Abstract
Synthetic biology is an area of biological research that combines science and engineering. Here, I merge the principles of synthetic biology and regulatory evolution to create a new species with a minimal set of known elements. Using preexisting transgenes and recessive mutations of Drosophila melanogaster, a transgenic population arises with small eyes and a different venation pattern that fulfils the criteria of a new species according to Mayr’s Biological Species Concept. The population described here is the first transgenic organism that cannot hybridize with the original wild type population but remains fertile when crossed with other identical transgenic animals. I therefore propose the term “synthetic species” to distinguish it from “natural species”, not only because it has been created by genetic manipulation, but also because it may never be able to survive outside the laboratory environment. The use of genetic engineering to design artificial species barriers could help us understand natural speciation and may have practical applications. For instance, the transition from transgenic organisms towards synthetic species could constitute a safety mechanism to avoid the hybridization of genetically modified animals with wild type populations, preserving biodiversity.
Introduction
It has been argued that reconstructing a system is the ultimate way of understanding it [1], [2]. In order to further comprehend the origin of new species, and to explore possible applications in modern biotechnology, I engineered reproductive isolation between populations of Drosophila melanogaster by generating a synthetic species boundary.
The use of Drosophila is justified because this model organism is leading the fields of regulatory evolution and speciation [3], [11]. For example, previous work in several species of Drosophila produced fundamental contributions regarding the genetics of speciation [7]–[10]. In addition, key studies of evolution in Drosophila have shown that novelty arises more readily from the recruitment of existing elements into new regulatory networks than from the development of completely new components [3]–[6].
However, speciation has been given no attention as a tool for biotechnology and in the context of genetically engineered organisms. Previous artificial speciation experiments produced “incipient species” that were not fully isolated or whose speciation genes were unknown [12]–[16]. Reproductive isolation has been brought about in plants for many decades through polyploidization, which creates individuals that in crosses to parents give rise to sterile progeny [13], and, more recently, in yeast [14]. For the animal kingdom, a parthenogenetic species of lizard was also generated [15], but, again, its speciation genetics are not understood, does not involve transgenesis nor synthetic design and its components can therefore not be reliably and predictably manipulated.
Unlike previous artificial speciation events, the synthetic species boundary described here is a genetic circuit based on the combination of 5 well known preexisting elements leading to reproductive isolation. Building components so well understood that can be reliably and predictably manipulated is one of the goals of synthetic biology. In this case it allows opening and closing speciation gates when desired.
Results
