Summary
Alterations of mitochondrial functions are linked to multiple degenerative or acute diseases. As mitochondria age in our cells, they become progressively inefficient and potentially toxic, and acute damage can trigger the permeabilization of mitochondrial membranes to initiate apoptosis or necrosis. Moreover, mitochondria have an important role in pro-inflammatory signaling. Autophagic turnover of cellular constituents, be it general or specific for mitochondria (mitophagy), eliminates dysfunctional or damaged mitochondria, thus counteracting degeneration, dampening inflammation, and preventing unwarranted cell loss. Decreased expression of genes that regulate autophagy or mitophagy can cause degenerative diseases in which deficient quality control results in inflammation and the death of cell populations. Thus, a combination of mitochondrial dysfunction and insufficient autophagy may contribute to multiple aging-associated pathologies.
Aging seems to be the only available way to live a long life.
Kitty O’Neill Collins
Introduction
There is a simple arithmetic of multicellular life that holds deep insights into health and disease—the rate of cell generation minus the rate of cell loss determines the growth or degeneration of a tissue. Somatic cells fall into two classes, those that are constantly renewed from proliferating stem cells, such as epithelial cells and leukocytes, and those that are rarely renewed after birth, such as neurons and cardiomyocytes. Although the former are programmed to undergo cell death and are replaced throughout life, the latter must endure until we expire. Excessive cell death in post-mitotic tissues precipitates degenerative states (1, 2), whereas the failure to execute timely programmed death in renovating tissues contributes to hyperplasia and cancer (3). As we age, cells accumulate errors in their nuclear and mitochondrial genomes and damage to organelles and macromolecules, and therefore quality control mechanisms determine the consequences of such accumulation for the cell and the organism. Recent evidence indicates that the interplay between mitochondria and autophagy links aging to health or disease.
Mitochondria are the evolutionary relics of aerobic bacteria that invaded the proto-eukaryotic cell about a billion years ago. As such, they have a separate genome and provide the oxygen consumption-driven synthesis of ATP (via oxidative phosphorylation, OXPHOS). However, as a side product of normal respiration, mitochondria produce reactive oxygen species (ROS) that must be detoxified. Moreover, as they replicate, mitochondrial genomes accumulate mutations that eventually compromise the efficiency of OXPHOS (4). Mitochondrial deficiency, excessive ROS, or both appear to be driving forces in aging because they reduce cellular fitness, inflict damage to other organelles, or cause mutations of the nuclear genome (4). Beyond their roles in the chronic process of cellular and organismal aging, mitochondria also mediate acute cell death. In many instances, developmental, homeostatic, and pathological cell death involves a critical step in which mitochondria release proteins that trigger the self-destructive enzymatic cascade that causes apoptosis (5).
Macroutophagy (herein, “autophagy”) occurs by the formation of autophagosomes, double-membraned vesicles that sequester organelles, proteins, or portions of the cytoplasm, which then fuse with lysosomes. As a result of this process, the sequestered contents are degraded by lysosomal enzymes, and recycled as a source of energy (6). Autophagy may occur either as a general phenomenon, for instance when cells lack nutrients and mobilize their energy reserves, or it can specifically target distinct cellular structures such as damaged mitochondria (“mitophagy”). Autophagy has cardinal roles in the cellular adaptation to stress, in innate immune responses, and as a quality control mechanism (7). Thus, autophagy represents an essential cytoprotective pathway and a potential anti-aging mechanism (8).
Inflammation is a critical organismal response to tissue damage and infection, in which various secreted mediators, such as cytokines, chemokines, and eicosanoids, coordinate defense and repair, but its long term consequences are a major cause of disease associated with aging (9). Both autophagy and apoptotic cell death tend to dampen inflammation, whereas necrotic cell death can promote it (10, 11). A key mechanism of inflammation is the activation of the “inflammasome,” a molecular platform that activates caspase-1, which in turn processes the inflammatory cytokines Interleukin-1 and Interleukin-18 and facilitates secretion or these and other inflammatory mediators. Mitochondria play a central role in the initiation of inflammasomes and other inflammatory pathways (discussed below), and as such integrate autophagy, cell death, and inflammation (Figure 1). Here, we discuss the functions of these organelles in aging as a consequence of the interplay of these three processes.
