Abstract
In mice and humans, IGF2 has multiple promoters to maintain its complex tissue- and developmental stage-specific imprinting and expression. IGF2 is also imprinted in marsupials, but little is known about its promoter region. In this study, three IGF2 transcripts were isolated from placental and liver samples of the tammar wallaby, Macropus eugenii. Each transcript contained a unique 5' untranslated region, orthologous to the non-coding exons derived from promoters P1–P3 in the human and mouse IGF2 locus. The expression of tammar IGF2 was predominantly from the P2 promoter, similar to humans. Expression of IGF2 was higher in pouch young than in the adult and imprinting was highly tissue and developmental-stage specific. Interestingly, while IGF2 was expressed throughout the placenta, imprinting seemed to be restricted to the vascular, trilaminar region. In addition, IGF2 was monoallelically expressed in the adult mammary gland while in the liver it switched from monoalleleic expression in the pouch young to biallelic in the adult. These data suggest a complex mode of IGF2 regulation in marsupials as seen in eutherian mammals. The conservation of the IGF2 promoters suggests they originated before the divergence of marsupials and eutherians, and have been selectively maintained for at least 160 million years.
Introduction
Insulin-like growth factor 2 (IGF2) is an important regulator of growth, metabolism and differentiation and was the first imprinted gene identified in eutherians and in marsupials [1], [2]. The coding region of IGF2 is highly conserved across all mammals, but the non-coding exons are variable. Human IGF2 has five promoters (huP1 and P0–P3) and transcription start sites (TSS) each adjoining distinct 5′ non-coding exons, while rodent IGF2 has only 4 (P0–P3) [3]–[12]. Preliminary investigations in the South American marsupial, Monodelphis domestica, detected a single product from total neonatal RNA using 5′ RACE and identified only one 5′ non-coding exon [13]. Thus, opossum neonates appear to produce only one IGF2 isotype from a single promoter.
Human IGF2 is imprinted in the fetal liver and is paternally expressed from P1, P2 and P3 (orthologous to mouse P1–P3 respectively) with dominant expression from the P2 promoter in the majority of tissues tested [14]. In the adult liver IGF2 has continued expression from P1 and P3 in addition to biallelic expression from huP1 (previously reported as P1) [14], [15]. There is no functional mouse homologue for huP1 which is located 20 kb upstream of P1–P3. In the human, reciprocal activation patterns of P2 and huP1 before and after birth respectively have been correlated to developmental-specific methylation at the two promoters [16].
In the mouse P0-derived transcripts are only expressed in the labyrinthine layer of the placenta, whereas transcripts from P1–P3 are found throughout the developing embryo and placenta with dominant expression from the P3 promoter [17]–[21]. Mouse Igf2 is paternally expressed in the embryo, but biallelically expressed in the choroid plexus and leptomeniges, which are the only major sites of Igf2 gene expression in the adult rodents [1], [22]–[24]. The human IGF2-P0 transcript is paternally expressed at high levels in fetal skeletal muscle, in the term placenta and at low levels in all adult tissues except the brain [12].
The mouse Igf2 domain has 3 differentially methylated regions (DMRs), DMR0 surrounding the area that codes for the placental specific promoter P0, DMR1 located 5′ of P1 and DMR2 located at the second coding exon [12], [25]. In the mouse, both DMR 1 and 2 are paternally hypermethylated. DMR1 is a mesodermal silencer. Therefore, deletion of this region results in biallelic expression of IGF2 in mesoderm-derived tissues (heart, kidney and lung) without disrupting H19 imprinting, whilst maintaining IGF2 imprinting in the endoderm-derived tissues (liver and placenta) [26]. Deletion of DMR2 has no effect on imprinting but reduces transcriptional activation of IGF2, therefore DMR2 is an activation site [27]. Humans lack DMR1 [18], but DMR2 is present and paternally hypermethylated [28]. In addition methylation of this region varies within tissues of patients with Beckwith-Wiedemann syndrome, suggesting a similar function to that seen in the mouse [28]. In humans, the promoter-specific methylation of P1–P3 spreads from the silenced maternal allele to the transcriptionally active paternal allele [29]. The increase in methylation on the paternal allele with age potentially regulates the level of expression from each promoter. This promoter specific methylation pattern is not seen in the mouse [30].
The presence of a potential functional equivalent of mouse DMR2 [28] and DMR0 [12] and the absence of DMR1 [18] in the human, coupled with the occurrence of parental allele-specific methylation in the promoter regions of the human, but not the mouse [29], [30], suggests a divergence of IGF2 control mechanisms amongst the eutherian mammals. Marsupials diverged from the eutherian mammals around 160 million years ago [31], yet IGF2 is highly conserved and imprinted. However, very little is known about the regulation of this gene in marsupials. Most marsupials have a short pregnancy supported by a much less invasive placenta than typically seen in eutherian mammals, and all give birth to a highly altricial neonate [32]. Therefore, conserved promoter elements between marsupials and eutherians are expected to be critical in the function and regulation of the IGF2 gene.
IGF2 is imprinted in the whole bodies of opossum neonates [2], [13]. In the tammar wallaby, IGF2 shows complete imprinting in the fetal liver, but has paternally biased expression in the placenta and brain and is biallelically expressed adult liver [33]–[35]. There is a 95 base pair non-coding exon upstream of the first protein coding exon of IGF2 in the neonatal opossum [13]. In monotemes only a single transcript has been characterized with no non-coding exons [36]. In the tammar, there are several different 5′ sequences produced from the IGF2 gene that may account for the biased expression observed in the head and the placenta [34]. However, as yet no one has identified the transcription start sites (TSSs) of tammar IGF2. This study investigates the evolution of the IGF2 regulatory regions in a marsupial, the tammar wallaby Macropus eugenii, and identified 3 promoters in the tammar IGF2 region.
Materials and Methods
Ethics Statement
All experiments and wild animal collection were approved by the University of Melbourne Animal Experimentation Ethics Committee and the animal handling and husbandry procedures were in accordance with the National Health and Medical Research Council of Australia (2004) guidelines. Animals were collected under approval from the South Australian Department of Environment and Natural Resources.
Animals
Tammar wallabies of Kangaroo Island, South Australia origin were held in our breeding colony in Melbourne. Pregnancy was initiated in females carrying an embryo in diapause by the removal of their pouch young (RPY) [37], [38]. Adult females carrying fetuses in the final third of gestation (day 19 to day 26 RPY of the 26.5 day pregnancy) or pouch young (day 0-day 350 post-partum) were either collected in the wild, or from our colony. The lactating mammary gland and adult and pouch young tissues were collected immediately after death and snap frozen in liquid nitrogen.
