Abstract
Streptococcus pneumoniae is the leading cause of vaccine-preventable deaths globally. The objective of this study was to determine the distribution and clonal type variability of three potential vaccine antigens: Pneumococcal serine-rich repeat protein (PsrP), Pilus-1, and Pneumococcal choline binding protein A (PcpA) among pneumococcal isolates from children with invasive pneumococcal disease and healthy nasopharyngeal carriers. We studied by Real-Time PCR a total of 458 invasive pneumococcal isolates and 89 nasopharyngeal pneumococcal isolates among children (total = 547 strains) collected in Barcelona, Spain, from January 2004 to July 2010. pcpA, psrP and pilus-1 were detected in 92.8%, 51.7% and 14.4% of invasive isolates and in 92.1%, 48.3% and 18% of carrier isolates, respectively. Within individual serotypes the prevalence of psrP and pilus-1 was highly dependent on the clonal type. pcpA was highly prevalent in all strains with the exception of those belonging to serotype 3 (33.3% in serotype 3 isolates vs. 95.1% in other serotypes; P<.001). psrP was significantly more frequent in those serotypes that are less apt to be detected in carriage than in disease; 58.7% vs. 39.1% P<.001. Antibiotic resistance was associated with the presence of pilus-1 and showed a negative correlation with psrP. These results indicate that PcpA, and subsequently Psrp and Pilus-1 together might be good candidates to be used in a next-generation of multivalent pneumococcal protein vaccine.
Introduction
Invasive disease caused by Streptococcus pneumoniae is responsible for more than 1.6 million childhood deaths worldwide every year [1]. In certain developed countries, including Spain, despite vaccination with a 7-valent conjugate vaccine against capsular polysaccharide (PCV-7), pneumococcal pneumonia remains a major cause of pediatric hospital admission [2], [3], [4]. PCV-7 is composed of capsular polysaccharide from serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F individually conjugated to diphtheria CRM197 and has proved to be effective in preventing pneumococcal disease caused by these serotypes in children [5]. PCV7 also prevents invasive pneumococcal disease (IPD) in adult and non-vaccinated children by an indirect effect (herd immunity) on pneumococcal transmission [5], [6]. Importantly, nowadays evidence exists of the emergence of non-vaccine serotypes in children and adults to occupy this vaccine-emptied niche, thereby partially eroding the benefit of PCV-7 [3], [7], [8], [9]. For example, in Spain disease caused by serotype 19A was responsible for 13.5% of pediatric IPD during the period 2000–2008, whereas in 2000, at the time of introduction of PCV-7, serotype 19A only accounted for 4.6% of pediatric infections [10]. The pneumococcus is also a primary cause of otitis media and PCV-7 only slightly reduces the rate of disease [11]. At present, more than 1,500,000 cases occur annually in the United States, with an estimated cost of 440 million U.S. dollars [12]. Thus, pneumococcal disease remains a major medical problem with an urgent need for an improved vaccine.
Due to these limitations, other conjugate vaccines with a larger number of serotypes have been recently commercialized. These include a 10-valent conjugate vaccine (PCV10), which includes the seven serotypes of PCV7 plus serotypes 1, 5 and 7F and PCV13 (PCV10 plus additional serotypes 3, 6A and 19A). These vaccines will most likely continue to reduce the burden of invasive pneumococcal disease and are becoming increasingly available in underdeveloped countries due to efforts of institutions such as The Bill and Melinda Gates Foundation through GAVI Alliance [13], [14]. However, due to the high cost of the conjugation process, these vaccines are limited in the number of serotypes that can be included in an affordable vaccine. The current cost for each dose of PCV13 is $100–125, with three immunizations recommended.
An alternate vaccine strategy is the use of a serotype-independent vaccine using conserved common pneumococcal protein antigens. These might stand alone, or replace the diphtheria toxoid in the conjugate vaccine and thereby enhance coverage of the existing vaccines. To date, numerous preclinical studies have shown that different pneumococcal proteins confer protection against pneumococcal challenge and that a combination of multiple proteins confers superior protection. The main advantage of a protein vaccine is that protection would not be serotype dependent and fewer antigen candidates could offer a high coverage with a lower cost of manufacturing. For these reasons, studies are warranted in determining if a next-generation of a multivalent protein vaccine against pneumococcus is feasible and desirable.
The objective of the present study was to determine the distribution and clonal type variability of three novel potential vaccine candidates: Pneumococcal serine-rich protein (PsrP), Pilus-1, and Pneumococcal choline binding protein A (PcpA). PsrP is a serine rich repeat protein (SRRP) previously demonstrated to be responsible for lung-cell attachment and in vivo biofilm formation [15], [16]. Pilus is a long organelle that, like PsrP, extends beyond the polysaccharide capsule and acts as an adhesin [17]. Finally, PcpA is a choline-binding protein with a role in pneumococcal adhesion and biofilm formation [18], [19]. Determining the prevalence and distribution of these proteins in strains that cause IPD and their correlation with disease and antibiotic resistance could be of great value for future vaccine formulations.
Methods
Clinical Isolates
All pediatric invasive pneumococcal isolates characterized by the Molecular Microbiology Department at University Hospital Sant Joan de Deu in Barcelona, Spain from January 2004 to December 2010 were included in this study. The department performs molecular surveillance of pneumococci in Catalonia, Spain. Clinical isolates were obtained from patients admitted to Sant Joan de Déu Hospital and, since 2009, from patients attended in 30 health centers throughout Catalonia region. In addition, we also included eighty-nine pneumococcal strains isolated from nasopharynx of healthy children during 2004–2008.
Serotyping and Antimicrobial Susceptibility
All isolates were serotyped by Quellung reaction at the National Pneumococcus Reference Centre (Majadahonda, Madrid). Pneumococcal isolates collected since 2009 were also serotyped by Real-Time PCR (RT-PCR) using published protocols [20]. Serotypes were classified according to coverage of the existing 7,10, and 13-valent conjugate vaccines and their attack rate according to the studies of Brueggemann et al. [21] and Sleeman et al. [22]. Serotypes with high attack rate (those that are less apt to be detected in carriage than in disease) included: 1, 4, 5, 7F, 9V, 14, 18C and 19A. Serotypes with low attack rate (that are less apt to be detected in disease than in carriage) included: 3, 6A, 6B, 8, 9N, 10A, 11A, 12F, 13, 15A, 15BC, 16F, 17F, 19F, 20, 21, 22F, 23A, 23B, 23F, 24F, 27, 31, 33F, 35B, 35F, 37 and 38. Agar dilution technique was used to determine the minimal inhibitory concentrations (MICs) of penicillin and other antibiotics. Antibiotic susceptibility was defined according to the 2008 meningeal breakpoints established by the Clinical Laboratory Standards Institute [23]. Isolates with intermediate or high level resistance were defined as non-susceptible.
Extraction of DNA
Genomic DNA was extracted from bacteria using Chelex-100 resin (BioRad Laboratories, Hercules, California, USA). Briefly, pneumococci scraped from blood agar plates were suspended in 100 µl of PBS-buffer; 50 µl were transferred to a new microcentrifuge tube and vigorously vortexed with 150 µl of 20% w/v Chelex-100 in PBS. The bacteria/resin suspensions were incubated for 20 minutes at 56°C followed by a 10-minute incubation at 100°C. After cooling and centrifugation, the supernatant was used as a DNA template in PCR reactions.
