Polymorphism data was submitted to the Database of Single Nucleotide Polymorphisms (dbSNP), National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland (dbSNP accession: ss102660140 – ss102660540).
Results
Mutations detected
Mutation results are summarized in Table 1 and Supp. Tables S1 and S2. Table 1 contains the summary of all mutations found in both the UDP and clinic referral populations. The clinical referral population includes patients in whom the specific clinical diagnosis could be ascertained by review of submitted records and referring documents, and patients for whom this information was incomplete (listed as B/DMD). A summary of the mutation distribution found solely among research subjects within the UDP, whose clinical diagnoses were recorded according to UDP prospectively defined criteria, are found in Supp. Table S1.
Table 1.
Summary of all mutations detected, by class and phenotype.
| MUTATION CLASS | DMD | IMD | BMD | Unknown (B/DMD) | Manifesting Carrier | Carrier (all phenotypes) | Total | % |
|---|---|---|---|---|---|---|---|---|
| DELETION | 284 | 15 | 55 | 106 | 3 | 14 | 477 | 42.9% |
| in | 30 | 2 | 36 | 17 | 0 | 2 | 87 | |
| out | 244 | 13 | 18 | 88 | 2 | 12 | 376 | |
| other | 10 | 0 | 1 | 2 | 1 | 0 | 14 | |
| STOP | 176 | 4 | 30 | 46 | 4 | 34 | 294 | 26.5% |
| UGA | 60 | 1 | 13 | 20 | 3 | 15 | 112 | |
| UAG | 71 | 0 | 11 | 13 | 0 | 4 | 99 | |
| UAA | 45 | 3 | 6 | 13 | 1 | 15 | 83 | |
| SUBEXONIC | 69 | 0 | 10 | 33 | 1 | 14 | 127 | 11.4% |
| FS Ins | 22 | 0 | 1 | 7 | 1 | 6 | 37 | |
| FS Del | 46 | 0 | 4 | 23 | 0 | 8 | 81 | |
| FS Ins/Del | 1 | 0 | 2 | 2 | 0 | 0 | 5 | |
| in-frame deletion | 0 | 0 | 3 | 1 | 0 | 0 | 4 | |
| DUPLICATION | 87 | 7 | 10 | 8 | 5 | 5 | 122 | 11.0% |
| SPLICE | 22 | 3 | 7 | 18 | 2 | 12 | 64 | 5.8% |
| MISSENSE | 2 | 1 | 6 | 6 | 0 | 0 | 15 | 1.4% |
| PSEUDOEXON | 0 | 2 | 2 | 0 | 0 | 2 | 6 | 0.5% |
| POTENTIAL | 2 | 0 | 0 | 3 | 0 | 1 | 6 | 0.5% |
| OTHER | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0% |
| TOTAL MUTATIONS | 642 | 32 | 120 | 220 | 15 | 82 | 1111 | 100.0% |
“Carrier” are obligate or asymptomatic carriers, whereas “Manifesting Carriers” display symptoms of significant myalgia and/or weakness. (Further characterization of carrier phenotypic class is found in Supp. Table S2, listing specific mutations detected.)
Mutations were detected in 1111 subjects, including 347 subjects who fall into 203 kindreds; the remainder of the subjects are not known to be related. We have identified 967 apparently unrelated mutations, which are listed in the online Supp. Table S1. Many of these are represented, via our own reports or those of others, within the Leiden Database of DMD mutations (www.dmd.nl). However, as summarized in Supp. Table S1, 187 mutations have not previously been reported, either in publications or in the Leiden Database.
Deletions account for nearly 43% of all dystrophinopathy mutations in this data set. This number is less than the approximately 65% usually reported (Dent, et al., 2005), but is likely due to selection bias, as our patient population was enriched for point mutations during those years when sequence analysis of the DMD gene was not widely available. All deletion mutations are listed in Supp. Table S1; Supp. Figure S1 represents those deletions that are either previously unreported, or associated with a phenotype that contradicts the reading frame rule (see below). The overall sensitivity and specificity of reading frame in predicting DMD is found in Table 2.
Table 2.
The value of mutational reading frame in predicting a phenotype of Duchenne Muscular Dystrophy.
| DMD | I/BMD | ||
|---|---|---|---|
| Exonic deletions only | |||
| truncating (out-of-frame) mutations | 254 | 32 | 88.8% Positive Predictive Value |
| non-truncating (in-frame) mutations | 30 | 38 | 55.9% Negative Predictive Value |
|
| |||
| sensitivity | 89.4% | - | |
| specificity | - | 54.3% | |
|
| |||
| All mutations a | |||
| truncating mutations | 519 | 79 | 86.8% Positive Predictive Value |
| non-truncating mutations | 37 | 63 | 63.0% Negative Predictive Value |
|
| |||
| sensitivity | 93.3% | ||
| specificity | - | 44.4% | |
“All mutations” includes exonic deletions as well as missense, nonsense, splice site, and pseudoexon mutations. The frame for splice site mutations is based on the assumption that mutations affecting a splice donor or acceptor site result in the exclusion of that exon from the mRNA, allowing prediction of the reading frame based on the surrounding exons. Duplications are excluded from this analysis, as discussed in the text.
Duplications account for 11.0% of all patient mutations. Among patients with duplications, one cannot make assumptions regarding the reading frame based upon the results of genomic testing. Although tandem duplications are common (Hu, et al., 1991), other types of non-tandem duplications may occur (White, et al., 2006), and determination of the orientation of the duplicated fragment requires mRNA analysis. For this reason, we have not included a prediction of reading frame among duplication patients. Consistent with results reported elsewhere (White, et al., 2006), the most common duplication noted is a single exon duplication of exon 2. We note a paucity of duplication mutations in the B/DMD group, but ascribe this to selection bias, because for several years the non-UDP testing protocol did not include duplication testing. Six subjects had complex duplication (or duplication/triplication) mutations. Three of these have been previously reported (Dent, et al., 2005; White, et al., 2006): patient 42391 (duplication exons 5–19, and duplication exons 38–41); patient 43000 (duplication exons 45–55, and duplication exons 65–79); and patient 43067 (duplication exons 5–18, triplication exons 19–41, duplication exon 42, triplication exons 43–44). Three others have not been previously reported: DR43699 (duplication exons 50–60, and duplication of exons 63–79); DC0111 (duplication exons 10–16 and 22–44); and DRVH43882 (duplication of exon 29 and duplication exon 45).