RNA and DNA Extraction and RT-PCR
Genomic DNA (gDNA) was extracted from approximately 20 mg of snap frozen tissue using a Wizard Genomic DNA Purification Kit (Promega). Total RNA was extracted from liver, placenta and brain tissues using Tri-Reagent (Ambion) as described by the manufacturer, and from mammary glands using RNeasy Lipid Tissue Mini Kit (QIAGEN), with a final elution of RNA in 50–80 µl of RNAsecure H2O (Ambion, Geneworks) in a dilution of 1/24. Total RNA was DNase treated (DNA-freeTM, Ambion) to remove contaminating DNA and tested by performing a housekeeping gene PCR. Total RNA quality was assessed by running samples on a gel (100%, 100 V for 30 min) and quantified with a nano-spectrometer (NanaDrop ND-1000 Spectrophotometer, NanoDrop Technologies Inc, Wilmington, DE, USA). cDNA was synthesised using Roche Transcriptor High Fidelity cDNA Synthesis Kit (Roche). Typically 4000 ng of total RNA was used in each cDNA synthesis reaction, with 1.1 µl of Oligo (dT)18 (50 µM), trilaminar yolk sac samples for qPCR analysis were amplified using random hexamer primers. All cDNA samples were diluted to 40 uL for a final concentration of 100 ug/ul. cDNA integrity was immediately assessed with GAPDH PCR.
All primers were designed using Primer3 (v. 0.4.0) [39] and synthesised by Sigma-Aldrich; (Table S1).
5′Race Amplification
To identify tammar IGF2 transcription start sites (TSS) we performed 5′RACE, using both the 5′ RACE System for Rapid amplification of cDNA ends, Version 2.0, (Invitrogen) and SMARTer RACE cDNA Amplification Kit (Clontech). Liver and placental RNA was used. Gene specific primers were designed in the first translated exon (Table S1) and were used in conjunction with the provided Abridged Anchor Primer to amplify the 5′ end of the transcript.
PCR products were subcloned in to the pGEM-T Easy vector and transformed in to JM109 competent cells (Promega). Plasmids were purified using the Wizard Plus SV Minipreps DNA Purification System (Promega) and sequences were obtained according to standard methods using ABI3130xl capillary genetic analysers, with BDV3.1 terminators.
2.5 Quantitative RT-PCR Analysis
Quantitative real-time polymerase chain reaction (qPCR) was used to quantify the relative expression from each of the IGF2 transcripts in the brain, liver, mammary gland and placenta. Each sample was extracted and prepared as described above. In the brain expression profile, 1 female and 3 male samples were used in the day 10 pouch young (PY) group and all 3 of the day 50 PY group were male. In the liver, 1 female and 3 male samples were used in the day 10 PY group and 2 female and 2 male samples were used in the day 75 PY group. All of the adult samples were female. As there were no obvious outliers in the data, we decided it was acceptable to use both sexes.
IGF2 qPCR primers were designed so the reverse primer encompassed the intron exon boundary between exon 2 and 3 and was used with the transcript specific primers (Table S1).
PCRs were carried out in triplicates in 20-µl volumes consisting of FastStart Universal SYBR Green Master (Rox) (Roche), forward and reverse IGF2 primers of an optimal concentration between 300–500 nM, and 1 µl cDNA template. Real-time PCR was carried out in an Stratagene Mx3000P™ Sequence Detector (integrated sciences) using the following conditions: 95°C for 10 min, followed by 45 cycles at 95°C for 15 sec and 61°C for 30 min and 72°C for 30 sec. A liver sample triplicate and a negative template triplicate were included on each plate as a calibrator and negative control respectively for each primer combination.
In addition to using the dissociation curve, PCR products from the first plate were run on a gel to confirm amplification of a single band and that the NTC wells were blank. The data was analysed in Microsoft Excel and R statistical package [40]. The amplification efficiency was calculated from the standard curve and Ct values corrected [41]. Paired t-tests were used in addition to one-way analysis of variance (ANOVA) and Tukey’s pair wise comparisons, to determine which results were significant. Means were considered different if P≤0.05.
Imprinting and Expression Analysis
To determine the presence or absence of IGF2 allelic expression sample specific genotypes were identified using PCR and direct sequencing of gDNA. Two single nucleotide polymorphisms [34] (Figure 1) were analysed by direct sequencing using common IGF2 primers for gDNA and transcript specific primers for cDNA (Table S1) similar PCR conditions were used for each sample.
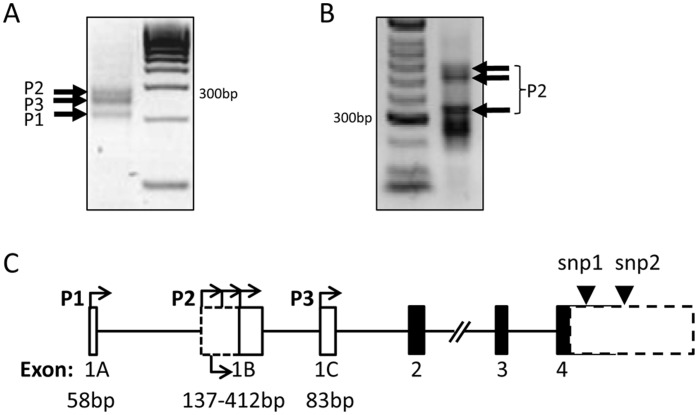
Figure 1. Identifying tammar IGF2 transcription start sites.
5′-RACE amplification was performed on (A) liver and (B) placenta RNA samples. (A) Three bands first identified in the liver (black arrows) were visible in both tissue types. (B) Additional exon 1B transcription start sites (TSS) were identified in the placenta. (C) Aligning the cloned products to the gDNA sequence identified the location of each of the non-coding exons 1A, 1B and 1C (white boxes) in relation to the coding exons 2, 3 and 4 (black boxes) followed by an untranslated region of unknown length (dashed box). TSSs are indicated by turned arrows. Two single nucleotide polymorphisms (black triangles) identified in the untranslated region were used for imprint analysis (snp1 and snp2). Diagram is not to scale.
Each PCR reaction contained approximately 4 ng/µL of template and 0.2 µM of each primer. For imprinting analysis Ex Taq DNA polymerase, Hot-Start Version (TaKaRa) was used to optimise reaction efficiency and only 30–35 cycles of PCR were used to prevent saturation. For expression analysis GoTaq Green Master Mix (Promega) was used. PCR cycles consisted of 94 or 96°C for 2 min, followed by a maximum of 35 cycles of 30 sec at 94 or 96°C, 30 sec-1 min at 58–63°C, and 30 sec-1 min at 72°C, and a final extension at 72°C for 5 min.
Approximately 9 pouch young, 13 adults and 17 placental samples were screened for IGF2 snps. Of these samples 2 adult livers, 3 mammary glands, 4 placentas and 2 pouch young brains and livers were analysed for IGF2 imprinting. PCR products from cDNA and gDNA were resolved by gel electrophoreses and bands extracted (QIAquick Gel Extraction Kit, QIAGEN). The purified product was then sequenced. Sequences were assessed by eye using the FinchTV (v.1.3.1) DNA sequence chromatogram trace viewer software. The relative peak height for each allele indicates biallelic (equal peak heights) or imprinted (unequal peak heights) expression.