Multilocus Sequence Typing (MLST)
Genetic characterization of pneumococci was performed using MLST. In brief, internal fragments of the aroE, gdh, gki, recP, spi, xpt and ddl genes were amplified by PCR using the primer pairs described by Enright and Spratt [24]. PCR products were sequenced using an ABI 3130xl GeneticAnalyzer (Applied Biosystems). The sequences at each of the seven loci were then compared with all of the known alleles at that locus. Sequences that are identical to a known allele were assigned the same allele number whereas those that differ from any known allele were assigned new allele numbers. The assignment of alleles at each locus was carried out using the software at the pneumococcal web page: www.mlst.net. The alleles at each of the seven loci define the allelic profile of each isolate and their sequence type (ST). Allelic profiles are shown as the combination of 7 alleles in the order aroE, gdh, gki, recP, spi, xpt and ddl. A clone is defined as a group of isolates with identical allelic profile or ST.
Real-Time PCR Assay
We analyzed the nucleotide sequence of psrP, pilus-1 subunit rrgC, and pcpA for primers in all publically available S. pneumoniae genomes available through the United States National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov/). The primers and probe selected for psrP detection were: forward primer: 5′-CTTTACATTTACCCCTTACGCTGCTA; reverse primer 3′ CTGAGAGTGACTTAGACTGTGAAAGTG and probe: FAM-CTGGTCGTGCTAGATTC (Quencher MGB). These primers identified a conserved region within Basic Region domain of PsrP. For pilus-1 detection the primers and probe were: forward primer: 5′-TTGTGACAAATCTTCCTCTTGGGA; reverse primer: 3′-GTCACCAGCTGATGATCTACCA and probe: FAM-CAGTGGCTCCACCTCC (Quencher MGB). These primers identified a conserved region within the structural subunit protein RrgC encoded in the rlrA islet of pilus type 1. For pcpA detection the primers and probe were: forward primer: 5′-GAAAAAGTAGATAATATAAAACAAGAAACTGATGTAGCTAAA; reverse primer: 3′-ACCTTTGTCTTTAACCCAACCAACT and probe: FAM-CTCCCTGATTAGAATTC (Quencher MGB). These primers identified a conserved region of N-terminal fragment of PcpA. Finally, as a positive control and to test PCR inhibitors and DNA quality, detection of ply gene by Real-Time PCR was performed as previously described in all strains [25]. Ply encodes the pneumolysin, a toxin found within all S.pneumoniae.
The reaction volume for each gene detected was a total of 25µl and contained 5µl of DNA extract from samples or controls and 12.5µl 2X TaqMan Universal Master Mix (Applied Biosystems), which includes dUTP and uracil-N-glycosylase; each primer was used at a final concentration of 900 nM. The TaqMan probes were used at a final concentration of 250 nM. DNA Amplification was done performing universal amplification conditions: incubation for 2 min at 50°C (uracil-N-glycosylase digestion) and 10 min denaturation at 95°C, 45 cycles of two-step amplification (15 s at 95°C, 60 s at 60°C). Amplification data were analyzed by SDS software (Applied Biosystems). The reporter dye was measured relative to the internal reference dye (ROX) signal to normalize for non-PCR related fluorescence fluctuations occurring from well to well. The cycle threshold (CT) value was defined as the cycle at which the reporting dye fluorescence first exceeds the background level.
Statistical Analysis
Statistical analysis was performed with the PASW software package (version 17.0). Continuous variables were compared using the t test (for approximately normally distributed data) or the Mann-Whitney U test (for skewed data) and described as mean values and standard deviations or median and interquartile range P25–P75 (IQR) according to the presence of normal distribution. Chi-square test or Fisher’s exact test (two-tailed) was used to compare categorical variables. Comparison between groups was performed by Kruskal-Wallis test. Statistical significance was set at a P value of <0.05.
Results
Strain Properties
Of the total 461 pediatric invasive pneumococcal isolates in our library, 3 of them could not be recovered from stocks and were thereby excluded from the study. As such, we examined a total of 458 invasive pneumococcal isolates and 89 nasopharyngeal pneumococcal isolates among children (total = 547 strains).
The clinical syndromes were: pneumonia 257 (111 of them with empyema), bacteremia 114, meningitis 68, arthritis 13, appendicitis 4, pericarditis 1 and peritonitis 1.
The most frequent serotypes detected among invasive isolates were serotype 1 (n = 134), 19A (n = 84), 7F (n = 35), 5 (n = 34) and 14 (n = 19). Among carriers the most frequent serotypes were 19A (n = 9), 6A (n = 9), 19F (n = 7), 15B (n = 6) and 23B (n = 6).
Among IPD isolates, the prevalence of serotypes included in the commercialized conjugate vaccines PCV7, PCV10 and PCV13 were 14.2% (65 isolates), 58.3% (267 isolates) and 83.6% (383 isolates) respectively. The prevalence of serotypes included in the three vaccines among isolates from the nasopharynx of healthy carriers was 23.6% (21 isolates), 27% (24 isolates) and 50.6% (45 isolates).
With respect to clonal properties the most frequent clonotypes among invasive isolates were ST306 (n = 107), ST191 (n = 31), ST1223 (n = 25), ST304 (n = 22), ST276 (n = 17). A high variety of clonotypes were detected in carriers (56 different clonotypes in 89 strains); the most frequent being ST2372 (n = 5), ST97 (n = 4), ST42 (n = 3), ST63 (n = 3), ST180 (n = 3), ST838 (n = 3) and ST2690 (n = 3). Finally, antibiotic susceptibility study was available in 543 of the 547 strains with 134 (24.5%) having diminished penicillin susceptibility (MIC ≥0.12). The percentage of isolates with diminished penicillin susceptibility was 23.4% (107 of 454) among invasive isolates and 30.3% (27 of 89) among carriers.
Overall Prevalence of PcpA, PsrP and Pilus-1
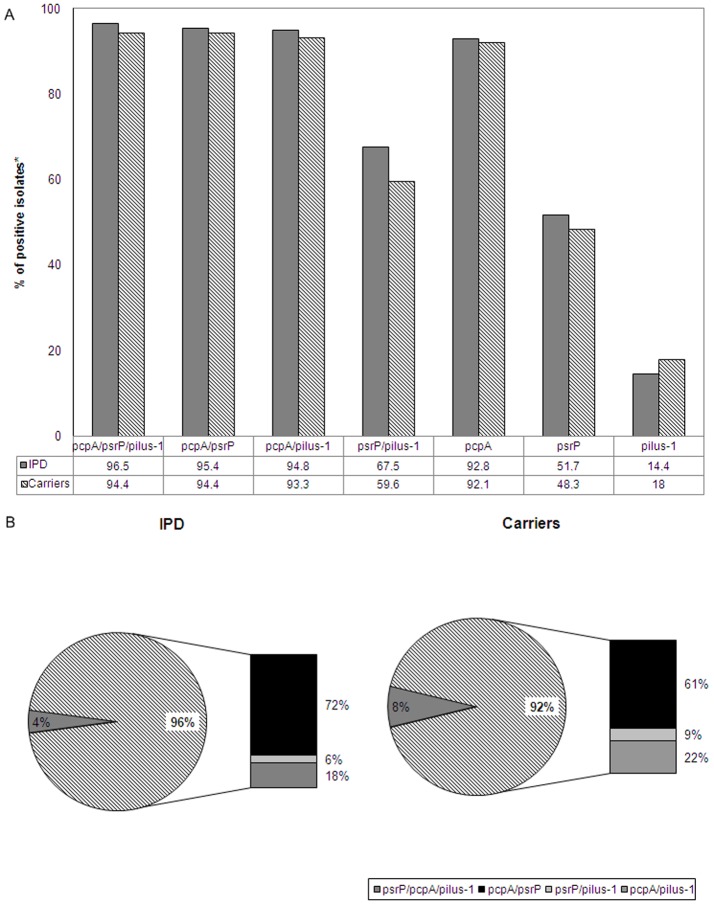
The individual prevalence of pcpA, psrP, and Pilus-1 in the 547 strains of our collection were 92.7%, 51.2% and 15% without significant differences occurring between invasive and carrier isolates: for pcpA 92.8% vs. 92.1%; P = 0.8, for psrP 51.7% vs. 48.3%; P = 0.5 and for pilus-1 14.4% vs. 18%; P = 0.3, respectively. Given the high prevalence of pcpA the potential coverage with at least one protein of a multivalent vaccine including these three candidates would be high: 96.5% among invasive isolates (442 of 458 isolates) and 94.4% among carriers (84 of 89 isolates). Figure 1A shows the prevalence for each protein alone and for at least 1 of the proteins in the specific combinations (PcpA/PsrP/Pilus-1, PcpA/PsrP, PcpA/Pilus-1 and PsrP/Pilus-1). Notably, in Figure 1B, we show that 96% of the invasive isolates carried at least two of the three proteins, whereas 92% of the carrier isolates did the same. Likewise, 6% of isolates carried all 3 proteins, (4% and 8% of the invasive and carrier isolates, respectively). Thus, the majority of individuals immunized with a vaccine composed of these three antigens would have antibodies for at least 2 of these 3 proteins.