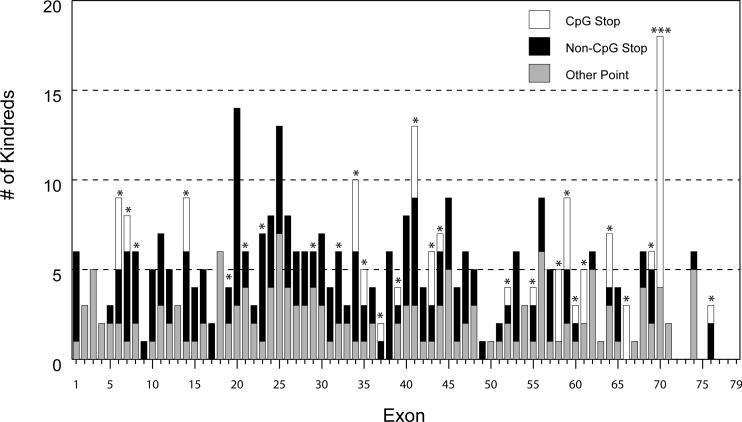
Point mutations account for 46% of all mutations in our cohort. As expected from previous reports, we did not find point mutation hotspots; point mutations were essentially evenly distributed across the exons of the DMD gene (Figure 1). Generally, multiple instances of a single point mutation are due to relatedness among subjects, with ascertainment bias thus introduced by testing of affected relatives after identifying the point mutation in a proband. Within our data set, however, there is one significant exception to this conclusion. We identified the mutation c.9G>A (p.Trp3>X) in six independent families with BMD, and from further genomic analysis have determined that it represents a true founder allele – the first reported in the DMD gene (Flanigan et al, manuscript in submission).
Figure 1.
Exon distribution of nonsense, splice and small insertion/deletion (indel) mutations by exon in unrelated kindreds. Exons containing CpG codons are marked by asterisks (exon 70 contains three CpG codons).
Mutation spectra and per nucleotide mutation rates
The mutational spectrum, target size, and observed mutation rates per nucleotide for the 397 observed point mutations from unrelated patients are shown in Table 3. Unrelatedness was determined by intragenic haplotype analysis of data derived from complete sequencing of the gene. Single base-pair substitutions are 2.5-fold more prevalent than small insertions/deletions that disrupt the reading frame or splice sites. The per nucleotide mutation rate of 15.1 × 10−9 (all substitutions) and 0.91 × 10−9 (all insertion/deletions) are similar to previous estimates of 24.6 × 10−9 and 0.56 × 10−9, respectively, from a meta-analysis of earlier DMD mutation studies (Kondrashov, 2003)
Table 3.
Mutation rates per nucleotide and number of observations for different mutation classes.
| Mutation Class | # of mutations | target size (nts.) | μx mutation rate (× 10−9) |
|---|---|---|---|
| Single base substitutions | 284 | 1,812 | 15.12 |
| Splice site substitutions | 41 | 312 | 12.54 |
| Nonsense substitutions | 243 | 1,500 | 15.46 |
| transitions at CpG sites | 62 | 29 | 204.03 |
| transversions at CpG sites | - | 7 | - |
| transitions at non-CpG sites | 105 | 380 | 26.37 |
| transversions at non-CpG sites | 76 | 1,091 | 6.65 |
| Small insertion/deletion | 113 | 11,370 | 0.95 |
| CDS insertion/deletion | 104 | 11,058 | 0.90 |
| Splice site insertion/deletion | 9 | 312 | 0.92 |
Target size is the number of nucleotides (nts.) in NM_004006 for each class of CDS mutation, and the number of canonical splice acceptor and donor nucleotides, flanking exon 1 (Dp427m) through exon 79. μx mutation rate is the per nucleotide per generation rate of mutations for a mutation class
The mutable target size for nonsense mutations caused by single base-pair substitutions is 1500 sense codons contained in the NM_004006.1 isoform, and the target size per codon and relative mutability for the 243 stop mutations from unrelated patients are shown in Table 4. As expected, G:C>A:T transitions are the most prevalent stop mutation class (68%), with 62 stop mutations (25%) due to C>T transitions at 23 of 29 CGA arginine codons (Table 4 and Figure 1). A 7.7-fold elevation of the per nucleotide mutation rate for transitions leading to stop codons at CpG versus non-CpG sites (204 × 10−9 versus 26.4 × 10−9, Table 3) was observed, presumably due to the spontaneous deamination of 5-methylcytosine to thymidine at methylated CpG dinucleotides (Cooper and Krawczak, 1989). This CpG versus non-CpG transition rate is directly comparable to and slightly lower than a 10.4-fold elevation measured in a previous study of 46 independent DMD stop codons (Buzin, et al., 2005). The per nucleotide mutation rates for CpG versus non-CpG transitions are also similar between this survey (204 × 10−9 versus 26.4 × 10−9) and a previous estimate (159 × 10−9 versus 15.3 × 10−9) (Buzin, et al., 2005).
Table 4.
Target sizes and single base substitutions for nonsense mutations.
