Abstract
Loss-of-function experiments in mice have yielded invaluable mechanistic insights into the pathogenesis of Marfan syndrome (MFS) and implicitly, into the multiple roles fibrillin-1 microfibrils play in the developing and adult organism. Unfortunately, neonatal death from aortic complications of mice lacking fibrillin-1 (Fbn1−/− mice) has limited the scope of these studies. Here we report the creation of a conditional mutant allele (Fbn1fneo) that contains loxP sites bordering exon1 of Fbn1 and an frt-flanked neo expression cassette downstream of it. Fbn1fneo/+ mice were crossed with FLPeR mice and the resulting Fbn1Lox/+ progeny were crossed with Fbn1−;CMV-Cre mice to generate Fbn1CMV−/− mice, which were found to phenocopy the vascular abnormalities of Fbn1−/− mice. Furthermore, mating Fbn1Lox/+ mice with Prx1-Cre or Osx-Cre mice revealed an unappreciated role of fibrillin-1 microfibrils in restricting osteoprogenitor cell recruitment. Fbn1Lox/+ mice are therefore an informative genetic resource to further dissect MFS pathogenesis and the role of extracellular fibrillin-1 assemblies in organ development and homeostasis.
Keywords: conditional gene inactivation, fibrillin-1, Marfan syndrome, osteogenesis
Fibrillin-1 assemblies (microfibrils) impart physical properties to a variety of tissues, communicate with resident cells through integrin receptors and store TGFβ and BMP complexes in the extracellular matrix (Ramirez and Rifkin, 2009). Mutations that alter the structure or decrease the production of fibrillin-1 cause Marfan syndrome (MFS), a common heritable disorder of connective tissue with predominant manifestations in the cardiovascular, ocular and musculoskeletal systems (Ramirez and Dietz, 2007). Mouse models of MFS have delineated key events in the life-threatening progression of vascular disease that have informed the development of new therapeutic strategies based on TGFβ antagonism (Pereira et al., 1999; Habashi et al., 2006; Carta et al., 2006; Nistala et al., 2010a; Holm et al., 2011). Unfortunately, neonatal demise of Fbn1−/− mice (a.k.a.: Fbn1mgN/mgN mice; Carta et al., 2006) due to ruptured aortic aneurysm has limited our ability to dissect organ-specific disease mechanisms in MFS and implicitly, to delineate the full spectrum of fibrillin-1 roles in the developing embryos and adult organism. To overcome this problem, here we report the creation of mice for Cre-mediated inactivation of the Fbn1 gene in specific tissues and at discrete stages of pre- and post-natal life.
The region of the Fbn1 locus that was selected for homologous recombination in mouse embryonic stem (mES) cells encompasses exon 1, which contains the 5’ untranslated region (5’UTR) and the ATG initiation codon (Fig. 1a) (Carta et al., 2006). The targeting vector included the insertion of an frt-flanked neo-expressing cassette 272 bp 3’ of exon 1 and loxP sequences immediately downstream of the neo cassette and within the 5’ UTR of exon 1 (Fig. 1b). In addition, the targeting vector contained a diagnostic BamHI cleavage sites within the neo cassette, as well as 16 kb and 2 kb of homology sequences 3’ and 5’ of exon 1 respectively (Fig. 1b). The linearized targeting vector was electroporated into mES cells that were selected for G418 resistance and whose BamHI-cleaved DNA was subsequently subject to Southern blot hybridizations to probes upstream or downstream of exon 1 in order to validate proper integration of the targeting vector into the Fbn1 locus. The screen resulted in the identification of 4/134 correctly targeted mES clones, two of which (clones D7 and H5) were randomly selected and microinjected into C57BL/6J blastocysts. The resulting chimeric mice were bred with C57BL/6J females to monitor germ line transmission of the targeted Fbn1 allele (Fbn1fneo allele), as evidenced by Southern blot analyses of tail DNA from the F1 progeny of the D7 or H5 chimeric mating (Fig. 1f and g).
FIG. 1.