Methylation Analysis
Using MethPrimer [42] several CpG sites were identified upstream of each of the non-coding exons. 1 ug of DNA was treated with a sodium bisulphite solution at 50°C for 4 hours before ethanol precipitating and eluting in 50 µl of TE. Approximately 20 ng of DNA was used as a template with 0.2 µM of each bisulphite (B/S) primer (Table S1) in a 25 µl reaction. To reduce the effects of PCR and cloning biases Takara Ex Taq DNA Polymerase (Hot-Start Version) was used to increase specificity, prevent background amplification, give higher yields and reduce sequence errors. PCR products generated with TaKaRa Ex Taq HS contain a mixture of 3'-A overhangs and blunt ends which allows >80% cloning efficiency in T-vectors. PCR cycles consisted of 96°C for 1 min, followed by 35 cycles of 30 sec at 96°C, 30 sec at 60°C, and 20 sec at 72°C, and a final extension at 72°C for 5 min. PCR products were cloned as described above and sequences were analysed using Quma quantification tool for methylation analysis [43].
Results and Discussion
The regulation of the imprinted IGF2 gene is highly complex in eutherian mammals. This study confirmed that marsupials also have complex regulation of IGF2, and identified three evolutionarily conserved promoters, each with three distinct non-coding exons that showed tissue specific imprinting and expression in the tammar wallaby.
Identification of Three Transcription Start Sites in the Tammar Wallaby IGF2 Locus
To determine how many different transcription start sites (TSS) there are in the tammar IGF2, 5′ RACE was performed on mRNA derived from two tissues, liver and placenta. Three distinct bands were amplified from the liver and placental samples, representing three distinct TSS (Figure 1A). As additional faint bands were detected in placenta, 5′ RACE was repeated with a more sensitive kit (Clontech) (Figure 1B). Each band was cloned, sequenced and aligned to the genomic sequence. Three non coding exons were identified and named 1A, 1B and 1C each associated with a separate promoter P1, P2 and P3 respectively (Figure 1C). Three additional P2 TSS were detected in the placenta. Therefore, tammar IGF2 may not have a liver specific promoter orthologous to human HuP1 or a placenta specific promoter orthologous to mouse P0.
Evolutionary Conservation of Three IGF2 Alternate Promoters
At the mouse and rat P1 there are no TATA, CAT or GC boxes, but there are direct and inverted repeats of various length in the region upstream of the P1 non-coding exon [17], [44]. Similarly, no TATA-box, initiator element (Inr) or downstream promoter element (DPE) were located around tammar exon 1A TSS, although some repeat sequences were identified (Figure S1A). Tammar P2 had multiple transcription start sites, and had multiple CCATT and GC-box/Sp1-sites. Both a TATA-box and an Inr element was identified around the forth P2 TSS (Figure S1B) and these elements are predicted to act synergistically since the distance between the TATA-box and Inr is between 25–30 bp [45]. Tammar P3 is predicted to be a Inr-DPE core promoter as the DPE was located at precisely +28 to +32 relative to the A+1 nucleotide in the Inr motif (where A+1 is the transcription start site) [46]–[48] (Figure S1C). A TATA-like sequence at -27 relative to the P3 A+1 nucleotide in the Inr motif, similar to the TATA-box observed at the mouse P3 [10], may have lost its consensus in marsupials after their divergence from the marsupial-eutherian ancestor. The conserved core promoter elements along with the arrangement of the three promoters suggests tammar promoters P1–P3 are orthologous to the human and mouse P1–P3.
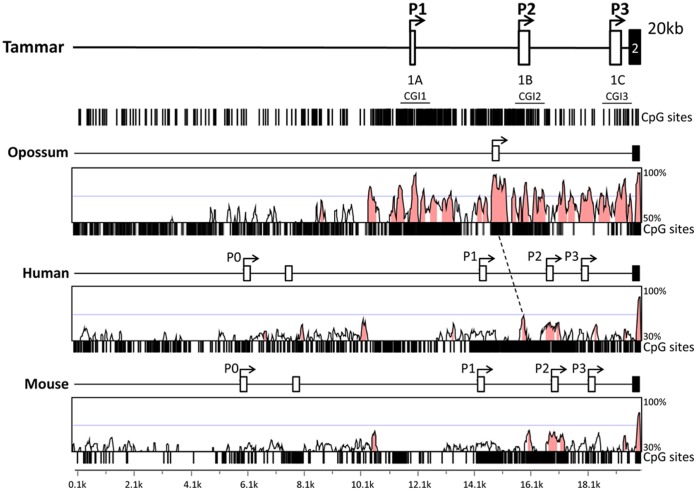
Using 20 kb genomic sequences of IGF2, pairwise alignments using a 100 bp sliding window were performed in VISTA [49] between tammar (clone MEKBa-346C2∶42205-62204) and the base genome of mouse (ch7∶149834890-149854889) human (Ch11∶2149386-2169385), and opossum (clone XX-223O16 ch5.102112- 821130). Peaks (pink) represent areas with over 70% sequence conservation between tammar and opossum and over 55% between tammar and human and mouse (Figure 2). A minimum conservation width of 100 bp was used for alignments between marsupial species and 50 bp for alignments between eutherians and marsupials. Relative exon location was conserved between species with dense CpG sites around the three non-coding exons (Figure 2). There was a highly conserved peak between P1 and P2 in both eutherians and marsupials, suggesting that this was the ancestral arrangement of the three promoters and non-coding exons. As further evidence, the tammar alternate promoter derived non-coding exons were highly conserved in the opossum. However, there was limited sequence similarity between the tammar, mouse and human non-coding exons. The opossum, (family Didelphidae) and the tammar (family Macropodidae) are predicted to have diverged approximately 80 million years ago, whereas marsupials and eutherians last shared a common ancestor over 160 million years ago [31]. As these exons are non-coding, sequence changes in these exons will not change the function of the peptide. Therefore, it was expected that there would be far less sequence conservation between the marsupial and eutherian species than between the two marsupial species.
Figure 2. Genomic structure of tammar IGF2 including 3 TSS and comparison with mouse, human and opossum genomes.
Opossum (clone XX-223O16 ch5∶102112-82113), human (ch11∶2156597-2176597) and mouse (ch7∶149841559-149861559), IGF2 base genomes were aligned to tammar IGF2 (clone MEKBa-346C2∶62540-82539). VISTA pairwise alignments using a 100 bp sliding window was performed between each species and tammar. Pink peaks represent areas of conservation of 70% over a minimum of 100 bp between tammar and opossum and 55% over a minimum of 50 bp between tammar and human or mouse. A dotted line indicates a region with 70% conservation between P1 and P2 in both marsupials and eutherians. A schematic of each IGF2 gene is located above the VISTA plot to show relative location of the non-coding exons (open box), including the non-coding exons transcribed with the P0 TSS [12], and coding exons (black box). Location of CpG sites are represented by vertical black bars below the VISTA plot.