| Substitution (sense > stop) | Independent mutations | Mutated sites | Total sites | Mutability | % Total Stop Mutations | Mutation type |
|---|---|---|---|---|---|---|
| CGA > TGA | 62 | 23 | 29 | 13.20 | 25.2% | Ts GC:AT at CpG |
| CAG > TAG | 50 | 44 | 174 | 1.77 | 20.3% | Ts GC:AT |
| CAA > TAA | 28 | 25 | 134 | 1.29 | 11.8% | Ts GC:AT |
| TGG>STOP | 26 | 21 | 72 | 2.23 | 10.6% | - |
| > TGA | 17 | - | - | 1.46 | - | Ts GC:AT |
| > TAG | 9 | - | - | 0.77 | - | Ts GC:AT |
| GAA > TAA | 18 | 17 | 237 | 0.47 | 7.3% | Tv GC:TA |
| GAG > TAG | 15 | 14 | 150 | 0.62 | 6.5% | Tv GC:TA |
| TCA>STOP | 12 | 10 | 51 | 1.45 | 5.3% | - |
| > TGA | 8 | - | - | 0.97 | - | Tv GC:CG |
| > TAA | 4 | - | - | 0.48 | - | Tv GC:TA |
| AAA > TAA | 9 | 8 | 154 | 0.36 | 3.7% | Tv AT:TA |
| TAC>STOP | 5 | 5 | 19 | 1.62 | 2.0% | - |
| > TAG | 3 | - | - | 0.97 | - | Tv GC:CG |
| > TAA | 2 | - | - | 0.65 | - | Tv GC:TA |
| TTA>STOP | 5 | 5 | 54 | 0.57 | 2.0% | - |
| > TAA | 3 | - | - | 0.34 | - | Tv AT:TA |
| > TGA | 2 | - | - | 0.23 | - | Tv AT:CG |
| AAG > TAG | 5 | 5 | 139 | 0.22 | 2.0% | Tv AT:TA |
| TTG > TAG | 3 | 3 | 94 | 0.20 | 1.2% | Tv AT:TA |
| TAT > TAA | 2 | 2 | 43 | 0.29 | 0.8% | Tv AT:TA |
| GGA > TGA | 2 | 2 | 47 | 0.26 | 0.8% | Tv GC:TA |
| AGA > TGA | 1 | 1 | 61 | 0.10 | 0.4% | Tv AT:TA |
| TGC > TGA | 0 | 0 | 21 | - | 0.0% | Tv GC:TA |
| TGT > TGA | 0 | 0 | 15 | - | 0.0% | Tv AT:TA |
| TCG > TAG | 0 | 0 | 6 | - | 0.0% | Tv GC:TA at CpG |
| Totals: | 243 | 185 | 1,500 |
The total sites for each triplet are based on Dp427m reference sequence from NM_004006, and are the number of codons for each triplet that can be mutated to a nonsense codon via a single base substitution.
The relative mutability of each sense codon is the ratio of the number of independent mutations to the expected number of mutations, a value calculated from the product of the total number of nonsense mutations (243) and the ratio of total triplet codon sites to the total nonsense target sites (1,500).
The transition rate at individual CpG dinucleotides is dependent both on germ-line methylation status and can be effected by local sequence context (Antonarakis, et al., 2000). To examine whether the distribution of observed mutations at individual CGA codons was sampled from a uniform CpG transition rate rather than different individual rates, mutation spectrum decomposition by the Simulation, Expectation, Maximization (SEM) classification approach as implemented in the CLUSTERM program was analyzed (Rogozin, et al., 2001). No evidence for different classes of CGA codons was observed and no significant deviation from the expected distribution was found. This observation contrasts with a previous report (Buzin, et al., 2005) that the exon 59 c.8713C>T (p.R2905X) yielded a statistically significant CpG hotspot, and this difference may be due to the larger sample size of independent CpG stop mutations examined in this study (n=62) versus the earlier study (n=16).
The 243 observed stop mutations occur in 15 of the 18 classes of sense codons (Table 4), with the three missing classes (TGC, TGT and TCG) representing transversions with the three smallest target sizes. This includes the lack of observed CpG transversions at six TCG serine codons and one TGC-G cysteine codon. The entire set of 243 stop mutations were found at 185 individual sites out of 1500 potential sites (Supp. Table S3), and the intersection of this set with other stop codon positions reported in the Leiden DMD mutation database (342 out of 1500 sites) reveals an overlap of 84 sites, with 101 unique stop sites observed here, and a total of 443 mutated sites out of 1500 potential sites observed in the joint set.
Polymorphisms and an idealized DMD transcript
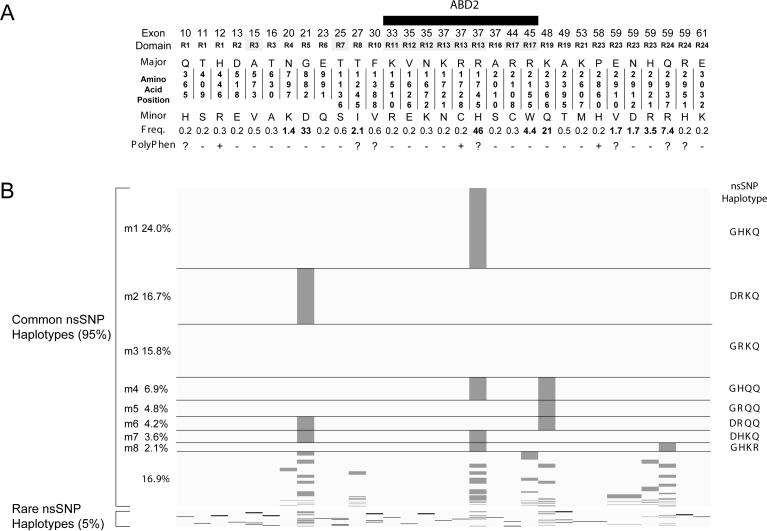
A detailed listing of exonic and flanking intronic polymorphisms in the DMD gene is included in Supp. Table S4 (diallelic SNPs) and Supp. Table S5 (VNTRs). In the subset of 698 patients that were fully resequenced, we noted the number of segregating sites at 395, including 51 coding region (31 nonsynonymous and 20 synonymous) polymorphisms, and 344 non-coding SNPs in flanking introns and UTRs. The 51 exonic SNPs were found in 157 cDNA haplotypes, and the 31 nonsynonymous SNPs (nsSNP) were found in 74 distinct protein haplotypes. The 31 nsSNPs all occur within central rod domain exons (R1 through R24, Figure 2A) and are predicted by PolyPhen analysis (Ramensky, et al., 2002) to be neutral (21 SNPs), possibly (7 SNPs) or probably (3 SNPs) protein damaging by physical and comparative considerations (Figure 2A). The most common nsSNP, exon 37 p.Arg1745His (rs1801187), has a minor allele frequency of 0.46 (p.His1745) in this study and is prevalent in three of the four HapMap populations (CEU 0.43, CHB 0.74, JPT 0.53, and YRI 0.01), although the p.Arg1745 allele appears to be ancestral in that it is found in all other sequenced vertebrate dystrophins. The 10 most common nonsynonymous SNPs (> 5% minor allele frequency) were observed as 45 distinct nonsynonymous haplotypes which encode subtly different protein isoforms (Figure 2B). The four most common nsSNPs (p.Gly882Asp, p.Arg1745His, p.Lys2366Gln and p.Gln2937Arg) define the eight most common nsSNP haplotypes and were observed at frequencies ranging from 24.0% (Dp427m1) through 2.1% (Dp427m8).