Regulatory evolution acts by using available preexisting genetic elements to generate novelty. Likewise, the synthetic genotype created here is achieved by implementing known elements, however their selection and specific arrangement establishes a previously unknown synthetic species barrier (Fig. 1).
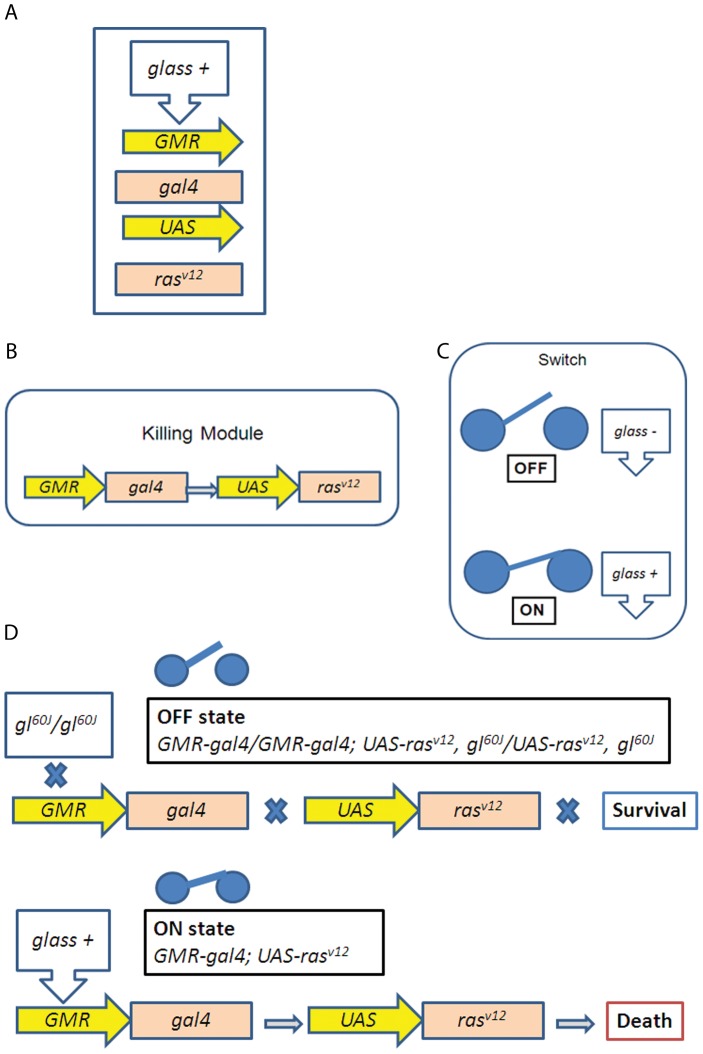
Figure 1. Design of a genetic circuit with selected components that form a synthetic species barrier.
(A) The 5 genetic elements used: transcription factor glass, enhancer GMR, transcription factor gal4, enhancer UAS and a constitutively activated form of ras. (B,C) Arrangement of the genetic elements in two modules, a killing module (B) composed by two independent transgenes, GMR-gal4 and UAS-rasv12,and a switch that depending on the presence or absence of the transcription factor glass can switch the killing module ON and OFF (C). (D) In the absence of Glass, activation of the killing module is not possible and the flies survive. However, in the presence of Glass, expression of the constitutively active form of ras kills the animal.
The first element consist of null mutations in the glass (gl) gene [19], [20]. The glass product is a transcription factor of 604 amino acids with five zinc-fingers. Mutations in gl specifically abolish photoreceptor cells resulting in blind, but viable flies [19], [20]. The gl60J allele is a spontaneous mutant caused by the insertion of 30 kb of unknown DNA into the gl locus and it is believed to be a null allele [19], [20]. Other alleles (gl3 and glBS1) have also been used for this study.
The second element is formed by the Glass Multimer Reporter (GMR) [20], [21], a heterologous promoter construct containing five tandem copies of a 27-bp glass-binding site normally present in the regulatory region of ninaE, the major rhodopsin gene in Drosophila. The GMR promoter can therefore drive glass-dependent expression in the photoreceptor cells of Drosophila eyes.
The yeast protein GAL4 as a third building block can activate transcription in Drosophila from promoters that bear GAL4 binding sites [22], [23]. In addition, the GMR sequence has been previously subcloned in front of gal4, thus driving Gal4 expression under the control of Glass (GMR-gal4) [21].
Fourth, a tandem array of five GAL4 binding sites (5×UAS, for Upstream Activation Sequence) is employed where GAL4 binds with high affinity to induce the transcription of a downstream located gene.
The fifth element is a rasv12 allele, a mutant form of the Drosophila ras gene [24], [25]. Conversion of the glycine residue at position 12 to valine constitutively activates the Ras protein. rasv12 has been previously subcloned behind GAL4 binding sites (UAS-rasv12), which permits activation only within cells where GAL4 is expressed [24].
The inherent logic of the design relies on a “killing module” (Fig. 1b) and a regulator to switch it ON and OFF (Fig. 1c):
The killing module is formed by GMR-gal4 and UAS-rasv12 whose activation is controlled by the presence or absence of the gene glass. When glass gene function is unperturbed, transcription of UAS-rasv12 driven by GMR-gal4 consistently kills 100% of the flies at any temperature from 17°C until 29°C (606/606 lethality at 17°C, 558/558 lethality at 23°C, 330/330 at 25°C, 110/110 at 29°C, Table 1). Pupae arrest at mid pupation and due to abnormal tissue lysis the pupal case ends up almost empty.