The opossum non-coding exon was highly conserved in the tammar genome and aligned to the tammar with 92% sequence identity approximately 300 bp upstream of the first tammar exon 1B TSS (Figure S2). This suggests that there may be another unidentified non-coding exon upstream of tammar exon 1B. There are no non-coding exons identified in monotreme IGF2 [36]. Therefore, the additional promoters and exons must have arisen after the therian-monotreme divergence. The high level of sequence conservation of the tammar exons 1A, 1B and 1C in the opossum sequence suggests that these non-coding exons and promoters existed in the therian common ancestor before the eutherian - marsupial divergence and that the opossums may have lost the function of these promoters, or they are yet to be identified.
Conserved Expression Patterns from Alternative IGF2 Promoters
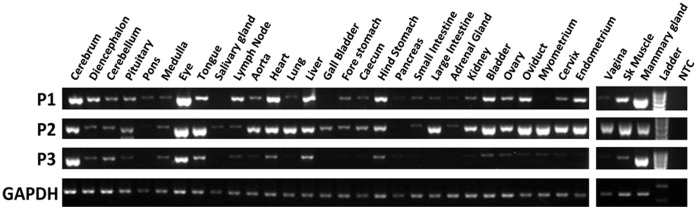
IGF2 is synthesized in most tissues and its production declines rapidly in rodents soon after birth [19], [50]. In humans, circulating plasma levels of IGF2 remain at relatively high concentrations throughout life [51]. However, total IGF2 transcription in the liver peaks shortly after birth [52]. Expression of IGF2 in the tammar is similarly developmental stage-specific; expression is higher in the pouch young, during the time of rapid growth and nutrient transfer from the mother, compared to both fetal and adult stages [53]. Transcription from P1 and P2 appeared to be expressed in almost all of the adult tissues tested while expression from P3 was predominantly seen in the brain, mammary gland and eye (Figure 3). As in the human and in contrast to the mouse [14], [54], [55], transcription in the tammar was higher from P2 compared to P1 and P3 in the pouch young liver (P<0.005) and brain (P<0.05) (Figure 4A & 4B), in the adult mammary gland (P<0.05) and in the bilaminar and trilaminar regions of the placenta (P<0.01) (Figure 4C, 4D & 4E).
Figure 3. IGF2 mRNA expression in the adult tammar.
RT-PCR expression profile after 35 cycles of each IGF2 mRNA transcript in a range of adult tammar tissues compared to GAPDH. The expression was strongest from P2, with expression of all three transcripts in the brain, eye, tongue, liver and mammary gland. No expression was detected in the No Template Control (NTC).
Figure 4. IGF2 mRNA expression from each promoter (P1, P2 and P3) relative to a reference gene in various wallaby tissues.
(A) IGF2 mRNA expression relative to GAPDH in the Liver, (B) IGF2 mRNA expression relative to GAPDH in the brain, (C) IGF2 mRNA expression relative to B-ACTIN in the mammary gland, (D) IGF2 mRNA expression relative to B-ACTIN in the bilaminar yolk sac and (E) IGF2 mRNA expression relative to 18S in the trilaminar yolk Sac. (A, B) Expression from P2 in the pouch young (PY) is significantly higher than expression from P1 and P3 (* P<0.05) in the liver (A) and brain (B). Except for P1 in the liver, IGF2 is expressed at a higher level in the pouch young then in the adult (a, P<0.01) in both the liver and the brain (if adult hypothalamus expression is representative of total adult brain expression). (C, D, E) P2-IGF2 is also the predominantly expressed transcript in the mammary gland (* P<0.05) (C) and in the bilaminar (D) and trilaminar (E) placenta (a,b,c have significantly different means, P<0.01). There was no difference in the expression levels of any of the transcripts between non-lactating and lactating mammary glands (C).
In the mouse, hepatic IGF2 mRNA peaks approximately 10 days after birth [19]. In the tammar, hepatic IGF2 expression peaks in the male pouch young between 70 and 100 days after birth and in females around 150 days [53]. As expected, expression in the tammar also varied with stage. In liver, P2 and P3 had significantly higher expression in the pouch young compared to the adult (P<0.01), but there was no significant difference in expression from P1 (Figure 4A). Expression of IGF2 in liver was predominantly monoallelic from each of the 3 promoters in pouch young aged 12 and 72 days, and biallelic in both adults tested (Figure 5). As H19 is not expressed in the adult liver, it is likely that the differentially methylated CTCF binding sites act as the imprinting control region in marsupials as it does in eutherians [33]. This epigenetic mark may be relaxed in the liver as the young grows, possibly by increased methylation on the maternal allele, allowing H19 downstream enhancers to activate maternal IGF2 promoters.
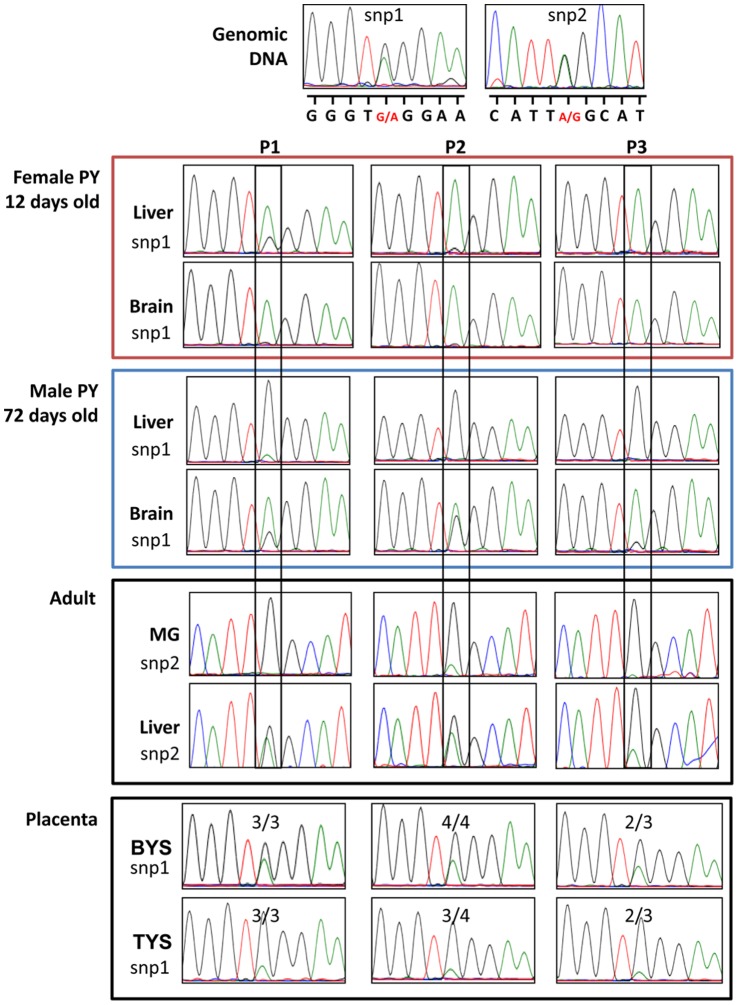
Figure 5. Allelic expression from the three promoters in various adult and pouch young tissues.