Figure 1. Prevalence of pcpA, psrP and pilus-1.
(A) Prevalence for pcpA, psrP and pilus-1 alone and for their combinations (isolates with al least one of the three combinations) in 458 pneumococcal isolates of patients with invasive pneumococcal disease (IPD) and in 89 pneumococcal isolates of healthy nasopharyngeal carriers. (B) Prevalence of strains that carry all three proteins, and two of possible protein combinations including pcpA and psrP, psrP and pilus-1, pcpA and pilus-1 among pneumococcal isolates of patients with IPD and healthy nasopharyngeal carriers.
Prevalence Based on Clinical Symptom and Antibiotic Resistance
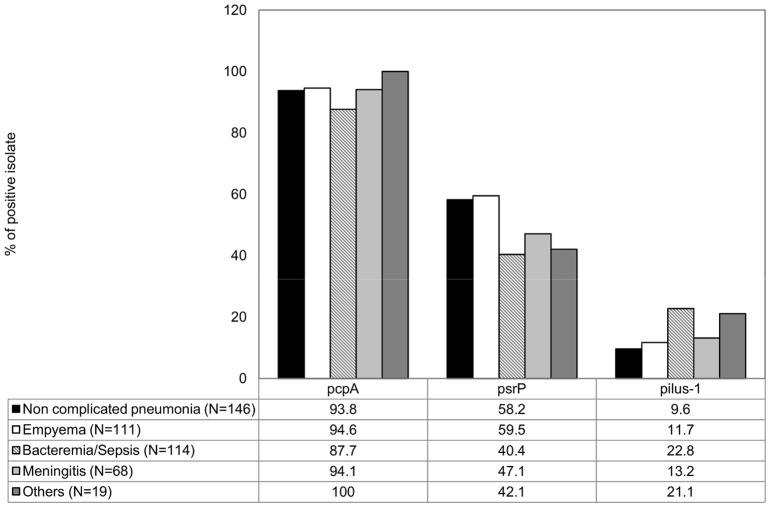
The prevalence of pcpA among all strains was too high to have any correlation with any clinical condition. In contrast, the prevalence of psrP was significantly higher in patients with non complicated pneumonia (58.2; %P<.001) or empyema (59.5%; P<.001) than in children with bacteremia (40.4%). Inversely, the prevalence of pilus-1 was greater in patients with bacteremia than in patients with non-complicated (22.8% vs. 9.6%; P = 0.005) and complicated pneumonia (11.7%; P = 0.04) (Figure 2). We also observed significant differences in the prevalence of psrP and pilus-1 according to susceptibility for different antimicrobials (Table 1). Overall psrP was significantly more frequently detected in penicillin, cefotaxime, erythromycin and tetracycline susceptible isolates while pilus-1 and, to a modest level pcpA, were more frequently detected in isolates non susceptible to these antimicrobials. In contrast, psrP was significantly more frequently detected in chloramphenicol non-susceptible isolates.
Figure 2. Prevalence of pcpA, psrP and pilus-1 according to clinical syndrome among pneumococcal invasive isolates.
Table 1. Prevalence of pcpA, psrP and pilus-1 according to antimicrobial susceptibility.
| Antimicrobial agent | pcpA | psrP | pilus-1 | |||||
| MIC | Isolates | % | %Positive | P | %Positive | P | %Positive | P |
| Penicillin | ||||||||
| ≤0.06 | 409 | 75.3 | 90.2 | <.000 | 62.6 | <.000 | 8.8 | <.001 |
| ≥0.12 | 134 | 24.7 | 100 | 17.9 | 34.3 | |||
| Cefotaxime | ||||||||
| ≤0.5 | 482 | 88.8 | 91.7 | 0.01 | 56.4 | <.000 | 9.3 | <.001 |
| ≥1 | 61 | 11.2 | 100 | 13.1 | 60.7 | |||
| Erythromicine | ||||||||
| ≤0.25 | 415 | 76.4 | 90.6 | 0.001 | 58.3 | <.000 | 10.8 | <.001 |
| ≥0.5 | 128 | 23.6 | 99.2 | 29.7 | 28.9 | |||
| Tetracycline* | ||||||||
| ≤2 | 409 | 75.9 | 91.4 | 0.07 | 58.9 | <.000 | 11.2 | 0.001 |
| ≥4 | 130 | 24.1 | 96.2 | 28.5 | 27.7 | |||
| Chloramphenicol** | ||||||||
| ≤4 | 515 | 95.2 | 92.8 | 0.4 | 50.7 | 0.02 | 15.7 | 0.09 |
| ≥8 | 26 | 4.8 | 88.5 | 73.1 | 3.8 | |||
The study was non-available in four* and six** isolates.
Prevalence of pcpA, psrP and Pilus-1 According to Serotype and Clonotype
Prevalence of these proteins was strongly associated with specific serotype and clonotypes. Table 2 shows significant differences in the prevalence of pcpA, psrP and pilus-1 according to serotype. pcpA is highly prevalent in almost all serotypes, the exception being serotype 3. pcpA was only detected in 7 of 21 isolates of serotype 3 (33.3%) vs. 500 of 526 non serotype 3 isolates (95.1%; P<.001). Interestingly, for certain serotypes the prevalence of psrP was high but occurred with an absence of pilus-1 or vice versa. For example, the prevalence of psrP among 136 strains tested of serotype 1 was 80.1% (109 isolates) but pilus-1 was not detected in any strain of serotype 1. This observation was also detected for serotype 5 where psrP was detected in 88.2% of the 34 strains but Pilus-1 was absent. In contrast, for serotypes 14 or 6B the prevalence of psrP was significantly lower than the prevalence of pilus-1 (5.3% vs. 84.2% among serotype 14 isolates (n = 19) and 35.7% vs. 57.1% among serotype 6B isolates (n = 14). Other serotypes without pilus-1 included serotype 7F (none of 36 strains) and serotype 3 (none of 21 strains). psrP was also very low in these serotypes (11.1% for serotype 7F and 9.5% for serotype 3). In fact, of all 547 strains tested, only 4.2%, tested positive for both psrP and pilus-1.