Table 1. Crosses between Drosophila synthetica and Drosophila melanogaster.
| Parental genotypes | F1 adult progeny | Number of dead pupae |
| ♂ GMR-gal4/GMR-gal4♀ UAS-rasv12/UAS-rasv12 | 0 (no survivors at any temperature from17°C to 29°C) | 606/606 lethality at 17°C 558/558 lethality at 23°C 330/330 lethality at 25°C 110/110 lethality at 29°C |
| ♂ GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/UAS-rasv12, gl60J♀ GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/UAS-rasv12, gl60J | >1000 at 17°C >1000 at 25°C | 0 at 17°C 20/100 at 25°C |
| ♂ GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/UAS-rasv12, gl60J♀ Oregon R | 0 (no survivors) | 293/293 lethality at 17°C |
| ♂ Oregon R♀ GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/UAS-rasv12, gl60J | 0 (no survivors) | 50/50 lethality at 17°C |
| ♂ & ♀ w; GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/ UAS-rasv12, gl60J (genotype 1)♂ & ♀ w/w (genotype 2) | >1000 of genotype 1>1000 of genotype 2 0hybrids (red normally sized eyes) | >1000 at 17°C |
| ♂ GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/UAS-rasv12, gl60J♀ tub-gal80/tub-gal80 | 71 at 25°C | 0 at 25°C |
| ♂ GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/UAS-rasv12, gl60J♀ gl60J/gl60J | 87 at 25°C | 0 at 25°C |
| ♂ & ♀ w; GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/ UAS-rasv12, gl60J (genotype 1)♂ & ♀ y,w/y,w (genotype 2) | 99 of genotype 1 165 of genotype 2 0hybrids (red normally sized eyes) | 140 at 17°C |
| ♂ & ♀ w; GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/ UAS-rasv12, gl60J (genotype 1)♂ & ♀ y,w,f/y,w,f (genotype 2) | 30 of genotype 1 11 of genotype 2 0hybrids (red normally sized eyes) | 24 at 17°C |
More than 20 other UAS transgenes were tested, including UAS-caudal [26], UAS-flowerLoseA [27] or UAS-eiger [27], but UAS-rasv12 was the only one that resulted in 100% lethality when driven by the presence of glass and the GMR-gal4 transgene at all temperatures (from 17°C to 29°C) (Table 1).
In the synthetic genotype GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/UAS-rasv12, gl60J (a glass60J mutant background, where no Glass protein is present) Rasv12 cannot be produced (OFF state, Fig. 1d). Surprisingly however, in addition to the small eye phenotype (Fig. 2a,b), those flies showed a different wing morphology, with lateral extra veins (Fig 2d, compare with the wt wing pattern shown in 2c). Other alleles (gl3 and glBS1) were also tested and yielded the same phenotype. Most likely the heat shock promoter (hsp70) of the GMR-gal4 construct is leaky, leading to very low activation of UAS-rasv12 and consequently to the phenotype [28].
Figure 2. Morphological traits of Drosophila synthetica.
(A–B) Scanning electronic microscopy (SEM) images of Drosophila synthetica flies. Eye is small due to lack of glass. (C–D) Wings of Drosophila synthetica show extraveins in the lateral regions of the wing (D) compared to the Drosophila melanogaster wing (C).
When hybrids between Drosophila melanogaster and the synthetic genotype are produced, the “killing module” GMR-gal4; UAS-rasv12 is triggered by the presence of the glass gene (Fig. 1d, Table 1, Fig. 3a). This genetic network, while still allowing normal reproduction among flies with the synthetic genotype, completely isolates GMR-gal4/GMR-gal4; UAS-rasv12, gl60J/UAS-rasv12, gl60J flies from normal D. melanogaster due to hybrid early pupal lethality (Fig. 3a, Table 1). Unlike with the other known and naturally occurring speciation mutations [8], the sex of the parents did not influence the lethality of the hybrids in this case (Table 1, Fig. 3a). Experiments were performed at 17°C because flies of the synthetic genotype grew better and because due to the temperature sensitiveness of the Gal4 it is likely to be the temperature at which the killing module may be less effective. Despite this, the killing module was 100% effective even at 17°C (Table 1, Fig 3a). In the initial population mutations in yellow (y1), which results in mild pigmentation, existed as a polymorphism in some individuals.