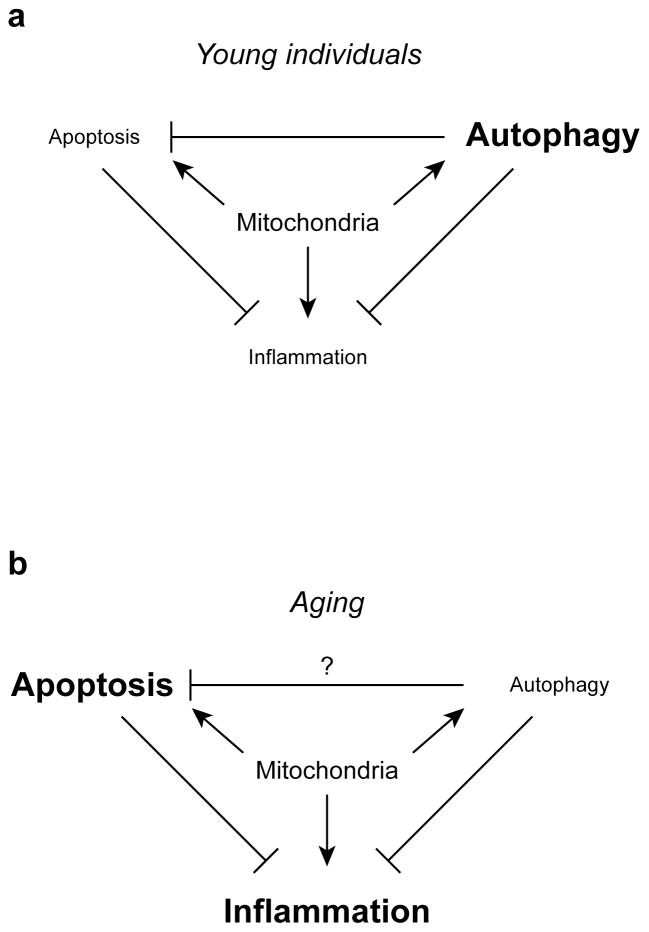
Figure 1. Interactions between mitochondria, cell death, autophagy, and inflammation in young individuals (a) and during aging (b).
In this scenario, loss of autophagy with age leads to accumulation of damaged mitochondria, which promote cell death and inflammation, both of which are otherwise limited by autophagy.
Damaging mitochondrial signals in inflammation and cell death
There are at least three major mechanisms through which mitochondria can damage or kill their host, namely through production of ROS or pro-inflammatory signals or through mitochondrial membrane permeabilization.
About 1 to 3% of the molecular oxygen is incompletely reduced during OXPHOS, hence generating the ROS superoxide anion. This fraction can increase when mitochondria are compromised by damage or mutation. ROS cause mutations in mitochondrial DNA (mtDNA), in turn compromising OXPHOS and initiating a vicious cycle of mitochondrial collapse. Mice in which the polymerase that replicates mtDNA has been mutated to increase the frequency of mtDNA mutations exhibit signs of premature aging, and this has been attributed to apoptotic death of stem cells (4).
Mitochondria participate in the detection of infectious microorganisms and cellular damage to activate innate immune responses. The pattern recognition receptors RIG-I and MDA-5 recognize viral RNA in the cytosol and interact with an adapter on the mitochondrial membrane, MAVS, to trigger a signal transduction cascade that drives the production of type I interferon (12). Similarly, IRGM, a protein that is required for the control of intracellular mycobacteria, translocates to mitochondria. Through the induction of mitochondrial fragmentation and mitochondrial outer membrane permeabilization (MOMP, see below), distinct IRGM isoforms can either activate autophagy (similarly required for defense against intracellular mycobacteria) or cell death (13).