Table 2. Prevalence of pcpA,psrP and Pilus-1 according to serotype of isolates.
| Serotype | Isolates | pcpA Pos | % | psrP Pos | % | pilus Pos | % |
| Overall | 547 | 507 | 92.7 | 280 | 51.2 | 82 | 15.0 |
| 1 | 136 | 132 | 97.1 | 109 | 80.1 | 0 | 0.0 |
| 19A | 93 | 90 | 96.8 | 44 | 47.3 | 26 | 28.0 |
| 7F | 36 | 36 | 100.0 | 4 | 11.1 | 0 | 0.0 |
| 5 | 34 | 28 | 82.4 | 30 | 88.2 | 0 | 0.0 |
| 6A | 22 | 21 | 95.5 | 11 | 50.0 | 4 | 18.2 |
| 3 | 21 | 7 | 33.3 | 2 | 9.5 | 0 | 0.0 |
| 19F | 20 | 16 | 80.0 | 13 | 65.0 | 6 | 30.0 |
| 14 | 19 | 19 | 100.0 | 1 | 5.3 | 16 | 84.2 |
| 6B | 14 | 14 | 100.0 | 5 | 35.7 | 8 | 57.1 |
| 15B | 13 | 13 | 100.0 | 10 | 76.9 | 2 | 15.4 |
| 9V | 12 | 12 | 100.0 | 3 | 25.0 | 9 | 75.0 |
| 23B | 12 | 12 | 100.0 | 1 | 8.3 | 0 | 0.0 |
| 24F | 10 | 10 | 100.0 | 1 | 10.0 | 0 | 0.0 |
| 23F | 10 | 10 | 100.0 | 1 | 10.0 | 0 | 0.0 |
| 10A | 9 | 9 | 100.0 | 2 | 22.2 | 1 | 11.1 |
| 23A | 6 | 6 | 100.0 | 4 | 66.7 | 0 | 0.0 |
| 18C | 6 | 6 | 100.0 | 5 | 83.3 | 0 | 0.0 |
| 15C | 6 | 6 | 100.0 | 5 | 83.3 | 1 | 16.7 |
| 38 | 6 | 3 | 50.0 | 2 | 33.3 | 2 | 33.3 |
| 21 | 5 | 5 | 100.0 | 3 | 60.0 | 0 | 0.0 |
| 4 | 5 | 3 | 60.0 | 4 | 80.0 | 4 | 80.0 |
| 15A | 4 | 4 | 100.0 | 1 | 25.0 | 0 | 0.0 |
| 24 | 4 | 4 | 100.0 | 2 | 50.0 | 0 | 0.0 |
| 35B | 3 | 3 | 100.0 | 1 | 33.3 | 1 | 33.3 |
| 22F | 3 | 3 | 100.0 | 3 | 100.0 | 0 | 0.0 |
| 16F | 3 | 3 | 100.0 | 3 | 100.0 | 0 | 0.0 |
| 12F | 3 | 3 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 9N | 2 | 2 | 100.0 | 1 | 50.0 | 0 | 0.0 |
| 37 | 2 | 2 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 34 | 2 | 1 | 50.0 | 0 | 0.0 | 1 | 50.0 |
| 31 | 2 | 2 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 29 | 2 | 2 | 100.0 | 1 | 50.0 | 0 | 0.0 |
| 28 | 2 | 1 | 50.0 | 0 | 0.0 | 0 | 0.0 |
| 27 | 2 | 1 | 50.0 | 0 | 0.0 | 0 | 0.0 |
| 22 | 2 | 2 | 100.0 | 1 | 50.0 | 0 | 0.0 |
| 16 | 2 | 2 | 100.0 | 2 | 100.0 | 0 | 0.0 |
| 6C | 1 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 35F | 1 | 1 | 100.0 | 1 | 100.0 | 0 | 0.0 |
| 33F | 1 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 24B | 1 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 17F | 1 | 1 | 100.0 | 1 | 100.0 | 0 | 0.0 |
| 11A | 1 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 47 | 1 | 1 | 100.0 | 0 | 0.0 | 1 | 100.0 |
| 39 | 1 | 1 | 100.0 | 1 | 100.0 | 0 | 0.0 |
| 17 | 1 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 13 | 1 | 1 | 100.0 | 1 | 100.0 | 0 | 0.0 |
| 11 | 1 | 1 | 100.0 | 1 | 100.0 | 0 | 0.0 |
| 10 | 1 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 8 | 1 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 2 | 1 | 1 | 100.0 | 0 | 0.0 | 0 | 0.0 |
Pos: positive detection.
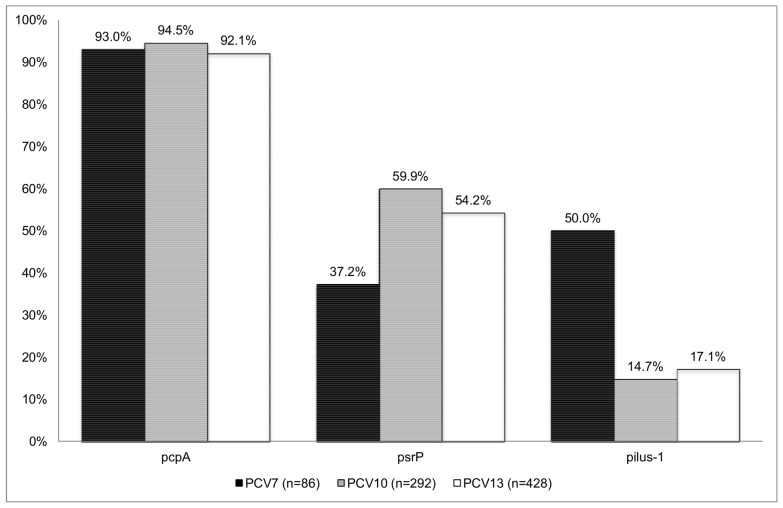
Using the designation of serotypes having high or low attack rate [21], [22] psrP was significantly more frequent in serotypes categorized as having high attack rate (those less apt to be detected in carriage than in disease) than in serotypes categorized as low attack rate (those less apt to be detected in disease than in carriage) (58.7% vs. 39.1%; P<.001). pcpA was also more frequently detected in serotypes with high attack rate (95.6% vs. 87.5%; P = 0.01). Pilus-1 distribution was similar in high and low attack rate serotypes (16.1% vs. 13.6%; P = 0.4). Considering only penicillin susceptible isolates, the prevalence of psrP between high and low attack rate serotypes was different (72.3% vs. 44.9%; P<.001). The distribution of pcpA among these susceptible isolates was also higher in high attack rate serotypes vs. low attack rate serotypes (94.3% vs. 81.9%; P = 0.01). Among penicillin susceptible isolates, the prevalence of Pilus-1 was higher in those expressing serotypes that are less apt to be detected in disease than in carriage (13.4% vs. 6.4%; P = 0.02). Figure 3 shows the prevalence of PcpA, PsrP and Pilus-1 according to serotypes within the commercialized conjugate vaccines. Pilus-1 was more frequent detected among PCV7 serotypes vs. non PCV7 serotype 50% vs. 8.5%; P<001). In contrast, psrP was more frequent detected among non PCV7 isolates vs PCV7 isolates (53.8% vs. 37.2%; P = 0.005).