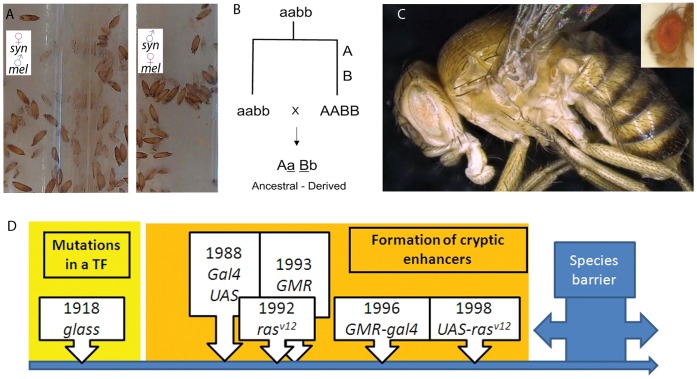
Figure 3. Creation of species boundaries by regulatory evolution.
(A) Hybrids between melanogaster and synthetica arrest in pupae and do not develop further, even at 17C. The sex of the parents did not affect the outcome. Pupae shown in the pictures are more than one month old. (B) Scheme of a classical Dobzhansky-Muller mechanism for speciation, where all mutations occur in one of the populations (“derived”), and the hybrids between the “ancestral” (aabb) and “derived” (AABB) populations are lethal. (C) High definition and depth of field images of Drosophila synthetica after several generations of coexistence with D. melanogaster. Image obtained with a Keyence VHX-600 microscope. Eyes are pale in addition to small. A D.melanogaster eye is shown for comparison in the upper right corner. (D) General model for the creation of species boundaries based on the modification of transcription factors and the subsequent appearance of cryptic enhancers. This could be a mechanism to create synthetic species and prevent hybridization of transgenic animals with natural populations. The case of Drosophila synthetica is shown. Years correspond to the first appearance of the mutation or transgene in a Drosophila laboratory.
It is often difficult to delineate “species boundaries” since they may carry identical mutations and are related to one another through common ancestors. However, most biologists agree on a very stringent definition for species, the Ernst Mayr’s Biological Species Concept, according to which species consist of populations of organisms that can reproduce with one another, but are reproductively isolated from other such groups [7] (Fig. 3b). This definition leads to a focus on the barriers to reproduction between species [8]–[14]. Such barriers represented one of the main problems for Darwin who wrote: “How can we account for species, when crossed, being sterile (…), whereas, when varieties are crossed, their fertility is unimpaired?” [11]. Because the postzygotically isolated population generated here conforms to the most stringent definition of species [8]–[14], it will subsequently be called Drosophila synthetica.
To further prove that the synthetic genetic network allowed zero gene flow with D. melanogaster, co-cultures of both populations were performed for 13 generations (using D.melanogaster white (w) mutants with white eyes) and not a single hybrid was recovered (Fig. 3c, Table 1). Hybrids would have been easily recognizable by normally sized red eyes, because they would carry a normal copy of gl and two w+ copies from the transgenes (Table 1), but D. synthetica behaved like a stable species, did not interbreed and maintained its characteristic eyes (Fig. 3c). Identical results were obtained when crossing D. synthetica with other melanogaster strains, including y,w flies and y,w,f flies (Table 1). In all cases synthetica and melanogaster did not interbreed (Table 1).