Figure 2.
Dystrophin amino acid polymorphisms and nsSNP haplotypes. (A) Position of non-synonymous variants based on the NM_004006.1 coding region is shown with exon number, spectrin repeat domain (R1 though R24), one-letter amino acid code, minor allele frequency and PolyPhen prediction indicated. (B) Protein haplotype structure for the nsSNPs from 698 fully resequenced patients sorted and scaled by haplotype frequency. Major and minor alleles are shaded by light and dark grey, respectively. The Dp427m5 nsSNP haplotype variant is identical to NM_004006.1 CDS and the four amino acid differences (p.G882D, p.R1745H, p.K2366Q and pQ2937R) observed in the other eight major haplotypes (Dp427m1-m8) are shown to the right of each nsSNP haplotype.
Our SNP analysis represents, to our knowledge, a unique data set. The depth of this set – the number of patients resequenced – allows us to calculate the molecular diversity metrics representative for this patient population. In comparison to the standard reference DMD sequence (NM_004006.1) we noted that an exact coding region (CDS) match to NM_004006.1 was only observed in 4.8% of patients that were fully sequenced. The mean pairwise difference between two individuals (π) for the coding region is 2.5 SNPs and the nucleotide diversity (averaged over CDS) is 2.24 × 10−4. Slightly higher diversity metrics were observed in the flanking intronic regions, with π = 35.6 and nucleotide diversity = 4.85 × 10−4. We observed further that each individual kindred could be assigned a unique genomic (exonic + intronic) SNP haplotype formed across the dystrophin gene. These unique individual kindred genomic haplotypes are most likely a consequence of the high recombination rate and high mean pairwise difference (π = 38 for CDS, UTRs and flanking intron regions) across the 4 cM dystrophin gene. Within the Dp427m coding region, the `idealized' DMD mRNA transcript in which each polymorphic nucleotide is represented by the major allele was observed in 9.0% of patients, although for the coding region, NM_004006.1 differs only by one nucleotide (c.7096A>Cp.Lys2366Gln) from this idealized version. Although our resequencing patient set is not made up of normal, asymptomatic individuals, it is reasonable to expect that the frequencies of these variants and haplotypes are representative of the population from which our patients were drawn.
Mono and Multiexon Skipping
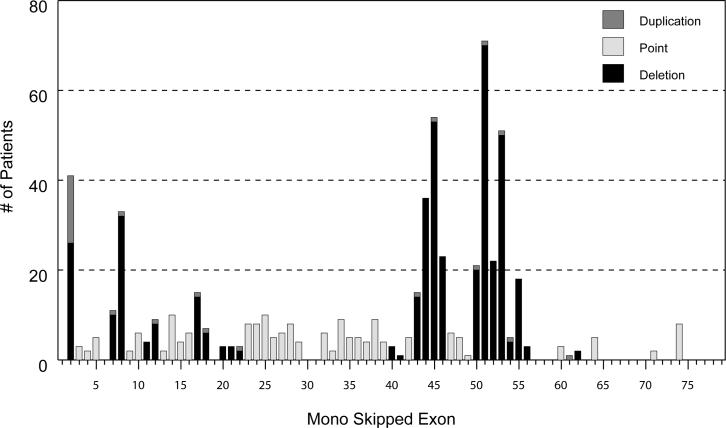
Antisense-mediated exon skipping to produce in-frame dystrophin mRNA has shown promise as a potential treatment for DMD patients (Aartsma-Rus, et al., 2003; Lu, et al., 2003), with one successful proof-of-principle clinical trial for single exon 51 skipping completed (van Deutekom, et al., 2007). To evaluate the number of patients in our study who may benefit from this approach, we calculated the number of patients with truncating mutations who would have their dystrophin frame restored by single exon skipping (mono-skipping). Figure 3 shows the distribution by mutation class, including single exon duplications, for a total of 864 patients with truncating mutations. Mono-skipping would potentially restore the reading frame in 515 (59.6%) patients, with the largest single fractions corresponding to the hotspot regions for deletions (exons 45–53) and duplications (exon 2). The highest number of patients would benefit from skipping of exon 51 (71 patients; 8.2%), followed by exon 45 (54 patients; 6.3%); the mutation distribution for each mono-exon skip is shown in Supp. Table S6. We note that these are the same exons identified in a recent large review of the Leiden DMD databse, in which exon 51 skipping was predicted to be of benefit for 13% of patients, and exon 45 for 8.1% (Aartsma-Rus, et al., 2009); the difference in values may be due to methods of ascertainment in the two groups. Recently it has been proposed that multi-exon skipping producing a “del45–55” (c.6439-8217del) dystrophin would treat 63% of deletion patients with DMD (Beroud, et al., 2007), and that a “del45–53” (c.6439-7872del) would treat 53.5% (Tuffery-Giraud, et al., 2009). In our set of 364 out-of-frame deletion patients, we observed that a similar level of patients would benefit from this multi-exon skipping approach (45 to 55 skipping = 62%, or 227 deletions, and 45 to 53 skipping = 53%, or 194 deletions) including 21 deletions not restorable by single exon skipping. In addition, exon 45–55 skipping would be predicted to benefit an additional 37 point mutation patients also not restorable by the single exon skipping.
Figure 3.
Suppression of truncating mutations by mono-exon skipping. The number of patients that can have their truncating mutation bypassed by single exon skipping is shown for each exon. The mutations found in each exon category are listed in Supp. Table S6.