Figure 3. Prevalence of pcpA, psrP and pilus-1 according to serotypes within the commercialized conjugate vaccines.
Finally, we observed stark and significant differences in prevalence of these proteins according to clonotype among isolates expressing the same serotype (Table 3). psrP was detected in almost all ST306 (106 of 109 isolates; 97.2%) while practically in none of the isolates with ST304 (1 of 22 isolates; 4.5%). Pilus-1 was totally absent in these clonotypes. The same phenomenon was observed for the penicillin susceptible clone ST1201: all isolates with this clone (n = 19) have psrP, while none have Pilus-1. The opposite was observed for multiresistant clone ST320, which all (n = 16) have pilus-1 yet lack psrP. Even in pcpA, which has a high prevalence within the entire collection, significant differences according to clonotype were detected in strains expressing the same serotype. For example, among isolates expressing serotype 3, pcpA was detected in 100% of strains with ST260, ST1220, ST1377 or ST2590 (6 isolates) while only in 6.6% of ST180 (1 of 15 isolates).
Table 3. Prevalence of pcpA. psrP and pilus-1 according to clonotypes (ST) detected in the study.
| pcpA | psrP | pilus-1 | ||||||
| ST | Isolates | Serotype | Positive | % Positive | Positive | % Positive | Positive | % Positive |
| 306 | 109 | 1 (n = 109) | 107 | 98.2 | 106 | 97.2 | 0 | 0.0 |
| 191 | 32 | 7F (n = 32) | 32 | 100.0 | 3 | 9.4 | 0 | 0.0 |
| 1223 | 25 | 5 (n = 25) | 23 | 92.0 | 21 | 84.0 | 0 | 0.0 |
| 304 | 22 | 1 (n = 22) | 22 | 100.0 | 1 | 4.5 | 0 | 0.0 |
| 1201 | 19 | 19A (n = 19) | 18 | 94.7 | 19 | 100.0 | 0 | 0.0 |
| 276 | 18 | 19A (n = 18) | 18 | 100.0 | 3 | 16.7 | 0 | 0.0 |
| 320 | 16 | 19A (n = 16) | 16 | 100.0 | 0 | 0.0 | 16 | 100.0 |
| 180 | 15 | 3 (n = 15) | 1 | 6.7 | 1 | 6.7 | 0 | 0.0 |
| 156 | 13 | 14 (n = 13) | 13 | 100.0 | 0 | 0.0 | 13 | 100.0 |
| 2013 | 13 | 19A (n = 13) | 13 | 100.0 | 2 | 15.4 | 0 | 0.0 |
| 2372 | 12 | 23B (n = 10) | 10 | 83.4 | 1 | 8.3 | 0 | 0.0 |
| 19A (n = 1) | 1 | 8.3 | 1 | 8.3 | 1 | 8.3 | ||
| 23F (n = 1) | 1 | 8.3 | 0 | 0 | 0 | 0 | ||
| 97 | 11 | 10A (n = 11) | 11 | 100.0 | 2 | 18.2 | 1 | 9.1 |
| 289 | 8 | 5 (n = 8) | 4 | 50.0 | 8 | 100.0 | 0 | 0.0 |
| 63 | 7 | 15A (n = 4) | 4 | 57.1 | 1 | 14.3 | 0 | 0.0 |
| 15B (n = 1) | 1 | 14.3 | 1 | 14.3 | 0 | 0.0 | ||
| 15C (n = 1) | 1 | 14.3 | 0 | 0.0 | 0 | 0.0 | ||
| 38 (n = 1) | 1 | 14.3 | 1 | 14.3 | 0 | 0.0 | ||
| 4677 | 6 | 24F (n = 6) | 6 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 2100 | 6 | 19F (n = 6) | 6 | 100.0 | 1 | 16.7 | 0 | 0.0 |
| 1167 | 6 | 19F (n = 5) | 1 | 16.7 | 5 | 83.3 | 4 | 66.6 |
| 19A (n = 1) | 0 | 0.0 | 1 | 16.7 | 1 | 16.7 | ||
| 838 | 6 | 9V (n = 6) | 6 | 100.0 | 0 | 0.0 | 6 | 100.0 |
| 230 | 6 | 24F (n = 3) | 3 | 50.0 | 0 | 0.0 | 0 | 0.0 |
| 24 (n = 2) | 2 | 33.3 | 0 | 0.0 | 0 | 0.0 | ||
| 24B (n = 1) | 1 | 16.7 | 0 | 0.0 | 0 | 0.0 | ||
| 202 | 6 | 19A (n = 6) | 5 | 83.3 | 2 | 33.3 | 5 | 83.3 |
| 113 | 6 | 18C (n = 6) | 6 | 100.0 | 5 | 83.3 | 0 | 0.0 |
| 199 | 5 | 19A (n = 4) | 4 | 80.0 | 4 | 80.0 | 0 | 0.0 |
| 15B (n = 1) | 1 | 20.0 | 1 | 20.0 | 0 | 0.0 | ||
| 42 | 5 | 23A (n = 5) | 5 | 100.0 | 4 | 80.0 | 0 | 0.0 |
| 1262 | 4 | 15B (n = 2) | 2 | 50.0 | 2 | 50.0 | 0 | 0.0 |
| 15C (n = 2) | 2 | 50.0 | 2 | 50.0 | 0 | 0.0 | ||
| 433 | 4 | 22 (n = 1) | 1 | 25.0 | 0 | 0.0 | 0 | 0.0 |
| 22F (n = 1) | 1 | 25.0 | 1 | 25.0 | 0 | 0.0 | ||
| 19A (n = 1) | 1 | 25.0 | 0 | 0.0 | 0 | 0.0 | ||
| 28 (n = 1) | 1 | 25.0 | 0 | 0.0 | 0 | 0.0 | ||
| 416 | 4 | 19A (n = 4) | 4 | 100.0 | 4 | 100.0 | 1 | 25.0 |
| 386 | 4 | 6B (n = 4) | 4 | 100.0 | 1 | 25.0 | 2 | 50.0 |
| 90 | 4 | 6A (n = 2) | 1 | 25.0 | 0 | 0.0 | 2 | 50.0 |
| 6B (n = 2) | 2 | 50.0 | 0 | 0.0 | 2 | 50.0 | ||
| 81 | 4 | 19A (n = 2) | 2 | 50.0 | 2 | 50.0 | 0 | 0.0 |
| 19F (n = 1) | 1 | 25.0 | 1 | 25.0 | 0 | 0.0 | ||
| 23F (n = 1) | 1 | 25.0 | 1 | 25.0 | 0 | 0.0 | ||
| 30 | 4 | 16 (n = 2) | 2 | 50.0 | 2 | 50.0 | 0 | 0.0 |
| 16F (n = 2) | 2 | 50.0 | 2 | 50.0 | 0 | 0.0 | ||
| 2690 | 3 | 29 (n = 2) | 2 | 66.7 | 1 | 33.3 | 0 | 0.0 |
| 21 (n = 1) | 1 | 33.3 | 0 | 0.0 | 0 | 0.0 | ||
| 1684 | 3 | 31 (n = 2) | 2 | 66.7 | 0 | 0.0 | 0 | 0.0 |
| 1 (n = 1) | 1 | 33.3 | 0 | 0.0 | 0 | 0.0 | ||
| 1143 | 3 | 6A (n = 3) | 3 | 100.0 | 3 | 100.0 | 1 | 33.3 |
| 310 | 3 | 38 (n = 2) | 0 | 0.0 | 0 | 0.0 | 2 | 66.7 |
| 34 (n = 1) | 0 | 0.0 | 0 | 0.0 | 1 | 33.3 | ||
| 280 | 3 | 9V (n = 2) | 2 | 66.7 | 2 | 66.7 | 0 | 0.0 |
| 9N (n = 1) | 1 | 33.3 | 1 | 33.3 | 0 | 0.0 | ||
| 224 | 3 | 6A (n = 3) | 3 | 100.0 | 0 | 0.0 | 0 | 0.0 |
| 193 | 3 | 21 (n = 2) | 2 | 66.7 | 2 | 66.7 | 0 | 0.0 |
| 15B (n = 1) | 1 | 33.3 | 1 | 33.3 | 0 | 0.0 | ||
| 101 | 3 | 15C (n = 2) | 2 | 66.7 | 2 | 66.7 | 0 | 0.0 |
| 15B (n = 1) | 1 | 33.3 | 0 | 0.0 | 0 | 0.0 | ||
| 72 | 3 | 24 (n = 2) | 2 | 66.7 | 2 | 66.7 | 0 | 0.0 |
| 24F (n = 1) | 1 | 33.3 | 1 | 33.3 | 0 | 0.0 | ||
Other ST detected with 2 isolates each: ST62, ST109, ST162, ST177, ST338, ST393, ST439, ST447, ST558, ST989, ST1011, ST1220, ST1377, ST1624, ST1692, ST2611, ST2948, ST4310, ST4828, ST5223, and ST5740.