Assembling synthetic species boundaries can have practical applications. For example, the use of recombinant DNA technology to alter organisms for a specific purpose has raised controversy [18] and is a growing problem due to the increasing number of transgenic organisms approved by regulatory agencies [16]–[18]. A new framework where safety mechanisms are genetically designed along with desired modification could help to gain public support for a technology with the potential to satisfy future medical and nutritional needs [16]–[18]. D. synthetica is the first transgenic organism that cannot reproduce with the original wildtype population. I therefore propose that synthetic species barriers may serve to compartmentalize dangers and protect natural species from interbreeding with emergent transgenic forms, therefore preserving natural biodiversity (Table 1).
Moreover, once a genetic network is identified, as is the case for the “ras-glass” synthetic boundary described here, opening or closing of the barrier can be controlled at will. In case the interbreeding of populations appears beneficial, targeted strategies can be implemented to reverse hybridization barriers. To test this experimentally, the GAL4-inhibitor GAL80 was expressed from a tubulin promoter (tub-gal80) [29] in D. melanogaster in order to remove hybrid lethality and traverse the species barrier. Males of D. synthetica hybridized successfully with tub-gal80 D. melanogaster females and produced viable hybrids (Table 1), as predicted because Gal80 can block the “killing module”.
Discussion
The postzygotically isolated population generated here in the genus Drosophila conforms to the most stringent definition of species [8]–[14], as well as to the principles of synthetic biology [1], [2], and it has been consequently named Drosophila synthetica. I propose the term “synthetic species” to distinguish it from “natural species”, not only because it has been created in the laboratory, but also because it may never be able to survive in the wild, unlike “natural species”. For example, because the flies created are blind and only survive at lower temperatures, they have potential fitness deficits and it could be argued that the changes could not be arrived at in concert because the “fitness valley” will not be traversed in the wild. However, blindness is a common adaptation in caves suggesting that fitness deficits are difficult to predict and depend on environmental conditions [7], [11], [27].
Interestingly, the generation of Drosophila synthetica matches the Dobzhansky-Muller theoretical model for postzygotic incompatibilities during naturally occurring speciation [7], [8], according to which an ancestral population splits into two independent populations that then accumulate mutations (Fig. 3b). Subsequent genetic interactions between those mutations cause hybrid incompatibilities. In particular, it conforms to a derived-ancestral incompatibility (Fig. 3b), in which all substitutions occur in the derived population.
I therefore propose that modifications in transcription factors and appearance of cryptic enhancers upstream of potentially lethal gene products can constitute a normal Dobzhansky-Muller mechanism for speciation (Fig. 3d). The appearance of those cryptic enhancers could be driven by the accumulation of point mutations in regulatory regions (Fig. 3d), in a manner similar to what has been described recently [30], but those enhancers will only be recognised by the ancestral transcription factor which is now missing (or modified) in the derived population (Fig. 3d). When hybridization between the derived and ancestral populations occurs, the genes with cryptic enhancers will be activated by the ancestral transcription factor, causing hybrid lethality and reproductive isolation (Figs. 1,2, Table 1). This could constitute a general mechanism through which regulatory evolution creates species boundaries (Fig. 3d) and may help to define concrete target genes mediating speciation.
One of the predictions of classical evolutionary theory is that organisms that connect two species must exist as part of the gradual divergence process [11]. Because we fully know the mutations forming a reproduction barrier between melanogaster and synthetica, it is feasible to dissect the process and move backwards, showing how populations of intermediate mutants can indeed interbreed with populations at either side of the evolutionary path towards postzygotic isolation (i.e., glass mutants can hybridize with both species (Table 1) and hence connect melanogaster with synthetica, as if they were a “missing link”) (Fig. 3).
The results shown here provide proof of principle for the transition from “transgenic animals” to “synthetic species”, as defined above, and should spur the debate for its use as a failsafe mechanism in biotechnology. Modifying the binding affinities of one transcription factor and the enhancers it recognises, could be used to engineer reproductive isolation in other living animals, not only in Drosophila. Moreover, the ability to open and close speciation gates when desired reflects one of the goals of synthetic biology –to build components that can be reliably and predictably manipulated–, and preserves flexibility while gaining control over the spread of genetically modified organisms.