Discussion
Mutation distribution
The distribution of mutation classes seen in this report differs from that previously reported in our own survey of unselected clinic patients (Dent, et al., 2005) and in other reports from referral laboratories (Prior and Bridgeman, 2005; Yan, et al., 2004). This is likely due to ascertainment bias. It is likely that our sample set has been particularly enriched by the enrollment of research subjects or referral of patients for whom no mutations had been detected using what was for many years the most commonly used diagnostic test, multiplex PCR. We postulate that this ascertainment bias accounts for our relatively high numbers of nonsense mutations (26.5%) and duplications (11.0%). This bias is likely shared with other reports from referral laboratories, in that the patients reported do not represent a population survey but represent only those patients who were sent for diagnostic purposes (Prior and Bridgeman, 2005; Yan, et al., 2004). Nevertheless, this represents one of the largest surveys to date on the mutational spectrum, and confirms the general distribution of mutation frequency.
Diagnostic algorithm/Subjects with undetected mutations
Our current diagnostic algorithm consists of MLPA analysis, followed by SCAIP analysis in a subset of patients. SCAIP is itself a two-step process. The first step in SCAIP is PCR amplification and visualization of amplicons from all 79 DMD exons, with deletions confirmed by PCR amplification using a second, independent set of primers. The second step in SCAIP is reserved for patients without deletions, and consists of direct sequence analysis of all 79 exons, and flanking intronic sequences. All samples without deletions or duplications by MLPA undergo SCAIP analysis for the detection of point mutations.
These modern methods of molecular diagnosis allow rapid and reliable detection of all deletions, duplications, and point mutations (including those affecting splice acceptor and splice donor sites at each exon). Among 858 patients in our UDP set were 51 asymptomatic females who underwent testing for carrier status but ultimately were found to not carry mutations from a related proband in their lymphocyte DNA. Among the remaining 807 patients – where enrollment was restricted to individuals with a presumably secure diagnosis of dystrophinopathy – were 14 patients (1.7%) in whom genomic mutational analysis led to reassessment of the phenotype, and reclassification as “not dystrophinopathy”. Of the remaining 793 symptomatic patients, there were 40 (5.0%) in whom no known mutation was detected by genomic analysis, consistent with earlier reports that found this range to be 4–7% (Dent, et al., 2005; Yan, et al., 2004). This patient population represents a diagnostic problem for clinicians. Improved molecular diagnostics were expected to obviate the need for muscle biopsy (Flanigan, et al., 2003). However, it is increasingly apparent that certain classes of mutations will continue to require mRNA analysis.
In 0.5% of patients, we identified pseudoexon mutations, an increasingly recognized category of mutation (Gurvich, et al., 2007; Tuffery-Giraud, et al., 2003) in which a point mutation within the introns results in the inclusion of intronic sequence in the final mRNA. Unfortunately, in most of the cases for whom no mutation was detected by analysis of genomic DNA from a blood sample, we were unable to obtain muscle for further analysis by reverse transcriptase PCR (RT-PCR) based analysis of the DMD gene transcript. In all cases except for one where we were able to obtain muscle for such analysis, a pseuodoexon mutation was detected. The three patients in whom no mutation has yet been detected have both clear X-linked histories of weakness consistent with DMD, and absent immunofluorescent staining for dystrophin protein. For each boy, extended analysis of the X-chromosome locus is underway, in a search for alterations in non-coding regions that may account for his syndrome.
Genotype/phenotype
We can make several observations regarding correlation of genotype to phenotype, although we are limited to some extent by the lack of standardized phenotypic information in the referred sample cohort. For non-UDP patients the diagnostic criteria used by the referring physician may differ, as opposed to the case in the UDP patients, where an agreed-upon criteria for diagnosis of dystrophinopathy subtype is used. This difficulty, of course, is inherent in interpreting the results from any large referral cohort.
The reading frame rule (Monaco, et al., 1988) states that BMD is due to mutations that preserve an open reading frame through the 3' end of the gene such that a carboxy-terminal encoding protein is translated, whereas DMD associated mutations result in an altered reading frame such that translational termination occurs prior to the carboxy-terminus. As noted in Table 2, among patients with deletions, truncating mutations result in DMD in 254 of 286 patients (89%), but non-truncating (in-frame) deletions result in BMD or IMD in 38 of 68 patients (56%). Adding point mutations (but excluding duplications, due to the uncertainty of the reading frame) alters this only slightly: truncating mutations result in DMD in 519 of 598 patients (87%), whereas non-truncating mutations result in BMD or IMD in 63 of 100 (63%). In our cohort, exceptions to the reading frame rule can in large part be explained by the effect of point mutations on exonic splicing control sequences [(Aartsma-Rus, et al., 2006; Disset, et al., 2006), and Flanigan et al, manuscript in preparation]. In patients with deletions, a low degree of baseline altered splicing, resulting in in-frame mRNA, may account for exceptions to the rule. Among patients with deletions, other molecular mechanisms underlying finer gradations in disease severity than those denoted by the classification of BMD versus DMD are unclear, but are the subject of ongoing research within the well-phenotyped UDP cohort. Possible influences include trans-acting polymorphisms (eg, within muscle performance genes, or within splicing factors), as well as cis-acting polymorphisms within the DMD locus itself.
Our data confirm the presence of hotspots for both deletion and duplication mutations. The increased frequency of exon 2 duplications has been reported previously (White, et al., 2006). Similarly, complex duplications are increasingly recognized (Janssen, et al., 2005; White, et al., 2006). Our point mutation data confirm the absence of a hotspot for point mutations, and reiterate that point mutations are frequently “private” mutations, occurring within families. The increased frequency of a handful of point mutations within our dataset reflects the enrollment of patients with unusual alleles for further study in our group, and disappears in our table of unique mutations. There is one exception to this: the c.9G>A (p.3Trp>X) mutation, which we have previously reported (Flanigan, et al., 2003; Howard, et al., 2004), was detected in six independent families. We have recently established this mutation, associated with a BMD phenotype, as the first founder allele described in the DMD gene (Flanigan et al, manuscript in submission).
These results serve as a reminder to the clinician that phenotype cannot be predicted from genotype alone. Clinical laboratories should therefore be cautious in the inclusion of language predicting phenotype in the interpretation section of mutation testing results.
Mutation rates and SNPs
Data on spontaneous nucleotide substitution mutation rates in humans derive mostly from locus-specific mutations of dominantly inherited diseases which may be biased in their mutational spectrum. Mutation rates more representative of the changes responsible for the DNA sequence evolution seen in the bulk of the genome can be obtained from direct measurements in X-linked recessive diseases (Kondrashov, 2003; Sommer, 1995), and our dataset contributes substantially in this area.