1 isolate each: ST9, ST66, ST88, ST94, ST110, ST124, ST143, ST176, ST179, ST205, ST217, ST228, ST245, ST260, ST274, ST311, ST315, ST327, ST343, ST392, ST404, ST425, ST446, ST450, ST460, ST494, ST557, ST614, ST876, ST994, ST1012, ST1064, ST1264, ST1475, ST1504, ST1577, ST1589, ST1611, ST1664, ST1844, ST1848, ST2319, ST2333, ST2376, ST2377, ST2467, ST2557, ST2590, ST2592, ST2594, ST2595, ST2618, ST2946, ST2947, ST2949, ST3254, ST3259, ST3436, ST3437, ST3438, ST3490, ST3609, ST3787, ST4306, ST4676, ST4796, ST4826, ST4832, ST4834, ST5224, ST5741, ST5825, ST5829, ST6006, ST6040, ST6394, ST6518 and ST6519.
Discussion
Among IPD isolates, the prevalence of disease caused by serotypes included in the commercialized conjugate vaccines increased from 14.2% in PCV7 to 83.6% in PCV13. In contrast, the overall prevalence of serotypes included in PCV13 in nasopharynx was only 50.6%. Thus, even though the newly introduced PCV13 vaccine had robust coverage against disease, its intermediate coverage of the current colonizing serotypes leaves open the possibility of serotype replacement by current invasive clones or continuing serotype shift. In the same way that an indirect effect of PCV7 preventing disease in adults and non-vaccinated children had been observed [5], [6], it is expected indirect protection offered by herd immunity using multivalent pneumococcal protein vaccines [26], [27].
PcpA was highly prevalent in our collection, suggesting that it is a conserved pneumococcal component. While previous studies, including our own, have examined the prevalence of psrP or pilus-1 alone among clinical isolates [28]–[31], to our knowledge no information exists on the prevalence of pcpA. As indicated PcpA is an adhesin, and immunization with recombinant protein has been demonstrated to reduce the number of bacteria in the lungs of mice challenged with S. pneumoniae and to increase survival time in a mouse sepsis model following intraperitoneal challenge [19]. Most recently, PcpA has been shown to be required for in vitro biofilm formation [32], upregulated in response to Zn(2+) [33], and capable of eliciting antibodies during human nasopharyngeal colonization and acute otitis media [34], but not during bacteremia in infants [35]. Our finding that pcpA was present in 500 of the 526 serotypes, excluding serotype 3 isolates, underlines the importance of this protein for pneumococcal biology and strongly supports its inclusion in any protein vaccine.
Surprisingly, pcpA was only present in 7 of the 21 serotype 3 isolates tested. The absence of adhesins in serotype 3 isolates is not unprecedented; Choline binding protein A (CbpA; also known as PspC), which binds to both polymeric immunoglobulin receptor and laminin receptor, and has been implicated in biofilm formation, has a low prevalence within serotype 3 isolates [36]. Serotype 3 isolates are distinct from most other pneumococcal serotypes in that they are exceedingly encapsulated, and therefore appear highly mucoid on blood agar plates. The absence of these adhesins and a distinct clinical profile suggest that serotype 3 isolates might have a pathogenesis dissimilar to other pneumococcal isolates, as numerous studies indicate that capsular polysaccharide inhibits bacterial adhesion, and serotype 3 isolates are frequently associated with necrotizing pneumonia. This suggests that a distinct protein vaccine formulation would be required for protection against serotype 3-mediated disease. This notion is supported by studies in experimentally infected mice, where a serotype 3 clinical isolate remained in the lungs but replicated to high titers, whereas clinical isolates of serotype 2 and 4 replicated to lower titers but caused disseminated disease [37].
PsrP is both an intraspecies and interspecies adhesin, mediating attachment to Keratin 10 on lung cells and promoting the presence of bacterial aggregates in vivo and biofilm formation in vitro [38]. Pilus also functions as an adhesin, having been demonstrated to mediate attachment to laminin and may also contribute to the invasiveness of strains [39].
Importantly, considerable evidence indicates that immunization of mice with either the basic region domain of PsrP or with individual components of Pilus-1 mediates protection [16], [40]. Using Real-Time PCR, we detected psrP in 51.2% of all clinical isolates, whereas we detected pilus-1 in 15% of all isolates. This was consistent with a past study where the prevalence of psrP in clinical isolates was found to be 52.4% and with studies of numerous other investigators where the prevalence of pilus-1 in clinical isolates was found to be between 10–30% [30], [31], [41], [42].
Our study expands on these past studies by providing the prevalence of these candidate vaccine antigens simultaneously. There by assessing the potential coverage of a multivalent vaccine composed of pcpA, psrP and pilus-1. In all, 96% of the strains examined carried at least 1 of these proteins, 96% carried 2, and 6% carried all 3. Our analysis determined that psrP and pilus-1 have a negative correlation in multiple serotypes raising the possibility that psrP and pilus-1 may have redundant roles, or that their production might be metabolically expensive and that an individual strain cannot support production of both of these extremely large proteins. Briefly, PsrP is a glycosylated surface protein that separates at a molecular weight >2000 kDa, whereas Pilus-1 is primarily composed of multiple repeats of the subunit RrgB. Both extend beyond the bacterial capsule to mediate adhesion. Interestingly, our study shows that psrP was found significantly among serotypes that are less apt to be detected in carriage than in disease, while Pilus-1 was not associated with these virulent serotypes. These data could suggest that PsrP is in part responsible for the increased virulence of high attack rate serotypes. Along this line, it is known that variation in virulence exists among isolates of the same serotype, due to the contribution of serotype-independent factors associated with clonal type [43]. The variability of the prevalence of pcpA, psrP and pilus-1 according to clonal type in strains expressing the same serotype confirms that the presence of these factors appears to be a clonal property. This fact has been reported for Pilus-1 by other authors [41].