ROS produced by mitochondria can activate an inflammasome composed of NLRP3, the adapter protein ASC, and caspase-1, a process that occurs at the interface between mitochondria and the endoplasmic reticulum (ER), the so-called mitochondria-associated ER membranes (MAMs) (14), possibly concomitant with the cytosolic release of mtDNA (10). Mitochondria are essential for NLRP3 inflammasome activation in response to various non-infectious agents, including uric acid and silica, and further, ROS-triggering OXPHOS inhibitors also activate this inflammasome. It is therefore likely that accumulation of damaged mitochondria is an important cause of inflammation.
In the intrinsic pathway leading to apoptotic death, MOMP results in the release of soluble mitochondrial intermembrane proteins that cause cell death by apoptosis (5). MOMP results from the interactions of proteins of the BCL-2 family that protect or disrupt the outer mitochondrial membrane. Alternatively, cell death can be triggered by the so-called mitochondrial permeability transition (MPT), which depends on the mitochondrial matrix protein cyclophilin D. MPT results in the instantaneous dissipation of the mitochondrial transmembrane potential (Δψm) and cessation of OXPHOS, thus triggering rapid necrotic cell death (15).
Mitochondria are highly dynamic organelles that can change their morphology, fragment by fission, or undergo fusion to generate highly interconnected tubular networks. These dynamics are also regulated by the BCL-2 proteins and undoubtedly affect the respiratory, ROS-generating, pro-inflammatory, and lethal signaling functions of mitochondria in ways that are only partially understood.
General versus mitochondrion-specific autophagy
In response to multiple forms of cellular stress including the shortage of growth factors, nutrients or oxygen, excessive ROS or DNA damage, general autophagy is stimulated through the coordinated activation of several multiprotein complexes (such as the complexes organized around the pro-autophagic protein kinase ULK1/2 and its upstream inhibitory kinase mammalian target or rapamycin (MTOR), as well as the lipid kinase PIK3C3/HVPS34 and its obligatory allosteric activator Beclin 1), a conjugation system that transfers the ubiquitin-like proteins ATG12 and MAP1LC3A/LC3 to their substrates (usually ATG5 and phosphatidylethanolamine, respectively), a dynein-dependent transport system that moves autophagosomes to lysosomes, and a subsequent fusion machinery (6). During general autophagy, cytoplasmic cargo including portions of the cytosol, mitochondria, and other organelles can be sequestered and digested. Nonetheless, mitochondria appear to have a major role in general autophagy, as they supply membranes for the biogenesis of autophagosomes during starvation (16). Disruption of MAMs by knockdown of MFN2 (an essential component of the mitochondrial fusion machinery) abolishes starvation-induced autophagy in human cancer cell lines (16), implying that MAMs (or perhaps MAM-unrelated functions of MFN2) are essential for phagophore formation. Knockout or inhibition of cyclophilin D also prevents starvation-induced autophagy in some cell types (17). However, it remains a matter of controversy (see Supplemental Discussion) whether MPT constitutes an essential step toward autophagy. Notably, cyclophilin D-deficiency has not been associated with premature aging or degeneration.
Low ATP production or enhanced ROS generation by mitochondria induces general autophagy (6, 7). Cells also have several distinct systems to specifically target mitochondria to autophagy (mitophagy) (Figure 2). One such system comes into action in red blood cell precursors, which eliminate mitochondria by overexpressing a BCL-2 family protein, BNIP3L/NIX. NIX associates with mitochondrial membranes to engage direct molecular interactions with LC3 (18) and/or causes Δψm dissipation (19) (which can suffice to target mitochondria to mitophagy, see below) (20). Similarly, BNIP3, a hypoxia-inducible BCL-2 family protein, is suggested to trigger mitophagy by competitively disrupting the inhibitory interaction between BCL-2 and Beclin 1 (7). Mitochondria can divide asymmetrically into functional progeny (with a high Δψm), which can reintegrate the mitochondrial network by fusion, and dysfunctional organelles (with a low Δψm), which are specifically destined for mitophagy (20).