If DMD nucleotide substitution rates are relatively unbiased, then the mutational spectrum we observed has broader utility in extrapolating this direct measurement to the general rate of germline mutation in the human population. Analogous, but indirect, estimations of nucleotide substitution rates have been derived from DNA sequence comparisons between human and chimpanzee, where genome-wide nucleotide divergence levels can be used to estimate germline mutations given assumptions regarding divergence times and effective population sizes. Both direct and indirect measurements suggest genome-wide mutation rates of about 1–2 × 10−8 per nucleotide site; our observed measurement of an overall base substitution rate of 1.51 × 10−8 is in close agreement with these estimates, and specifically with a prior estimate of 1.78 × 10−8 from a meta-analysis averaged across 20 human disease loci (Kondrashov, 2003), suggesting that the point mutation rate at DMD gene is a good estimate of unbiased germline substitution rates.
Specific comparisons of our substitution rate (1.51 × 10−8) with a prior DMD meta-analysis substitution rate (2.46 × 10−8) reveals a lower rate in our data that is primarily due to a lack of observed transversions at CpG sites. As expected from transitions caused by spontaneous deamination of methylated cytosines, the CpG to TpG transitions within CGA arginine codons accounted for 25% of the observed nonsense mutations even though CGA codons comprise only 2% of the target codons. Our observed CpG to non-CpG mutation ratio of 7.7 is in substantial agreement with measurements from human-chimpanzee divergence where the mutation rate for bases in a CpG dinucleotide are 10-fold higher than for other bases (Consortium, 2005). We also noted that there is no evidence for differential hypermutability at CGA codons, as had been previously suggested (Buzin, et al., 2005), perhaps due to the larger number of events (62 here, versus 16 mutations) that we observed. There were also no apparent hotspots for small insertion/deletion mutations within the 11 kb Dp427m coding region, and deletions (average size = 4.2 nts) outnumbered insertions (average size = 2.8 nts) by 2 to 1. The overall insertion/deletion substitution rate of 0.95 × 10−9 observed from our set of 113 such mutations within the 11 kb coding region is also consistent with previous direct estimates of insertion/deletion mutation rates excluding hotspots.
Our exonic-centric SNP data highlight the degree of variation across the very large DMD locus. Just as our point mutation data show that essentially all point mutations are “private mutations”, our haplotype data (using exonic and intronic SNPs) show that there are essentially individual haplotypes in unrelated families across the locus. As is the case with point mutations, shared background haplotypes have been only found in the case of related individuals, and these unique genomic haplotypes are the result of the large size and high recombination rate across the DMD gene. More limited diversity is observed with the restricted number of haplotypes based on exonic SNPs. The identification of eight major nsSNP haplotypes in Dp427m sub-isoforms provides the opportunity to begin to utilize them in genotype/phenotype correlation studies to look for cis effects on dystrophin function, and we therefore suggest that laboratories determine on which sub-isoform a given point mutation is found.
On the basis of the `long-range haplotype' methods, it has been recently demonstrated that the strongest positive selection signal in the genomes of the HapMap African sample (120 Yoruba people in Ibadan, Nigeria) resides within the LARGE gene, a glycosylase that post-translationally modifies α-dystroglycan (Sabeti, et al., 2007). It was also noted that the DMD rs80540 SNP, located 500 nucleotides 5'of exon 13, demonstrated a significant `long-range haplotype' signal of selection in the Yoruba sample. Prior evidence from a sampling of a 2.4 kb segment in DMD intron 7 indicated positive directional selection in a small set of individuals (10) from Africa (Nachman and Crowell, 2000). α-dystroglycan is the cellular receptor for Lassa fever virus and other arenaviruses, and the evidence for strong signatures of recent selective pressure in populations where the virus is endemic may indicate that there is joint selection for functional variation in LARGE and DMD that modulates α-dystroglycan in these populations. The pattern of SNP allelic diversity observed in our affected patient population should allow further examination of this viral-mediated selection hypothesis and DMD functional variation by correlating the geographical distribution of the selected DMD SNPs and haplotypes with arenavirus endemicity. As a caveat, the majority of our population is North American and European, and the allele frequencies we report may not be represented at the same frequency worldwide.
Summary
In addition to expanding the mutational spectrum of the dystrophinopathies, these results establish improved SNP frequency data for the DMD gene and lead us to propose a novel classification of sub-isoforms of the muscle isoforms of dystrophin. The significance of these variants requires further study. Whether these variants play a role in phenotype amelioration is a question amenable to study in our patient population, which also serves as a catalogue of genotyped patients for future clinical trials.
Online Resources.
We have established an online resource for public access to this data at the Utah Genome Center website, using two sites that will be updated regularly (http://www.genome.utah.edu/DMD/mutationtables, and http://www.genome.utah.edu/DMD/dystrophysnps). In addition, the UDP has established the UDP Online Duchenne and Becker Muscular Dystrophy Patient Registry (http://www.dystrophin.org). At this site, patients can self-report a core set of demographic and phenotypic information, which will allow further studies in genotype/phenotype correlation, and will further expand cohorts for clinical trials. This database is open to individuals who have had genotyping at other laboratories, and curation of outside genetic testing results are performed by staff of the UDP.
Supplementary Material
Acknowledgments
The authors wish to thank all referring physicians; thank A. Bringard and J. Tyce for administrative assistance; and to acknowledge the study coordinator assistance of K. Hart, C. Moural and K. Hak and the technical assistance of L. Zhao, T. Tuohy, L. Taylor, O. Gurvich, B. Duval, C. Hamil, M. Mahmoud, and A. Aoyagi. This work is supported by the National Institute of Neurologic Diseases and Stroke (R01 NS043264 [KMF, MTH, RBW]; the National Center for Research Resources (M01-RR00064, to the University of Utah, Dr. L. Betz, P.I.); by the Association Francaise Contre les Myopathies (KMF); and by the Parent Project Muscular Dystrophy (PS).