Antibiotic resistance was associated with the presence of pilus-1 and showed a negative correlation with psrP. The association of pilus-1 with antibiotic resistance has been reported previously, but the reasons for this association are not clear. It could be that the rrlA islet and specific resistance genes might be recombined together. Moschioni et al. suggest that pilus aid in adhesion during colonization of the nasopharynx and that pilus expressing strains could be selected as a result of antibiotic treatment [44]. The reason for negative association of psrP with resistant strains is unknown. Interestingly, psrP had greater correlation with strains isolated from individuals with pneumonia, both uncomplicated and complicated, whereas Pilus-1 had a predilection for strains associated with bacteremia. This observation is consistent with the known roles of PsrP as a lung cell adhesin and Pilus-1 as a mediator of invasive disease [17].
A limitation of the study is that the absence or presence of these genes/proteins is based on PCR results of wellknown and published genes [15], [18], [44] but potential primer divergence could implied that a PCR negative result is not necessary equivalent of the absence of the protein and viceversa.
In summary, our results indicate that pcpA is highly prevalent and its addition to a multivalent pneumococcal protein vaccine would result in considerable coverage. In contrast, psrP and pilus-1 have less robust individual coverage but, since psrP is present in high attack rate strains and pilus-1 in antibiotic resistant strains, could be added in an effort to reduce the likelihood of disease. The inverse correlation of these proteins suggests that they could be paired as part of a multi-valent vaccine to compensate for each other. This notion is highlighted by the fact that 96% of all strains carried pcpA and either psrP or pilus 1. Future studies are planned to determine the protective efficacy of this trivalent vaccine against invasive disease caused by multiple clinical isolates.
Acknowledgments
We thank Drs, Juan J. García-García, Iolanda Jordán, Susanna Hernandez-Bou, Asuncion Fenoll and Amadeu Gené for their contribution in taking care of patients and/or microbiological studies. We thank Pedro Brotons for statistical analysis. We also thank the availability of the public MLST database, which is located at Imperial College of London and the Catalan Study Group of Invasive Pneumococcal Disease.
Members and (centers) of the Catalan Study Group of Invasive Pneumococcal Disease are as follows: P Ciruela, S Hernandez (General Directorate of Public Health of Government of Catalonia, Barcelona); F Marco (Hospital Clinic-IDIBAPS, Barcelona); A Martinez-Roig (Hospital del Mar, Barcelona); J Gomez (Hospital del Mar, Barcelona, Hospital de Sant Celoni, Hospital de la Esperanza, Barcelona); A Díaz (Hospital de Nens, Barcelona); R Bartolomé, F Moraga (Hospital del Vall d’Hebron, Barcelona); E Palacin, JM Gairi (Institut Universitari Dexeus, Barcelona); M Sierra, P Sala (Hospital de Barcelona); M Curriu (Hospital Sant Bernabe, Berga); C Galles, A Puig, E Corrales (Hospital Sant Jaume, Calella); C Esteva, L Selva, S Hernandez-Bou, MF de Sevilla, M Iñigo, E del Amo, T Juncosa, A Gene, I Jordan, JJ Garcia-Garcia, C Muñoz-Almagro (Hospital Sant Joan de Deu, Esplugues); P Gassiot (Hospital de Figueras,Figueras); J Batlle (Hospital Josep Trueta, Girona); C Martí, L Masiques (Hospital General, Granollers); C Alonso-Tarrés (Hospital Dos de Maig y Hospital General, Hospitalet de Llobregat); M Morta, JL Lopez-Madrid (Althaia, Xarxa Asistencial, Manresa); G Sauca, L Garcia (Hospital de Mataro, Mataro); A Gassos, MJ Comesias (Hospital de Martorell, Martorell); A Gonzalez-Cuevas (Hospital de Sant Boi); E Sanfeliu (Hospital Sant Jaume, Olot); F.Ballester, I Pujol (Hospital Sant Joan, Reus); Montse Olsina, JL Arimany (Hospital General de Catalunya, Sant Cugat del Valles); F Corcoy, A Fenollosa (Hospital de Sant Camil, Sant Pere de Ribes); Xavier Raga, X Cliville (Hospital Sant Pau i Santa Tecla, Tarragona); F Gómez-Bertomeu, A Soriano (Hospital Joan XXIII, Tarragona); MO Pérez-Moreno (Hospital Verge de la Cinta,Tortosa); M Navarro, A Vilamala (Hospital de Vic, Vic).
Funding Statement
This work was supported by a grant from the Caja Navarra Foundation and by Agency for Management of University and Research Grants (AGAUR) (expedient number 2009/SGR 136). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, et al. (2009) Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374: 893–902. [DOI] [PubMed] [Google Scholar]
- 2. Obando I, Muñoz-Almagro C, Arroyo LA, Tarrago D, Sanchez-Tatay D, et al. (2008) Pediatric parapneumonic empyema, Spain. Emerg Infect Dis 14: 1390–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li ST, Tancredi DJ (2010) Empyema hospitalizations increased in US children despite pneumococcal conjugate vaccine. Pediatrics125: 26–33. [DOI] [PubMed] [Google Scholar]
- 4. Byington CL, Korgenski K, Daly J, Ampofo K, Pavia A, et al. (2006) Impact of the pneumococcal conjugate vaccine on pneumococcal parapneumonic empyema. Pediatr Infect Dis J 25: 250–254. [DOI] [PubMed] [Google Scholar]
- 5. Pilishvili T, Lexau C, Farley MM, Hadler J, Harrison LH, et al. (2010) Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis 201: 32–41. [DOI] [PubMed] [Google Scholar]
- 6. Pulido M, Sorvillo F (2010) Declining invasive pneumococcal disease mortality in the United States, 1990–2005. Vaccine 28: 889–892. [DOI] [PubMed] [Google Scholar]
- 7. Eastham KM, Freeman R, Kearns AM, Eltringham G, Clark J, et al. (2004) Clinical features, aetiology and outcome of empyema in children in the north east of England. Thorax 59: 522–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Muñoz-Almagro C, Jordan I, Gene A, Latorre C, Garcia-Garcia JJ, et al. (2008) Emergence of invasive pneumococcal disease caused by non-vaccine serotypes in the era of 7-valent conjugate vaccine. Clin Infect Dis 46: 174–182. [DOI] [PubMed] [Google Scholar]
- 9. Ardanuy C, Tubau F, Pallares R, Calatayud L, Domínguez MA, et al. (2009) Epidemiology of invasive pneumococcal disease among adult patients in Barcelona before and after pediatric 7-valent pneumococcal conjugate vaccine introduction, 1997–2007. Clin Infect Dis 48: 57–64. [DOI] [PubMed] [Google Scholar]
- 10. Tarragó D, Aguilar L, García R, Gimenez MJ, Granizo JJ, et al. (2011) Evolution of clonal and susceptibility profiles of serotype 19A Streptococcus pneumoniae among invasive isolates from children in Spain, 1990 to 2008. Antimicrob Agents Chemother 55: 2297–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grijalva CG, Poehling KA, Nuorti JP, Zhu Y, Martin SW, et al. (2006) National impact of universal childhood immunization with pneumococcal conjugate vaccine on outpatient medical care visits in the United States. Pediatrics 118: 865–873. [DOI] [PubMed] [Google Scholar]
- 12. Huang SS, Johnson KM, Ray GT, Wroe P, Lieu TA, et al. (2011) Healthcare utilization and cost of pneumococcal disease in the United States. Vaccine 29: 3398–3412. [DOI] [PubMed] [Google Scholar]
- 13.Pneumonia. Strategy overview. Bill and Melinda Gates Foundation. Available: http://www.gatesfoundation.org/global-health/Documents/pneumonia-strategy.pdf Accessed 2011 Dec 1..