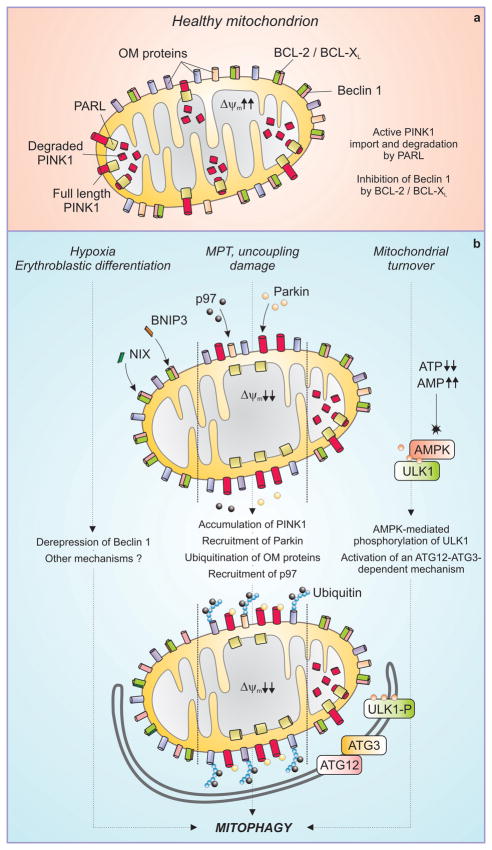
Figure 2. Mechanisms of mitophagy.
In healthy mitochondria (a), PINK1 is actively imported by a mitochondrial transmembrane potential (Δψm)-dependent mechanism and degraded by the inner mitochondrial membrane protease PARL. BCL-2 and BCL-XL bind to and inhibit Beclin 1. Different triggers can stimulate distinct pathways to mitophagy (b). The BH3-only proteins NIX and BNIP3 are activated during erythroblast differentiation and under hypoxia, respectively, and may cause mitophagy by displacing Beclin 1 from inhibitory interactions with BCL-2 and BCL-XL. In response to uncoupling, mitochondrial damage or the mitochondrial permeability transition (MPT), the Δψm is dissipated and full length PINK1 accumulates at the outer mitochondrial membrane (OM). This allows for the recruitment of the AAA ATPase p97 and of Parkin, which together render mitochondria a palatable substrate for the autophagic machinery. Mitochondrial turnover can also be mediated by accumulation of AMP, leading to the phosphorylation of ULK1 by AMPK and possibly involving a ATG12-ATG3 conjugate that functions specifically in mitophagy.
The kinase PINK1 is imported into healthy mitochondria, dependent on Δψm, where it is degraded by the protease PARL (20). On the surface of mitochondria with low Δψm, PINK1 accumulates, leading to the recruitment of the ubiquitin ligase Parkin, which ubiquitinylates outer membrane proteins including BCL-2 (21), VDAC1 (22), MFN1 and MFN2 (23). This may de-repress the HVPS34/Beclin 1 complex (21) (with which Parkin may also directly interact) (24). By favoring the proteasomal degradation of MFN1 and MFN1 through a mechanism that requires the AAA-type ATPase p97/VCM, Parkin suppresses mitochondrial fusion and promotes mitophagy (23). This can be further stimulated by histone deacetylase 6, which is recruited to mitochondria by ubiquitinylated proteins and catalyzes pro-autophagic cytoplasmic deacetylation reactions (25). In cells that lack Parkin, Δψm dissipation does not induce rapid removal of mitochondria, although other mechanisms almost certainly ensure a mitochondrial quality control by mitophagy. Another, possibly redundant, mechanism of mitophagy involves the activation of adenosine monophosphate-activated protein kinase (AMPK) under conditions where AMP is produced (such as when ATP concentrations decline). AMPK phosphorylates and thereby activates ULK1, one of the initiators of autophagy, and this engages autophagic removal of mitochondria (26). This appears to require the formation of conjugates between ATG3 and ATG12, because cells carrying a mutant ATG3 that binds LC3 but not ATG12 can activate autophagy but do not remove mitochondria (27). How the different mechanisms of mitophagy (Figure 2) interact is not known, although defects in mitophagy (for example, that is caused by loss of PINK1) can lead to increased general autophagy (22), possibly as a compensatory mechanism triggered by ROS-mediated damage.