Direct sequencing of two single nucleotide polymorphisms (snp1 and snp2) was used to determine the allelic expression of each of the IGF2 transcripts in adult and pouch young (PY) tissues. In a 12 day old female pouch young monoallelic expression was detected from all 3 promoters in the brain and liver, except P1 in the liver was expressed from both alleles. In a 72 day old male pouch young, biased to biallelic expression was detected in the brain while the liver expression was similar to the day 12 pouch young. In the adult, both alleles were detected from all three TSS in two liver samples and monoallelic expression was detected in three mammary gland (MG) samples. Placental samples had biallelic expression in the bilaminar region (BYS) in all samples tested for P1 and P2 and 2 of the 3 samples tested for P3. Biased to monoalleleic expression was observed in the trilaminar region (TYS) in all samples tested for P1, 3 of the 4 samples for P2, and 2 of the 3 samples for P3.
In the human overall IGF2 mRNA expression is higher in the fetal brain compared to the adult [55], [56]. In tammar, expression from all three promoters in the adult hypothalamus was significantly lower than in the whole pouch young brain (P<0.01) (Figure 4B). IGF2 RT-PCR amplification was detected in several regions within the brain including the cerebral cortex, cerebellum, diencephalon and pituitary (Figure 3), similar to the expression in the human brain [55], [57], [58]. In rodents, expression in the choroid plexus and leptomeniges are the only major sites of Igf2 gene expression in the adult. In both humans and rodents, IGF2 is biallelically expressed in a region-specific manner in the brain [1], [54], [55], [59]. As in the liver, imprinting in the tammar brain may be developmental stage-specific. Expression was monoallelic in the day 12 female PY but in the day 72 male there was expression from both alleles (Figure 5). This could suggest that the imprinting mark is relaxed with age or, as in eutherians, some regions in the brain may maintain monoallelic expression while other regions, such as the choroid plexus, may express IGF2 biallelically. In the tammar pouch young liver, IGF2 expression levels vary between males and females with a significant difference at day 70 post partum [53]. Thus, another possibility, although less likely, is that IGF2 imprinting in the brain is sex specific, as observed at the Interleukin 18 locus in the mouse [60].
There was no significant variation in the expression of the three different transcripts in the mammary glands between early and mid lactation (Figure 4C). Both P1 and P3 were monoallelically expressed while P2 showed some expression from the silenced allele (Figure 5). All three mammary glands tested produced the same result. If imprinting is maintained in organs that influence maternal nutrient transfer, monoallelic expression of IGF2 may be a selective advantage in the adult mammary gland [35], but not in the liver.
Imprinting in the Placenta
The pattern of expression from the three promoters appeared to be conserved in both the trilaminar and bilaminar regions of the chorio-vitelline placenta with the highest expression from P2 compared to both P1 and P3. The imprint status was analysed in 4 different placentas, aged between days 21–26 of pregnancy. Interestingly, the expression of the three IGF2 promoters in the vascular trilaminar region was predominantly from one allele while both alleles were expressed in the bilaminar yolk sac placenta (Figure 5).
Expression of IGF2 is significantly higher in the trilaminar yolk sac compared to the bilaminar [61]. The avascular bilaminar yolk sac serves primarily in the uptake of nutrients from the uterine secretions, while the vascular trilaminar yolk sac is important for respiration [62], [63]. IGF2 has been implicated in several aspects of placental development, including blood vessel formation [64] and gaseous transfer [20], [21], [65]. Therefore, monoallelic expression of IGF2 may be selectively advantageous for vascular growth specifically in the vascular trilaminar placenta.
The tammar promoters P1–P3 were imprinted in the placenta and pouch young with selectively maintained imprinted expression in adult tissues, including the mammary gland. In mouse and human, these three promoters are mainly expressed in the fetus and placenta with expression levels dropping off, and in some cases becoming biallelically expressed, in the adult [1], [14], [15]. In the tammar, the duration of development of the young in the pouch is equivalent to fetal stages of eutherians, so the tammar IGF2 promoters would be expected to have similar expression patterns and imprint patterns in the pouch young as they do in the eutherian fetus. However, expression was maintained in the adult tammar, albeit at lower levels, suggesting that the regulation of these promoters is more similar to that in humans than in the rodents.
DNA Methylation in the Promoter Regions of Tammar IGF2
Promoter specific methylation in adult humans increases with age as expression levels decrease [16], [29]. In the tammar, CpG islands (CGI) were located around each of the transcription start sites as well as across exon 3 (CGI4, the location of eutherian DMR2) (Figure 6). While it is still unknown whether DNA methylation at promoters is the primary cause or consequence of gene silencing, the expression of tammar P2-IGF2 significantly decreased in the adult liver in absence of increased TSS methylation (Figure 6). Therefore, it is unlikely that methylation at the P2 TSS controls the expression levels in the liver. The presence of multiple Sp1 sites (Figure S1) may explain the lack of methylation observed at this promoter. Deletion or mutation of mouse Spl sites within the unmethylated Aprt CpG island causes de novo methylation [66], [67]. However, the region between P1 and P2 is very CpG rich (Figure 6) so methylation further upstream of the P2 may regulate expression. The P3 TSS was highly methylated without any obvious allelic bias in young or adult liver and mammary gland (Figure 6). As P3-IGF2 transcript number is usually lower in the adult tissues relative to P1- and P2- IGF2 expression, methylation may function to regulate the expression levels at this promoter.
Figure 6. Methylation at the tammar IGF2 transcription start sites.
Location of CpG sites are represented by black bars below the schematic of the tammar IGF2 gene. Bisulphite sequencing of the IGF2 CpG islands (CGI) at each of the transcription start sites (CGI1, CGI2 and CGI3) and at exon 3 (CGI4) in the pouch young and adult liver, in the mammary gland, whole yolk sac placenta and in the bilaminar and trilaminar regions of the yolk sac placenta. Black circles represent a methylated CpG site and open circles represent an unmethylated CpG site. Each row represents the methylation pattern of a separate DNA fragment from the same sample. Only CGI3 and CGI4 were significantly methylated in all tissues. CGI4 appears to be differentially methylated in the placenta. However, sequence polymorphisms that might distinguish parental copies of CGI4 were not found among the available stocks of tammar placental samples.