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
United Dystrophinopathy Project: Other Investigators and Members The University of Utah, Salt Lake City, Utah: Mark Bromberg, MD, PhD; Kathy Swoboda, MD; Lynne Kerr, MD, PhD; Kim Hart, MS; Cybil Moural, MS; Kate Hak, BS Nationwide Medical Center, Columbus, Ohio: Laurence Viollet, PhD; Susan Gailey, MS Washington University, St. Louis, Missouri: Glenn Lopate, MD; Paul Golumbek MD, PhD; Jeanine Schierbecker MHS, PT; Betsy Malkus MHS, PT; and Catherine Siener MHS, PT University of Iowa, Iowa City, Iowa: Kris Baldwin, LPT Children's Hospital/University of Pennsylvania, Philadelphia, Pennsylvania: Allan M. Glanzman, PT, DPT, PCS, ATP; Jean Flickinger, RPT Cincinnati Children's Hospital, Cincinnati, Ohio: University of Minnesota, Minneapolis, Minnesota: Cameron E. Naughton
References
- Aartsma-Rus A, Fokkema I, Verschuuren J, Ginjaar I, van Deutekom J, van Ommen GJ, den Dunnen JT. Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat. 2009;30(3):293–9. doi: 10.1002/humu.20918. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Janson AA, Kaman WE, Bremmer-Bout M, den Dunnen JT, Baas F, van Ommen GJ, van Deutekom JC. Therapeutic antisense-induced exon skipping in cultured muscle cells from six different DMD patients. Hum Mol Genet. 2003;12(8):907–14. doi: 10.1093/hmg/ddg100. [DOI] [PubMed] [Google Scholar]
- Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34(2):135–44. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- Abbs S, Bobrow M. Analysis of quantitative PCR for the diagnosis of deletion and duplication carriers in the dystrophin gene. J Med Genet. 1992;29(3):191–6. doi: 10.1136/jmg.29.3.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Krawczak M, Cooper DN. Disease-causing mutations in the human genome. Eur J Pediatr. 2000;159(Suppl 3):S173–8. doi: 10.1007/pl00014395. [DOI] [PubMed] [Google Scholar]
- Beggs AH, Koenig M, Boyce FM, Kunkel LM. Detection of 98% of DMD/BMD gene deletions by polymerase chain reaction. Hum Genet. 1990;86(1):45–8. doi: 10.1007/BF00205170. [DOI] [PubMed] [Google Scholar]
- Bennett RR, Dunnen J, O'Brien KF, Darras BT, Kunkel LM. Detection of mutations in the dystrophin gene via automated DHPLC screening and direct sequencing. BMC Genet. 2001;2(1):17. doi: 10.1186/1471-2156-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroud C, Tuffery-Giraud S, Matsuo M, Hamroun D, Humbertclaude V, Monnier N, Moizard MP, Voelckel MA, Calemard LM, Boisseau P, et al. Multiexon skipping leading to an artificial DMD protein lacking amino acids from exons 45 through 55 could rescue up to 63% of patients with Duchenne muscular dystrophy. Hum Mutat. 2007;28(2):196–202. doi: 10.1002/humu.20428. [DOI] [PubMed] [Google Scholar]
- Buzin CH, Feng J, Yan J, Scaringe W, Liu Q, den Dunnen J, Mendell JR, Sommer SS. Mutation rates in the dystrophin gene: a hotspot of mutation at a CpG dinucleotide. Hum Mutat. 2005;25(2):177–88. doi: 10.1002/humu.20132. [DOI] [PubMed] [Google Scholar]
- Chamberlain JS, Gibbs RA, Ranier JE, Caskey CT. Multiplex PCR for the diagnosis of Duchenne muscular dystrophy. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. Academic Press; San Francisco: 1990. pp. 272–281. [Google Scholar]
- Consortium CSaA. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437(7055):69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet. 1989;83(2):181–8. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- Curtis A, Haggerty D. Deletion and Duplication Analysis in Males Affected with Duchenne or Becker Muscular Dystrophy. In: Bushby K, Anderson LV, editors. Muscular Dystrophy: Methods and Protocols. Humana Press; Totowa, New Jersey: 2001. pp. 53–84. [Google Scholar]
- Den Dunnen JT, Grootscholten PM, Bakker E, Blonden LA, Ginjaar HB, Wapenaar MC, van Paassen HM, van Broeckhoven C, Pearson PL, van Ommen GJ. Topography of the Duchenne muscular dystrophy (DMD) gene: FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet. 1989;45(6):835–47. [PMC free article] [PubMed] [Google Scholar]
- Dent KM, Dunn DM, von Niederhausern AC, Aoyagi AT, Kerr L, Bromberg MB, Hart KJ, Tuohy T, White S, den Dunnen JT, et al. Improved molecular diagnosis of dystrophinopathies in an unselected clinical cohort. Am J Med Genet A. 2005 doi: 10.1002/ajmg.a.30617. [DOI] [PubMed] [Google Scholar]
- Disset A, Bourgeois CF, Benmalek N, Claustres M, Stevenin J, Tuffery-Giraud S. An exon skipping-associated nonsense mutation in the dystrophin gene uncovers a complex interplay between multiple antagonistic splicing elements. Hum Mol Genet. 2006;15(6):999–1013. doi: 10.1093/hmg/ddl015. [DOI] [PubMed] [Google Scholar]
- Dolinsky LC, de Moura-Neto RS, Falcao-Conceicao DN. DGGE analysis as a tool to identify point mutations, de novo mutations and carriers of the dystrophin gene. Neuromuscul Disord. 2002;12(9):845–8. doi: 10.1016/s0960-8966(02)00069-x. [DOI] [PubMed] [Google Scholar]
- Flanigan KM, von Niederhausern A, Dunn DM, Alder J, Mendell JR, Weiss RB. Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet. 2003;72(4):931–9. doi: 10.1086/374176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisso G, Carsana A, Tinto N, Calcagno G, Salvatore F, Sacchetti L. Direct detection of exon deletions/duplications in female carriers of and male patients with duchenne/becker muscular dystrophy. Clin Chem. 2004;50(8):1435–8. doi: 10.1373/clinchem.2004.033795. [DOI] [PubMed] [Google Scholar]