Autophagy for the avoidance of cell death
Although autophagy often precedes apoptosis or necrosis, it rarely constitutes a suicidal mechanism and it probably reflects failed attempts of cells to adapt to stress (28). Usually, autophagy has a cytoprotective rather than cytocidal function, (although there are exceptions). When MOMP is induced and post-mitochondrial caspase activation is blocked or disrupted, permeabilized mitochondria are removed by autophagy (29, 30), and inhibition of mitophagy can accelerate cell death (30). Mitochondria that have not undergone MOMP can repopulate the mitochondrial pool and rescue the cell (29, 31).
Induction of general autophagy before cells are stressed with otherwise lethal stimuli can protect them against cell death. This has been correlated with the relative resistance of residual mitochondria to MOMP or MPT and may be explained by the removal of mitochondria that have a rather low threshold for permeabilization (mitochondrial “purging”) (20). Alternatively, occurrence of MOMP or MPT in a fraction of mitochondria may activate repair or recycling pathways that activate autophagic sequestration of depolarized mitochondria (2, 20). In this case, the intensity of the autophagic flow might set the threshold beyond which partial MOMP becomes lethal. However, evidence that forced induction of mitophagy would confer cytoprotection in these circumstances is scarce, and it is possible that general autophagy (as opposed to specific mitophagy) may reduce the cell’s propensity to engage in lethal signal transduction cascades.
How can general autophagy confer cytoprotection and interrupt signals that lead to MOMP? Autophagy facilitates the maintenance of high concentrations of ATP (which reduces the propensity of mitochondria to undergo MPT) and furnishes basic building blocks for the adaptive synthesis of proteins including potential apoptosis inhibitors (6, 7). It may also lead to the elimination of potentially toxic protein aggregates and help limit the accumulation of ubiquitinylated proteins that otherwise would inhibit proteasome function (32). Induction of autophagy affects the progression of the cell cycle (33) (and vice versa), suggesting that it can impinge on the propensity of cells to succumb to cell cycle-dependent cell death inducers. Moreover, as a correlate of autophagy induction, anti-apoptotic proteins like BCL-2 and the caspase-8 inhibitor FLIP may be liberated from inhibitory interaction with autophagic effectors (such as Beclin 1 and ATG3, respectively) (7). Multiple mechanisms exist through which autophagy can intercept lethal signaling before or at the level of mitochondria (Figure 3). Thus, induction of autophagy might affect the circuitry through which lethal signals are relayed at mitochondria.
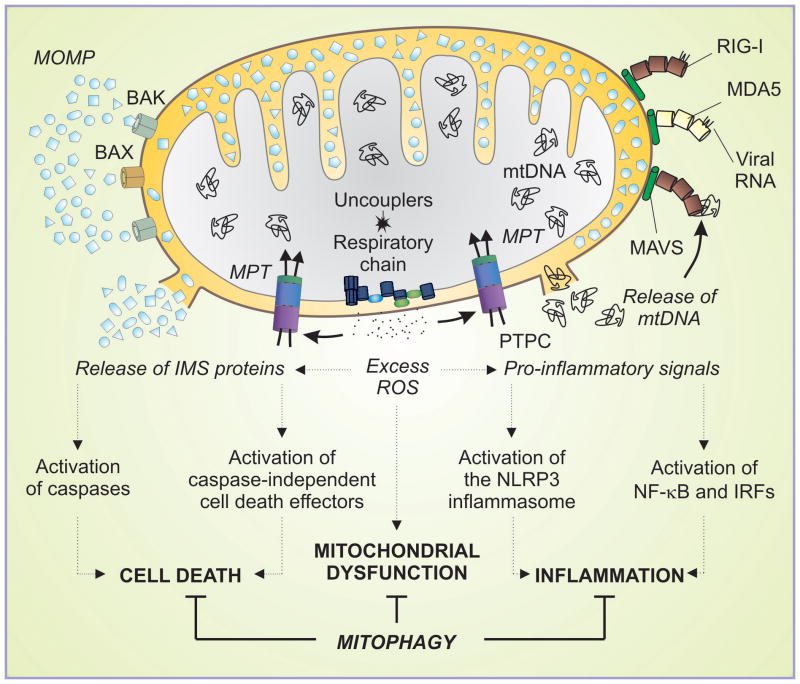
Figure 3. Mitophagy exerts cytoprotective effects by intercepting lethal signals before or at the level of mitochondria.