In the mouse, methylation at DMR1 and DMR2 on the expressed paternal allele correlates significantly with the level of overall IGF2 expression [19]. Differential methylation was not detected at CGI4, in either the mammary gland or liver and a similar level of methylation was detected at the P3 TSS (Figure 6). There was, however, a possible DMR in the placenta at the CGI4, with a higher level of methylation in the trilmainar region compared to the bilaminar region (Figure 6). This suggests that methylation at this site may regulate either the expression levels and/or the imprint status of IGF2 in the placenta. Sequence polymorphisms that might distinguish parental copies of CGI4 were not found among the available samples. However, the presence of hypo- and hyper- methylated alleles is indicative of a DMR. In eutherians, the DMR2 is predicted to be an enhancer [27], so in the tammar this region may also be a tissue or cell specific activation site.
Conclusion
The expression and imprinting of IGF2 is controlled in a developmental- and tissue-specific manner in both eutherians and marsupials. One differentially methylated TSS has previously been reported in the opossum [13]. While not differentially methylated, at least three promoters appear to be functional in the tammar. The conserved core-promoter elements in addition to the conserved sequence between P1 and P2 in both eutherians and marsupials suggest these promoters were present in the eutherian-marsupial ancestor. The expression pattern of these three transcripts mirrors that which is seen in humans, with predominant expression from the P2 promoter and continued expression into adulthood, despite their differences in reproductive strategies. Interestingly, unlike the human and opossum, tammar IGF2 lacks promoter specific DMRs, similar to that which has been reported in the mouse [29], [30]. While marsupial IGF2 imprinting is likely to be regulated by the H19 ICR, as in eutherians [33], the differences in the promoter methylation patterns between human, mouse, opossum and tammar demonstrate divergence in some epigenetic mechanisms. However, the existence of three orthologous promoters and a putative DMR across exon 3 in the tammar, human and mouse provides evidence of a conserved origin of imprinting mechanisms for this gene between marsupials and eutherians.
Supporting Information
Nucleotide sequence of the three tammar IGF2 gene promoters. The DNA sequences of IGF2 promoters P1, P2 and P3 are illustrated in A, B, and C, respectively. Transcription start sites (TSSs) identified by 5′RACE are indicated by the arrows. ggaggag repeats are in boxes, CTCCCAAG repeats are highlighted in yellow and TGTCCC repeats are highlighted in green. TATA-boxs (TATAWAAR) are emboldened and red, CCATT and GC-box/Sp1-sites are underlined, Inr elements (YYANWYY) are emboldened and blue and DPE (RGWYV) sequences are emboldened and green. Exon sequences are in capitals. P1 is characterised by multiple repeat sequences. P2 has multiple TSSs and a high number of CCATT and GC-box/Sp1-sites. A TATA-Inr core promoter was identified around the fourth TSS. P3 is an Inr-DPE core promoter. Degenerate nucleotides represented using IUPAC codes.
(PDF)
Alignment of tammar exon 1B region with opossum exon 1 region. Opossum non-coding exon = 1–420 Tammar exon 1B = 725–1104. Alignments were performed using ClustalW and were highlighted using BOXshade 3.31.
(PDF)
Primers used in this study.
(PDF)
Acknowledgments
We thank Alison Bradfield and Scott Brownlees for assistance with the animals. We thank Helen Clark and Bonnie Dopheide for technical assistance, Takashi Kohda for the use of his Pearl script to draw the CpG density maps and all the members of the Tammar Research Group for their help in collecting tissue.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Australian Research Council (ARC) Centre of Excellence in Kangaroo Genomics; an ARC Federation Fellowship to MBR and a National Health and Medical Research Council R.D. Wright Fellowship to AP and the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad to SS. JMS received an Australian Postgraduate Award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 2.O’Neill MJ, Ingram RS, Vrana PB, Tilghman SM. Allelic expression of IGF2 in marsupials and birds. Dev Genes Evol. 2000;210:18–20. doi: 10.1007/pl00008182. [DOI] [PubMed] [Google Scholar]
- 3.Bell GI, Gerhard DS, Fong NM, Sanchez-Pescador R, Rall LB. Isolation of the human insulin-like growth factor genes: insulin-like growth factor II and insulin genes are contiguous. Proceedings of the National Academy of Sciences of the United States of America. 1985;82:6450–6454. doi: 10.1073/pnas.82.19.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Irminger JC, Rosen KM, Humbel RE, Villa-Komaroff L. Tissue-specific expression of insulin-like growth factor II mRNAs with distinct 5' untranslated regions. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:6330–6334. doi: 10.1073/pnas.84.18.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Pagter-Holthuizen P, Hoppener JWM, Jansen M. Chromosomal localization and preliminary characterization of the human gene encoding insulin-like growth factor II. Human Genetics. 1985;69:170–173. doi: 10.1007/BF00293291. [DOI] [PubMed] [Google Scholar]
- 6.de Pagter-Holthuizen P, Jansen M, van der Kammen RA, van Schaik FMA, Sussenbach JS. Differential expression of the human insulin-like growth factor II gene Characterization of the IGF-II mRNAs and an mRNA encoding a putative IGF-II-associated protein. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1988;950:282–295. doi: 10.1016/0167-4781(88)90124-8. [DOI] [PubMed] [Google Scholar]
- 7.de Pagter-Holthuizen P, Jansen M, van Schaik FMA, van der Kammen R, Oosterwijk C, et al. The human insulin-like growth factor II gene contains two development-specific promoters. FEBS Letters. 1987;214:259–264. doi: 10.1016/0014-5793(87)80066-2. [DOI] [PubMed] [Google Scholar]
- 8.de Pagter-Holthuizen P, van Schaik FMA, Verduijn GM, van Ommen GJB, Bouma BN, et al. Organization of the human genes for insulin-like growth factors I and II. FEBS Letters. 1986;195:179–184. doi: 10.1016/0014-5793(86)80156-9. [DOI] [PubMed] [Google Scholar]
- 9.Holthuizen P, van der Lee FM, Ikejiri K, Yamamoto M, Sussenbach JS. Identification and initial characterization of a fourth leader exon and promoter of the human IGF-II gene. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1990;1087:341–343. doi: 10.1016/0167-4781(90)90010-y. [DOI] [PubMed] [Google Scholar]
- 10.Rotwein P, Hall LJ. Evolution of Insulin-Like Growth Factor II: Characterization of the Mouse IGF-II Gene and Identification of Two Pseudo-Exons. DNA and Cell Biology. 1990;9:725–735. doi: 10.1089/dna.1990.9.725. [DOI] [PubMed] [Google Scholar]