- Gurvich OL, Tuohy TM, Howard MT, Finkel RS, Medne L, Anderson CB, Weiss RB, Wilton SD, Flanigan KM. DMD pseudoexon mutations: splicing efficiency, phenotype, and potential therapy. Ann Neurol. 2007 doi: 10.1002/ana.21290. [DOI] [PubMed] [Google Scholar]
- Hofstra RM, Mulder IM, Vossen R, de Koning-Gans PA, Kraak M, Ginjaar IB, van der Hout AH, Bakker E, Buys CH, van Ommen GJ, et al. DGGE-based whole-gene mutation scanning of the dystrophin gene in Duchenne and Becker muscular dystrophy patients. Hum Mutat. 2004;23(1):57–66. doi: 10.1002/humu.10283. [DOI] [PubMed] [Google Scholar]
- Howard MT, Anderson CB, Fass U, Khatri S, Gesteland RF, Atkins JF, Flanigan KM. Readthrough of dystrophin stop codon mutations induced by aminoglycosides. Ann Neurol. 2004;55(3):422–6. doi: 10.1002/ana.20052. [DOI] [PubMed] [Google Scholar]
- Hu XY, Ray PN, Worton RG. Mechanisms of tandem duplication in the Duchenne muscular dystrophy gene include both homologous and nonhomologous intrachromosomal recombination. EMBO J. 1991;10(9):2471–7. doi: 10.1002/j.1460-2075.1991.tb07786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen B, Hartmann C, Scholz V, Jauch A, Zschocke J. MLPA analysis for the detection of deletions, duplications and complex rearrangements in the dystrophin gene: potential and pitfalls. Neurogenetics. 2005;6(1):29–35. doi: 10.1007/s10048-004-0204-1. [DOI] [PubMed] [Google Scholar]
- Kondrashov AS. Direct estimates of human per nucleotide mutation rates at 20 loci causing Mendelian diseases. Hum Mutat. 2003;21(1):12–27. doi: 10.1002/humu.10147. [DOI] [PubMed] [Google Scholar]
- Lampe AK, Dunn DM, von Niederhausern AC, Hamil C, Aoyagi A, Laval SH, Marie SK, Chu ML, Swoboda K, Muntoni F, et al. Automated genomic sequence analysis of the three collagen VI genes: applications to Ullrich congenital muscular dystrophy and Bethlem myopathy. J Med Genet. 2005;42(2):108–20. doi: 10.1136/jmg.2004.023754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QL, Mann CJ, Lou F, Bou-Gharios G, Morris GE, Xue SA, Fletcher S, Partridge TA, Wilton SD. Functional amounts of dystrophin produced by skipping the mutated exon in the mdx dystrophic mouse. Nat Med. 2003;9(8):1009–14. doi: 10.1038/nm897. [DOI] [PubMed] [Google Scholar]
- Mendell JR, Buzin CH, Feng J, Yan J, Serrano C, Sangani DS, Wall C, Prior TW, Sommer SS. Diagnosis of Duchenne dystrophy by enhanced detection of small mutations. Neurology. 2001;57(4):645–50. doi: 10.1212/wnl.57.4.645. [DOI] [PubMed] [Google Scholar]
- Monaco AP, Bertelson CJ, Liechti Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2(1):90–5. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL. Contrasting evolutionary histories of two introns of the duchenne muscular dystrophy gene, Dmd, in humans. Genetics. 2000;155(4):1855–64. doi: 10.1093/genetics/155.4.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior TW, Bridgeman SJ. Experience and strategy for the molecular testing of Duchenne muscular dystrophy. J Mol Diagn. 2005;7(3):317–26. doi: 10.1016/S1525-1578(10)60560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RG, Barby TF, Manners E, Bobrow M, Bentley DR. Direct detection of dystrophin gene rearrangements by analysis of dystrophin mRNA in peripheral blood lymphocytes. Am J Hum Genet. 1991;49(2):298–310. [PMC free article] [PubMed] [Google Scholar]
- Rogozin I, Kondrashov F, Glazko G. Use of mutation spectra analysis software. Hum Mutat. 2001;17(2):83–102. doi: 10.1002/1098-1004(200102)17:2<83::AID-HUMU1>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449(7164):913–8. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M, Duno M. Improved molecular diagnosis of dystrophin gene mutations using the multiplex ligation-dependent probe amplification method. Genet Test. 2004;8(4):361–7. doi: 10.1089/gte.2004.8.361. [DOI] [PubMed] [Google Scholar]
- Sommer SS. Recent human germ-line mutation: inferences from patients with hemophilia B. Trends Genet. 1995;11(4):141–7. doi: 10.1016/s0168-9525(00)89028-9. [DOI] [PubMed] [Google Scholar]
- Tuffery-Giraud S, Beroud C, Leturcq F, Yaou RB, Hamroun D, Michel-Calemard L, Moizard MP, Bernard R, Cossee M, Boisseau P, et al. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum Mutat. 2009;30(6):934–45. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- Tuffery-Giraud S, Saquet C, Chambert S, Claustres M. Pseudoexon activation in the DMD gene as a novel mechanism for Becker muscular dystrophy. Hum Mutat. 2003;21(6):608–14. doi: 10.1002/humu.10214. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357(26):2677–86. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- White S, Kalf M, Liu Q, Villerius M, Engelsma D, Kriek M, Vollebregt E, Bakker B, van Ommen GJ, Breuning MH, et al. Comprehensive detection of genomic duplications and deletions in the DMD gene, by use of multiplex amplifiable probe hybridization. Am J Hum Genet. 2002;71(2):365–74. doi: 10.1086/341942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SJ, Aartsma-Rus A, Flanigan KM, Weiss RB, Kneppers AL, Lalic T, Janson AA, Ginjaar HB, Breuning MH, den Dunnen JT. Duplications in the DMD gene. Hum Mutat. 2006;27(9):938–45. doi: 10.1002/humu.20367. [DOI] [PubMed] [Google Scholar]
- Yan J, Feng J, Buzin CH, Scaringe W, Liu Q, Mendell JR, den Dunnen J, Sommer SS. Three-tiered noninvasive diagnosis in 96% of patients with Duchenne muscular dystrophy (DMD) Hum Mutat. 2004;23(2):203–4. doi: 10.1002/humu.10307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.