In response to lethal stimuli, mitochondria can undergo BAX- or BAK-mediated mitochondrial outer membrane permeabilization (MOMP), or activate the permeability transition pore complex (PTPC), driving the mitochondrial permeability transition (MPT). In both instances, intermembrane space proteins (IMS) are released into the cytosol where they activate caspase-dependent and -independent mechanisms that mediate cell death. One MPT trigger is represented by reactive oxygen species (ROS), which can be generated upon respiratory chain uncoupling. Production of mitochondrial ROS can trigger the NALP3 inflammasome via an unknown mechanism. In some settings, ROS-mediated MPT may favor the release of mitochondrial DNA (mtDNA), which can activate stimulate pro-inflammatory signaling via RIG-I and MDA5, both of which function as viral RNA sensors and interact with mitochondria through the adaptor MAVS. Activated RIG-I and MDA5 promote the activation of NF- κB and interferon regulatory factors (IRFs).
Autophagy and aging
Autophagy appears to decline with age, and the expression of several key players in the autophagic pathway (for example ATG5 and ATG7) show decreased expression in the brains of aging individuals (34). Conditions that promote autophagy, such as caloric restriction and exercise, delay aging-associated degeneration (2), suggesting that autophagy counteracts the aging process. Stimulation of autophagy can increase the healthy lifespan in multiple model organisms including mice and primates (8) (Supplemental Table 1).
Experimental inactivation of genes required for the execution of autophagy is lethal at the whole-body level, whereas tissue-specific knockouts induce organ-specific degenerative changes (28). Major neurodegenerative diseases affecting humans have been linked to defects in mitochondria and autophagy. Parkinson disease (PD) is caused by the selective loss of dopaminergic neurons, and such cell loss can be experimentally induced by mitochondrial toxins including 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and the complex I inhibitor rotenone. Several mutations that cause hereditary PD affect genes including those encoding PINK1, Parkin, or DJ1, whose products function in mitophagy (20, 22–24), suggesting that deficient mitochondrial quality control may contribute to PD. In both sporadic and familial PD, intraneuronal inclusions called Lewy bodies have been detected. Lewy bodies contain α-synuclein, a protein that impairs autophagy when overexpressed in cells or mice (35).
Huntington disease is a monogenic degeneration affecting striatal neurons that is caused by an extended polyglutamine stretch in the Huntingtin protein. In mice, a Huntington-like disease can be provoked by systemic injection of mitochondrial uncouplers. Moreover, Huntingtin can interact with mitochondria, and patients’ lymphocytes manifest mitochondrial dysfunction, while whole-body energy expenditure is affected by signs of inefficient OXPHOS (36). Although Huntingtin itself may be an autophagic substrate, it also regulates autophagy. Thus, complete deletion of the polyglutamine tract within Huntingtin increases baseline autophagy (and augments longevity) in mice (37). Moreover, expression of mutant Huntingtin in mice results in deficient sequestration of autophagic cargoes (38).
In hereditary Alzheimer’s disease, deficient Presenilin-1 activity has been thought to induce the accumulation of the β-amyloid peptide, which can mediate mitochondrial toxicity (1). However, Presenilin-1 also acts as a chaperone for one subunit of the lysosomal proton pump, and its loss-of-function mutation results in defective lysosomal acidification and impaired autophagosome clearance, which are reflected in increased autophagic vacuolization (39). The decline of the expression of autophagy genes with age is exacerbated in the brains of Alzheimer’s patients (34).