- 11.Vu TH, Hoffman AR. Promoter-specific imprinting of the human insulin-like growth factor-II gene. Nature. 1994;371:714–717. doi: 10.1038/371714a0. [DOI] [PubMed] [Google Scholar]
- 12.Monk D, Sanches R, Arnaud P, Apostolidou S, Hills FA, et al. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum Mol Genet. 2006;15:1259–1269. doi: 10.1093/hmg/ddl041. [DOI] [PubMed] [Google Scholar]
- 13.Lawton B, Carone B, Obergfell C, Ferreri G, Gondolphi C, et al. Genomic imprinting of IGF2 in marsupials is methylation dependent. BMC Genomics. 2008;9:205. doi: 10.1186/1471-2164-9-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohlsson R, Hedborg F, Holmgren L, Walsh C, Ekstrom TJ. Overlapping patterns of IGF2 and H19 expression during human development: biallelic IGF2 expression correlates with a lack of H19 expression. Development. 1994;120:361–368. doi: 10.1242/dev.120.2.361. [DOI] [PubMed] [Google Scholar]
- 15.Ekstrom TJ, Cui H, Li X, Ohlsson R. Promoter-specific IGF2 imprinting status and its plasticity during human liver development. Development. 1995;121:309–316. doi: 10.1242/dev.121.2.309. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Gray SG, Flam F, Pietsch T, Ekstrm TJ. Developmental-dependent DNA methylation of the IGF2 and H19 promoters is correlated to the promoter activities in human liver development. The International journal of developmental biology. 1998;42:687–693. [PubMed] [Google Scholar]
- 17.Ikejiri K, Furuichi M, Ueno T, Matsuguchi T, Takahashi K, et al. The presence and active transcription of three independent leader exons in the mouse insulin-like growth factor II gene. Biochimica et biophysica acta. 1991;1089:77. doi: 10.1016/0167-4781(91)90087-3. [DOI] [PubMed] [Google Scholar]
- 18.Moore T, Constancia M, Zubair M, Bailleul B, Feil R, et al. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber M, Milligan L, Delalbre A, Antoine E, Brunel C, et al. Extensive tissue-specific variation of allelic methylation in the Igf2 gene during mouse fetal development: relation to expression and imprinting. Mechanisms of Development. 2001;101:133–141. doi: 10.1016/s0925-4773(00)00573-6. [DOI] [PubMed] [Google Scholar]
- 20.Constancia M, Hemberger M, Hughes J, Dean W, Ferguson-Smith A, et al. Placental-specific IGF-II is a major modulator of placental and fetal growth. Nature. 2002;417:945–948. doi: 10.1038/nature00819. [DOI] [PubMed] [Google Scholar]
- 21.Sibley CP, Coan PM, Ferguson-Smith AC, Dean W, Hughes J, et al. Placental-specific insulin-like growth factor 2 (Igf2) regulates the diffusional exchange characteristics of the mouse placenta. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8204–8208. doi: 10.1073/pnas.0402508101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soares MB, Turken A, Ishii D, Mills L, Episkopou V, et al. Rat insulin-like growth factor II gene : A single gene with two promoters expressing a multitranscript family. Journal of Molecular Biology. 1986;192:737–752. doi: 10.1016/0022-2836(86)90025-2. [DOI] [PubMed] [Google Scholar]
- 23.Gray A, Tam AW, Dull TJ, Hayflick J, Pintar J, et al. Tissue-Specific and Developmentally Regulated Transcription of the Insulin-Like Growth Factor 2 Gene. DNA. 1987;6:283–295. doi: 10.1089/dna.1987.6.283. [DOI] [PubMed] [Google Scholar]
- 24.Stylianopoulou F, Herbert J, Soares MB, Efstratiadis A. Expression of the insulin-like growth factor II gene in the choroid plexus and the leptomeninges of the adult rat central nervous system. Proceedings of the National Academy of Sciences. 1988;85:141–145. doi: 10.1073/pnas.85.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feil R, Walter J, Allen ND, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994;120:2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- 26.Constancia M, Dean W, Lopes S, Moore T, Kelsey G, et al. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat Genet. 2000;26:203–206. doi: 10.1038/79930. [DOI] [PubMed] [Google Scholar]
- 27.Murrell A, Heeson S, Bowden L, Constância M, Dean W, et al. An intragenic methylated region in the imprinted Igf2 gene augments transcription. EMBO reports. 2001;2:1101–1106. doi: 10.1093/embo-reports/kve248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneid H, Seurin D, Vazquez MP, Gourmelen M, Cabrol S, et al. Parental allele specific methylation of the human insulin-like growth factor II gene and Beckwith-Wiedemann syndrome. Journal of Medical Genetics. 1993;30:353–362. doi: 10.1136/jmg.30.5.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Issa JP, Vertino PM, Boehm CD, Newsham IF, Baylin SB. Switch from monoallelic to biallelic human IGF2 promoter methylation during aging and carcinogenesis. Proceedings of the National Academy of Sciences. 1996;93:11757–11762. doi: 10.1073/pnas.93.21.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki H, Jones PA, Chaillet JR, Ferguson-Smith AC, Barton SC, et al. Parental imprinting: potentially active chromatin of the repressed maternal allele of the mouse insulin-like growth factor II (Igf2) gene. Genes & Development. 1992;6:1843–1856. doi: 10.1101/gad.6.10.1843. [DOI] [PubMed] [Google Scholar]
- 31.Luo Z-X, Yuan C-X, Meng Q-J, Ji Q. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature. 2011;476:442–445. doi: 10.1038/nature10291. [DOI] [PubMed] [Google Scholar]
- 32.Renfree MB. Review: Marsupials: Placental Mammals with a Difference. Placenta. 2010;31:S21–S26. doi: 10.1016/j.placenta.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 33.Smits G, Mungall AJ, Griffiths-Jones S, Smith P, Beury D, et al. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet. 2008;40:971–976. doi: 10.1038/ng.168. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki S, Renfree MB, Pask AJ, Shaw G, Kobayashi S, et al. Genomic imprinting of IGF2, p57 (KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mech Dev. 2005;122:213–222. doi: 10.1016/j.mod.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 35.Stringer JM, Suzuki S, Pask AJ, Shaw G, Renfree MB. Selected imprinting of INS in the marsupial. Epigenetics & Chromatin In Press. 2012. [DOI] [PMC free article] [PubMed]
- 36.Killian JK, Nolan CM, Stewart N, Munday BL, Andersen NA, et al. Monotreme IGF2 expression and ancestral origin of genomic imprinting. J Exp Zool. 2001;291:205–212. doi: 10.1002/jez.1070. [DOI] [PubMed] [Google Scholar]
- 37.Renfree MB, Tyndale-Biscoe H. Intrauterine development after diapause in the marsupial Macropus eugenii. Developmental Biology. 1973;32:28–40. doi: 10.1016/0012-1606(73)90217-0. [DOI] [PubMed] [Google Scholar]
- 38.Tyndale-Biscoe H, Renfree MB. Monographs on Marsupial Biology: Reproductive physiology of marsupials: Cambridge University Press. 1987.