One potential caveat could be that this barrier is not irreversible since it can be overturned quite simply if a spontaneous mutation was to arise in any of the components. However, if we think in terms of engineering (or synthetic biology), having fail safe mechanisms in a machine makes it safer; despite they may stop working. The solution is to add more fail safe devices. Identically, adding more synthetic speciation barriers will increase safety. Other transcription factors and enhancers could be easily used to create those extra barriers, because the concept goes beyond any particular element. Importantly, modification of the binding properties, instead of complete elimination of the transcription factor, could also be implemented, reducing the constraint of not finding enough non-essential transcription factors to build several barriers.
In summary, the synthetic species boundary described here can be reliably and predictably manipulated, allowing opening and closing speciation gates when desired, and isolates for the first time a transgenic animal from the original wild-type population.
Methods
High definition and depth of field photographs were obtained with a Keyence VHX-600 microscope. Flies were frozen at −20°C overnight before imaging. For SEM, adults were fixed in 2.5% glutaraldehyde in PBS overnight at 4°C, post-fixed in 1% osmium for 2 h at 4°C, washed, dehydrated in ethanol and with Hexamethyldisilazane until evaporation of the solvent. Samples were coated with 30 nm of gold and observed with a 440 Leica microscope under 20 kV tension.
The fly stocks used were obtained from the Bloomington Stock Center except where indicated. The following stocks were used: GMR-gal4, UAS-rasv12, glass60J, UAS-Dpp, UAS-wg-HA, UAS-egr, UAS-brk (G.Campbell), UAS-hepCA, UAS-fweLose-A and UAS-fweLose-B, UAS-hid (H. Steller), tub-GAL80.
For the balancing of the different transgenes the following stocks were used:
ywhs-FLP;If/CyO; MKRS/TM6b.
w1118; PasSC1 gl3/TM6B, glBS1 Tb1.
w1118; If/CyO; MKRS/TM6B, glBS1 Tb1.
C(1)DX,y1,f1,hs-hid.
Nomenclatural Acts
The electronic version of this document does not represent a published work according to the International Code of Zoological Nomenclature (ICZN), and hence the nomenclatural acts contained in the electronic version are not available under that Code from the electronic edition. Therefore, a separate edition of this document was produced by a method that assures numerous identical and durable copies, and those copies were simultaneously obtainable (from the publication date noted on the first page of this article) for the purpose of providing a public and permanent scientific record, in accordance with Article 8.1 of the Code. The separate print-only edition is available on request from PLoS by sending a request to PLoS ONE, Public Library of Science, 1160 Battery Street, Suite 100, San Francisco, CA 94111, USA along with a check for $10 (to cover printing and postage) payable to “Public Library of Science”. The online version of this work is archived and available from the following digital repositories: PubMed Central, LOCKSS.
Acknowledgments
I thank M. Calleja, H.Stocker and the Bloomington stock centre for materials. M. Martínez and M. Stoffel for help with the Keyence VHX-600 microscope and for performing SEM microscopy. S.Noselli, T. Ochsenreiter, P.Leopold, C.Rhiner and JP. Vincent for discussions. O. Gerlitz, S. Noselli and C. Rhiner for critically reading the manuscript and the academic editor for insightful discussions.
Footnotes
Competing Interests: The author has declared that no competing interests exist.
Funding: The author has no funding or support to report.
References
- 1.Benner SA, Sismour AM. Synthetic biology. Nat Rev Genet. 2005;6:533–543. doi: 10.1038/nrg1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purnick PE, Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 3.Prud’homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci U S A. 2007;15:8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sucena E, Delon I, Jones I, Payre F, Stern DL. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature. 2003;424:935–938. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- 5.Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature. 2005;433:481–487. doi: 10.1038/nature03235. [DOI] [PubMed] [Google Scholar]
- 6.McGregor AP, Orgogozo V, Delon I, Zanet J, Srinivasan DG, et al. Morphological evolution through multiple cis-regulatory mutations at a single gene. Nature. 2007;448:587–590. doi: 10.1038/nature05988. [DOI] [PubMed] [Google Scholar]
- 7.Coyne JA, Orr HA. Speciation. Sinauer Associates, Inc., Sunderland, Massachusetts. ISBN 0-87893-091-4. 2004.