The aforementioned examples (and others, Supplemental Table 2) follow a common leitmotif. After the identification of pathogenic mutations, the disease etiology was initially ascribed to primary mitochondrial effects of the mutant proteins. More recently, however, it has been discovered that the disease-causing gene product subverts autophagy. It remains to be determined whether the mitochondrial alterations simply result from deficient quality control or whether the genetic defects act as ‘dual hits’ and simultaneously disrupt mitochondrial and autophagic functions. It is also possible that environmental factors (such as mitochondrial toxins) and hereditary perturbations in mitophagy cooperate in disease pathogenesis. Irrespective of these incognita, it appears that pharmacological induction of autophagy can postpone the manifestation of neurodegenerative diseases at least in some animal models of hereditary neurodegeneration including Huntington disease (40).
Autophagy may also have a major cardioprotective role. Ischemic preconditioning, the transient reduction of coronary blood flow, protects the heart against subsequent ischemic necrosis through a massive induction of autophagy (2). This correlates with an increased mitochondrial resistance to MPT, perhaps because mitochondria with an elevated threshold for MPT induction have been selected. Exercise and caloric restriction stimulate autophagy in most tissues including the myocardium (2), and it is possible that autophagy constitutes (one of) the mechanisms through which physical activity and leanness confer cardioprotection.
Autophagy can mitigate inflammatory reactions through several mechanisms. Autophagy in dying cells is required for optimal macrophage-mediated clearing of apoptotic corpses, thus reducing inflammatory reactions (28). Beyond its contribution to the control of intracellular microorganisms, autophagy can inhibit signaling via RIG-I-like receptors by directly conjugating the receptors to ATG5-ATG12 complexes and through elimination of dysfunctional mitochondria (12). Autophagy can also inhibit NLRP3 activation by removing permeabilized or ROS-producing mitochondria (10, 14). Because neurodegenerative processes and pathological aging are accompanied by chronic inflammation, these anti-inflammatory effects of autophagy may mediate additional health benefits.
Open questions and perspectives
Undoubtedly, inhibition of autophagy can participate in the pathogenesis of major diseases including neurodegeneration, and stimulation of autophagy may mediate cytoprotective and anti-inflammatory effects that at least partially can be ascribed to the removal of dysfunctional mitochondria, as this has been shown in the context of several pathologies. It remains to be seen if the mechanisms of autophagy-driven longevity are the inverse of those accounting for age-related disease - for example, declining autophagy, progressive mitochondrial dysfunction, and ensuing cell death and inflammation - and whether behavioral or pharmacological measures destined to induce general autophagy can be broadly used to improve human health. In rodents, one day of nutrient deprivation reportedly suffices to halve mitochondrial mass in various tissues (2). Therefore intermittent fasting may invoke drastic recycling of the mitochondrial pool while improving longevity to the same extent as does continuous caloric restriction (1). Nonetheless, it not clear if the longevity-extending and health-protective effects of specific induction of mitophagy would be as positive as those of general autophagy. For the development of specific mitophagy-inducing gene therapies and drugs, it will be important to resolve questions as to how mitochondria and autophagy crosstalk in molecular terms: Does the deconstruction of mitochondrial membranes induced by BCL-2 family proteins directly relate to the construction of autophagic membranes? How does the mitochondrial fusion-fission cycle impact on autophagy? How is the autophagic membrane generated at MAMs and how does this interface with metabolic signaling, ion fluxes and lethal signals, and in turn, inflammatory disease?
Supplementary Material
Acknowledgments
We apologize to all colleagues whose work we could not cite owing to space limitations. DRG receives grants from the NIH and from the American Lebanese Syrian Associated Charities. LG is financed by the EU (APO-SYS). GK is supported by ANR, ARC, AXA, FRM, INCA, EU and LNC (équipe labelisée).