- 14.Pneumococcal vaccine support. Gavi Alliance. Available: http://www.gavialliance.org/support/nvs/pneumococcal/. Accessed 2011 Dec 1..
- 15. Obert C, Sublett J, Kaushal D, Hinojosa E, Barton T, et al. (2006) Identification of a candidate Streptococcus pneumoniae core genome and regions of diversity correlated with invasive pneumococcal disease. Infect Immun 74: 4766–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rose L, Shivshankar P, Hinojosa E, Rodriguez A, Sanchez CJ, et al. (2008) Antibodies against PsrP, a novel Streptococcus pneumoniae adhesin, block adhesion and protect mice against pneumococcal challenge. J Infect Dis198: 375–383. [DOI] [PubMed] [Google Scholar]
- 17. Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, et al. (2006) A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A 103: 2857–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sánchez-Beato AR, López R, García JL (1998) Molecular characterization of PcpA: a novel choline-binding protein of Streptococcus pneumoniae . FEMS Microbiol Lett 164: 207–214. [DOI] [PubMed] [Google Scholar]
- 19. Glover DT, Hollingshead SK, Briles DE (2008) Streptococcus pneumoniae Surface Protein PcpA Elicits Protection against Lung Infection and Fatal Sepsis. Infect and Immunity 76: 2767–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tarrago D, Fenoll A, Sanchez-Tatay D, Arroyo LA, Muñoz-Almagro C, et al. (2008) Identification of pneumococcal serotypes from culture-negative clinical specimens by novel Real-Time PCR. Clin Microbiol Infect 14: 828–834. [DOI] [PubMed] [Google Scholar]
- 21. Brueggemann AB, Griffiths DT, Meats E, Peto T, Crook DW, et al. (2003) Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J Infect Dis 187: 1424–1432. [DOI] [PubMed] [Google Scholar]
- 22. Sleeman KL, Griffiths D, Shackley F (2006) Capsular serotype-specific attack rates and duration of carriage of Streptococcus pneumoniae in a population of children. J Infect Dis 194: 682–688. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards (NCCLS) (2008) Performance Standards for antimicrobial susceptibility testing: Eighteenth informational supplement. CLSI document M100-S18 (ISBN 1-5-56238-653-0) Clinical and laboratory standard institute. Wayne Pa.
- 24. Enright MC, Spratt BG (1998) A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144: 3049–3060. [DOI] [PubMed] [Google Scholar]
- 25. Muñoz-Almagro C, Gala S, Selva L, Jordan I, Tarragó D, et al. (2011) DNA bacterial load in children and adolescents with pneumococcal pneumonia and empyema. Eur J Clin Microbiol Infect Dis 30: 327–335. [DOI] [PubMed] [Google Scholar]
- 26. Briles DE, Ades E, Paton JC, Sampson JS, Carlone GM, et al. (2000) Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae . Infect Immun 68: 796–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tai SS (2006) Streptococcus pneumoniae Protein Vaccine Candidates: Properties, Activities and Animal Studies. Crit Rev Microbiol 32: 139–53. [DOI] [PubMed] [Google Scholar]
- 28. Muñoz-Almagro C, Selva L, Sanchez CJ, Esteva C, de Sevilla MF, et al. (2010) PsrP, a protective pneumococcal antigen, is highly prevalent in children with pneumonia and is strongly associated with clonal type. Clin Vaccine Immunol 17: 1672–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Imai S, Ito Y, Ishida T, Hirai T, Ito I, et al. (2011) Distribution and clonal relationship of cell surface virulence genes among Streptococcus pneumoniae isolates in Japan. Clin Microbiol Infect 17: 1409–1414. [DOI] [PubMed] [Google Scholar]
- 30. Vainio A, Kaijalainen T, Hakanen AJ, Virolainen A (2011) Prevalence of pilus-encoding islets and clonality of pneumococcal isolates from children with acute otitis media. Eur J Clin Microbiol Infect Dis 30: 515–519. [DOI] [PubMed] [Google Scholar]
- 31. Moschioni M, De Angelis G, Melchiorre S, Masignani V, Leibovitz E, et al. (2010) Prevalence of pilus-encoding islets among acute otitis media Streptococcus pneumoniae isolates from Israel. Clin Microbiol Infect 16: 1501–1504. [DOI] [PubMed] [Google Scholar]
- 32. Moscoso M, García E, López R (2006) Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol 188: 7785–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kloosterman TG, Witwicki RM, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP (2008) Opposite effects of Mn2+ and Zn2+ on PsaR-mediated expression of the virulence genes pcpA, prtA, and psaBCA of Streptococcus pneumoniae . J Bacteriol 190: 5382–5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kaur R, Casey JR, Pichichero ME (2011) Serum antibody response to five Streptococcus pneumoniae proteins during acute otitis media in otitis-prone and non-otitis-prone children. Pediatr Infect Dis J 30: 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hagerman A, Posfay-Barbe KM, Grillet S, Ochs MM, Brookes RH, et al. (2011) Failure to elicit seroresponses to pneumococcal surface proteins (pneumococcal histidine triad D, pneumococcal choline-binding protein A, and serine proteinase precursor A) in children with pneumococcal bacteraemia. Clin Microbiol Infect. In press. doi: 10.1111/j.1469–0691.2011.03629.x. [DOI] [PubMed] [Google Scholar]
- 36. Brooks-Walter A, Briles DE, Hollingshead SK (1999) The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect Immun 67: 6533–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Orihuela CJ, Gao G, McGee M, Yu J, Francis KP, et al. (2003) Organ-specific models of Streptococcus pneumoniae disease. Scand J Infect Dis 35: 647–652. [DOI] [PubMed] [Google Scholar]
- 38. Sanchez CJ, Shivshankar P, Stol K, Trakhtenbroit S, Sullam PM, et al. (2010) The pneumococcal serine-rich repeat protein is an intra-species bacterial adhesin that promotes bacterial aggregation in vivo and in biofilms. PLoS Pathog 6: e1001044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hilleringmann M, Giusti F, Baudner BC, Masignani V, Covacci A, et al. (2008) Pneumococcal pili are composed of protofilaments exposing adhesive clusters of Rrg A. PLoS Pathog. 4: e1000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Harfouche C, Filippini S, Gianfaldoni C, Ruggiero P, Moschioni M, et al. (2012) RrgB321, a fusion protein of the three variants of the pneumococcal pilus backbone RrgB, is protective in vivo and elicits opsonic antibodies. Infect Immun 80: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aguiar SI, Serrano I, Pinto FR, Melo-Cristino J, Ramirez M (2008) The presence of the pilus locus is a clonal property among pneumococcal invasive isolates. BMC Microbiol 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Regev-Yochay G, Hanage WP, Trzcinski K, Rifas-Shiman SL, Lee G, et al. (2010) Re-emergence of the type 1 pilus among Streptococcus pneumoniae isolates in Massachusetts, USA. Vaccine 28: 4842–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harvey RM, Stroeher UH, Ogunniyi AD, Smith-Vaughan HC, Leach AJ, et al. (2011) A variable region within the genome of Streptococcus pneumoniae contributes to strain-strain variation in virulence. PLoS One 6: e19650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Moschioni M, Donati C, Muzzi A, Masignani V, Censini S, et al. (2008) Streptococcus pneumoniae contains 3 rlrA pilus variants that are clonally related. J Infect Dis197: 888–896. [DOI] [PubMed] [Google Scholar]