- 8.Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, et al. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- 9.Ortiz-Barrientos D, Grealy A, Nosil P. The genetics and ecology of reinforcement: implications for the evolution of prezygotic isolation in sympatry and beyond. Ann N Y Acad Sci. 2009;1168:156–182. doi: 10.1111/j.1749-6632.2009.04919.x. [DOI] [PubMed] [Google Scholar]
- 10.Barbash DA. Ninety years of Drosophila melanogaster hybrids. Genetics. 2010;186:1–8. doi: 10.1534/genetics.110.121459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray. 1859. [PMC free article] [PubMed]
- 12.Rice WR, Salt GW. Speciation via disruptive selection on habitat preference: experimental evidence. The American Naturalist. 1988;131:911–917. [Google Scholar]
- 13.Mayrose I, Zhan SH, Rothfels CJ, Magnuson-Ford K, Barker MS, et al. Recently formed polyploid plants diversify at lower rates. Science. 2011;333:1257–1258. doi: 10.1126/science.1207205. [DOI] [PubMed] [Google Scholar]
- 14.Greig D. Reproductive isolation in Saccharomyces. Heredity. 2009;102:39–44. doi: 10.1038/hdy.2008.73. [DOI] [PubMed] [Google Scholar]
- 15.Lutes AA, Baumann DP, Neaves WB, Baumann P. Laboratory synthesis of an independently reproducing vertebrate species. Proceedings of the National Academy of Sciences. 2011;108:302–304. doi: 10.1073/pnas.1102811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kling J. First US approval for a transgenic animal drug. Nat. Biotechnol. 2009;27:302–304. doi: 10.1038/nbt0409-302. [DOI] [PubMed] [Google Scholar]
- 17.Marris E. Transgenic fish go large. Nature. 467, 259. 2010. [DOI] [PubMed]
- 18.Pardo R, Engelhard M, Hagen K, Jorgensen RB, Rehbinder E, et al. The role of means and goals in technology acceptance A differentiated landscape of public perceptions of pharming. EMBO reports. 2009;10:1069–1075. doi: 10.1038/embor.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moses K, Ellis MC, Rubin GM. The glass gene encodes a zinc-finger protein required by Drosophila photoreceptor cells. Nature. 1989;340:531–536. doi: 10.1038/340531a0. [DOI] [PubMed] [Google Scholar]
- 20.Moses K, Rubin GM. Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev. 5, 583–593. 1991. [DOI] [PubMed]
- 21.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87, 651–660. 1996. [DOI] [PubMed]
- 22.Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature 332, 853–865. 1988. [DOI] [PubMed]
- 23.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. 1993. [DOI] [PubMed]
- 24.Karim FD, Rubin GM. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 125, 1–9. 1998. [DOI] [PubMed]
- 25.Fortini ME, Simon MA, Rubin GM. Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature. 1992;355:559–561. doi: 10.1038/355559a0. [DOI] [PubMed] [Google Scholar]
- 26.Moreno E, Morata G. Caudal is the Hox gene that specifies the most posterior Drosophila segment. Nature 400, 873–877. 1999. [DOI] [PubMed]
- 27.Rhiner C, Lopez-Gay JM, Soldini D, Casas-Tintó S, Martín FA, Moreno E. Flower forms an extracellular code that reveals the fitness of a cell to its neighbors in Drosophila. Dev Cell. 18, 882–883. 2010. [DOI] [PubMed]
- 28.Brunner D, Oellers N, Szabad J, Biggs III WH, Zipursky SL, et al. A gain-of-function mutation in Drosophila MAP kinase activates multiple receptor tyrosine kinase signaling pathways. Cell. 1994;76:875–888. doi: 10.1016/0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 29.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 30.Frankel N, Erezyilmaz DF, McGregor AP, Wang S, Payre F, et al. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]