References
- 1.Mattson MP, Gleichmann M, Cheng A. Neuron. 2008 Dec 10;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gottlieb RA, Mentzer RM. Annu Rev Physiol. 2010 Mar 17;72:45–59. doi: 10.1146/annurev-physiol-021909-135757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. N Engl J Med. 2009 Oct 15;361:1570–1583. doi: 10.1056/NEJMra0901217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wallace DC, Fan W, Procaccio V. Annu Rev Pathol. 2010;5:297–348. doi: 10.1146/annurev.pathol.4.110807.092314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tait SW, Green DR. Nat Rev Mol Cell Biol. 2010 Sep;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 6.He C, Klionsky DJ. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kroemer G, Marino G, Levine B. Mol Cell. 2010 Oct 22;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madeo F, Tavernarakis N, Kroemer G. Nat Cell Biol. 2010 Sep;12:842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 9.Perry VH, Cunningham C, Holmes C. Nat Rev Immunol. 2007 Feb;7:161–167. doi: 10.1038/nri2015. [DOI] [PubMed] [Google Scholar]
- 10.Nakahira K, et al. Nat Immunol. 2011 Mar;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kono H, Rock KL. Nat Rev Immunol. 2008 Apr;8:279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitoh T, Akira S. J Cell Biol. 2010 Jun 14;189:925–935. doi: 10.1083/jcb.201002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh SB, et al. Nat Cell Biol. 2010 Dec;12:1154–1165. doi: 10.1038/ncb2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou R, Yazdi AS, Menu P, Tschopp J. Nature. 2011 Jan 13;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 15.Kroemer G, Galluzzi L, Brenner C. Physiol Rev. 2007 Jan;87:99–163. doi: 10.1152/physrev.00013.2006. [DOI] [PubMed] [Google Scholar]
- 16.Hailey DW, et al. Cell. 2010 May 14;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carreira RS, Lee Y, Ghochani M, Gustafsson AB, Gottlieb RA. Autophagy. 2010 May 19;6:462–472. doi: 10.4161/auto.6.4.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novak I, et al. EMBO Rep. 2010 Jan;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandoval H, et al. Nature. 2008 Jul 10;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Youle RJ, Narendra DP. Nat Rev Mol Cell Biol. 2011 Jan;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D, et al. J Biol Chem. 2010 Dec 3;285:38214–38223. doi: 10.1074/jbc.M110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geisler S, et al. Nat Cell Biol. 2010 Feb;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka A, et al. J Cell Biol. 2011 Dec 27;191:1367–1380. [Google Scholar]
- 24.Michiorri S, et al. Cell Death Differ. 2010 Jun;17:962–974. doi: 10.1038/cdd.2009.200. [DOI] [PubMed] [Google Scholar]
- 25.Lee JY, Nagano Y, Taylor JP, Lim KL, Yao TP. J Cell Biol. 2010 May 17;189:671–679. doi: 10.1083/jcb.201001039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Egan DF, et al. Science. 2010 Jan 28;331:456–461. [Google Scholar]
- 27.Radoshevich L, et al. Cell. 2010 Aug 20;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine B, Kroemer G. Cell. 2008 Jan 11;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colell A, et al. Cell. 2007 Jun 1;129:983–997. doi: 10.1016/j.cell.2007.03.045. [DOI] [PubMed] [Google Scholar]
- 30.Xue L, Fletcher GC, Tolkovsky AM. Curr Biol. 2001 Mar 6;11:361–365. doi: 10.1016/s0960-9822(01)00100-2. [DOI] [PubMed] [Google Scholar]
- 31.Tait SW, et al. Dev Cell. 2010 May 18;18:802–813. doi: 10.1016/j.devcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC. Mol Cell. 2009 Feb 27;33:517–527. doi: 10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipinski MM, et al. Dev Cell. 2010 Jun 15;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lipinski MM, et al. Proc Natl Acad Sci U S A. 2010 Aug 10;107:14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winslow AR, et al. J Cell Biol. 2010 Sep 20;190:1023–1037. doi: 10.1083/jcb.201003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bossy-Wetzel E, Petrilli A, Knott AB. Trends Neurosci. 2008 Dec;31:609–616. doi: 10.1016/j.tins.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng S, et al. PLoS Genet. 2010;6:e1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martinez-Vicente M, et al. Nat Neurosci. 2010 May;13:567–576. doi: 10.1038/nn.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JH, et al. Cell. 2010 Jun 25;141:1146–1158. [Google Scholar]
- 40.Moreau K, Luo S, Rubinsztein DC. Curr Opin Cell Biol. 2010 Apr;22:206–211. doi: 10.1016/j.ceb.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.