- 39.Rozen S, Skaletsky HJ. Primer3. Bioinformatics Methods and Protocols: Methods in Molecular Biology. 2000. pp. 365–386. [DOI] [PubMed]
- 40.R_Development_Core_Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. 2011.
- 41.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L-C, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 43.Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Research. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueno T, Takahashi K, Matsuguchi T, Endo H, Yamamoto M. A new leader exon identified in the rat insulin-like growth factor II gene. Biochemical and Biophysical Research Communications. 1987;148:344–349. doi: 10.1016/0006-291x(87)91116-8. [DOI] [PubMed] [Google Scholar]
- 45.O’shea-greenfield A, Smale ST. Roles of TATA and Initiator elements in determining the start site location and direction of RNA Polymerase-II transcription. Journal of Biological Chemistry. 1992;267:1391–1402. [PubMed] [Google Scholar]
- 46.Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 47.Smale ST, Kadonaga JT. The RNA Polymerase II core promoter. Annual Review of Biochemistry. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 48.Kutach AK, Kadonaga JT. The Downstream Promoter Element DPE Appears To Be as Widely Used as the TATA Box in Drosophila Core Promoters. Molecular and Cellular Biology. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frazer KA PL, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res(Web Server issue) W273–9. 2004. [DOI] [PMC free article] [PubMed]
- 50.Brown AL, Graham DE, Nissley SP, Hill DJ, Strain AJ, et al. Developmental regulation of insulin-like growth factor II mRNA in different rat tissues. Journal of Biological Chemistry. 1986;261:13144–13150. [PubMed] [Google Scholar]
- 51.Nissley SP, Rechler MM. Insulin-like growth factors: biosynthesis, receptors and carrier proteins. Hormones, Proteins and Peptides. 1986;12:127–203. [Google Scholar]
- 52.Li X, Cui H, Sandstedt B, Nordlinder H, Larsson E, et al. Expression levels of the insulin-like growth factor-II gene (IGF2) in the human liver: developmental relationships of the four promoters. Journal of Endocrinology. 1996;149:117–124. doi: 10.1677/joe.0.1490117. [DOI] [PubMed] [Google Scholar]
- 53.Menzies BR, Shaw G, Fletcher TP, Pask AJ, Renfree MB. Maturation of the growth axis in marsupials occurs gradually during post-natal life and over an equivalent developmental stage relative to eutherian species. Molecular and Cellular Endocrinology. 2012;349:189–194. doi: 10.1016/j.mce.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 54.Pedone PV, Cosma MP, Ungaro P, Colantuoni V, Bruni CB, et al. Parental imprinting of rat insulin-like growth factor II gene promoters is coordinately regulated. Journal of Biological Chemistry. 1994;269:23970–23975. [PubMed] [Google Scholar]
- 55.Pham NV, Nguyen MT, Hu J-F, Vu TH, Hoffman AR. Dissociation of IGF2 and H19 imprinting in human brain. Brain Research. 1998;810:1–8. doi: 10.1016/s0006-8993(98)00783-5. [DOI] [PubMed] [Google Scholar]
- 56.Sandberg A-C, Engberg C, Lake M, von Holst H, Sara VR. The expression of insulin-like growth factor I and insulin-like growth factor II genes in the human fetal and adult brain and in glioma. Neuroscience Letters. 1988;93:114–119. doi: 10.1016/0304-3940(88)90022-5. [DOI] [PubMed] [Google Scholar]
- 57.Haselbacher GK, Schwab ME, Pasi A, Humbel RE. Insulin-like growth factor II (IGF II) in human brain: regional distribution of IGF II and of higher molecular mass forms. Proceedings of the National Academy of Sciences. 1985;82:2153–2157. doi: 10.1073/pnas.82.7.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hartmann W, Koch A, Brune H, Waha A, Schuller U, et al. Insulin-Like Growth Factor II Is Involved in the Proliferation Control of Medulloblastoma and Its Cerebellar Precursor Cells. Am J Pathol. 2005;166:1153–1162. doi: 10.1016/S0002-9440(10)62335-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu JF, Vu TH, Hoffman AR. Differential biallelic activation of three insulin-like growth factor II promoters in the mouse central nervous system. Molecular Endocrinology. 1995;9:628–636. doi: 10.1210/mend.9.5.7565809. [DOI] [PubMed] [Google Scholar]
- 60.Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-Specific Parent-of-Origin Allelic Expression in the Mouse Brain. Science. 2010;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ager E, Pask A, Shaw G, Renfree MB. Expression and protein localisation of IGF2 in the marsupial placenta. BMC Developmental Biology. 2008;8:17. doi: 10.1186/1471-213X-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renfree MB. The composition of fetal fluids of the marsupial Macropus eugenii. Developmental Biology. 1973;33:62–79. doi: 10.1016/0012-1606(73)90165-6. [DOI] [PubMed] [Google Scholar]
- 63.Freyer C, Renfree MB. The mammalian yolk sac placenta. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2009;312B:545–554. doi: 10.1002/jez.b.21239. [DOI] [PubMed] [Google Scholar]
- 64.Han VKM, Carter AM. Spatial and Temporal Patterns of Expression of Messenger RNA for Insulin-Like Growth Factors and their Binding Proteins in the Placenta of Man and Laboratory Animals. Placenta. 2000;21:289–305. doi: 10.1053/plac.1999.0498. [DOI] [PubMed] [Google Scholar]
- 65.Lopez MF, Dikkes P, Zurakowski D, Villa-Komaroff L. Insulin-like growth factor II affects the appearance and glycogen content of glycogen cells in the murine placenta. Endocrinology. 1996;137:2100–2108. doi: 10.1210/endo.137.5.8612553. [DOI] [PubMed] [Google Scholar]
- 66.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes & Development. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 67.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, et al. Spl elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide sequence of the three tammar IGF2 gene promoters. The DNA sequences of IGF2 promoters P1, P2 and P3 are illustrated in A, B, and C, respectively. Transcription start sites (TSSs) identified by 5′RACE are indicated by the arrows. ggaggag repeats are in boxes, CTCCCAAG repeats are highlighted in yellow and TGTCCC repeats are highlighted in green. TATA-boxs (TATAWAAR) are emboldened and red, CCATT and GC-box/Sp1-sites are underlined, Inr elements (YYANWYY) are emboldened and blue and DPE (RGWYV) sequences are emboldened and green. Exon sequences are in capitals. P1 is characterised by multiple repeat sequences. P2 has multiple TSSs and a high number of CCATT and GC-box/Sp1-sites. A TATA-Inr core promoter was identified around the fourth TSS. P3 is an Inr-DPE core promoter. Degenerate nucleotides represented using IUPAC codes.
(PDF)
Alignment of tammar exon 1B region with opossum exon 1 region. Opossum non-coding exon = 1–420 Tammar exon 1B = 725–1104. Alignments were performed using ClustalW and were highlighted using BOXshade 3.31.
(PDF)
Primers used in this study.
(PDF)