Generation of Fbn1Lox/− mice. The diagrams show (a) Fbn1 genomic locus with exon 1 and 5’ UTR region (gray box); 5’ and 3’ Southern hybridization probes are depicted (striped boxes) along with restriction sites and expected band sizes (B, BamHI and E, EcoRI) ; (b) Fbn1 targeting vector including the neo-cassette and engineered BamHI site within the 3’ loxP site for Southern hybridization genotyping with expected band sizes of BamHI digestion; (c) Fbn1fneo allele with indicated forward (F) and reverse (R) amplification primers for PCR genotyping; (d) Fbn1Lox allele following FLPeR-mediated recombination of Fbn1fneo with expected BamHI band sizes and indicated location of genotyping PCR primers; (e) Fbn1Cmv− allele following CMV-Cre-mediated recombination of Fbn1Lox with indicated location of genotyping PCR primers; (f) Southern blot hybridizations to the 3’ probe of BamHI-digested tail DNA from mice of the indicated genotypes; (g) Southern hybridizations to the 5’ probe of BamHI-digested tail DNA from mice of the indicated genotypes; and (h) PCR genotyping using 3F and 3R primers of tail DNA purified from mice of the indicated genotypes.
Fbn1fneo/fneo mice from Fbn1fneo/+ intercrosses were born at the expected Mendelian ratio but they died from vascular complications by post-natal day 16 (P14). This result indicated that retention of the neo cassette interferes with Fbn1fneo expression similarly to what has been previously reported for other Fbn1 mutant alleles (Pereira et al., 1997 and 1999; Lima et al., 2010). Indeed quantitative (qPCR) analyses of Fbn1 transcripts in Fbn1fneo/fneo tissues confirmed loss of targeted gene expression (data not shown). Fbn1fneo/+ mice were therefore bred with FLPeR mice (Farley et al., 2000) to excise the neo cassette in the germ line and generate Fbn1Lox/+ mice (Fig. 1d), as evidenced by Southern blot analyses of BamHI-cleaved tail DNA (Fig. 1g). Fbn1Lox/+ mice were subsequently crossed with Fbn1+/− mice carrying the CMV-Cre transgene (Schwenk et al., 1995) to delete exon 1 ubiquitously and generate FbnCMV−/− mice, as evidenced by PCR-based genotyping of tail DNA (Fig. 1h).
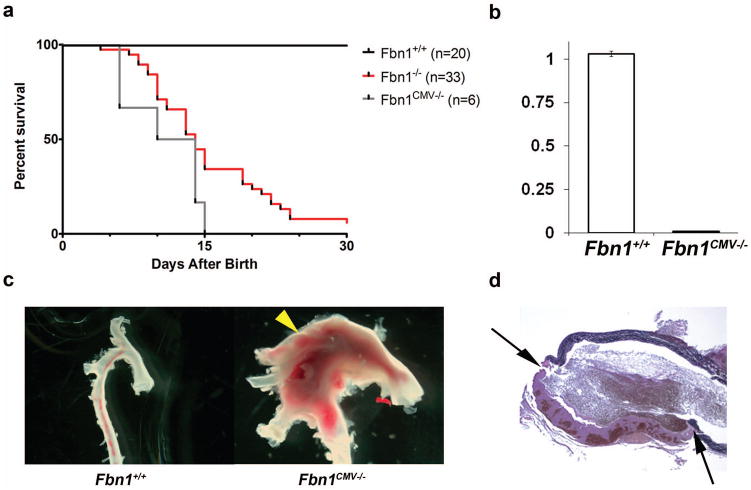
Fbn1CMV−/− mice were born at the expected Mendelian frequency and died by P16 like Fbn1−/−mice (Fig. 2a) (Carta et al., 2006). Post-mortem examination of Fbn1CMV−/− mice revealed pathological features of Fbn1−/− mice (Carta et al., 2006), most notably dilated ascending aortas with hemothorax or hemopericardium that histological analyses correlated with breaching of the aortic wall (Fig. 2c and d). Moreover, qPCR estimates related the vascular phenotype to loss of Fbn1 gene expression (Fig. 2b). Incidentally, Fbn1CMV−/− mice derived from mES clones D7 or H5 displayed identical survival curves, pathological features and lack of Fbn1 gene expression. Altogether these findings therefore equated conditional inactivation with germ line ablation of Fbn1 gene expression and implicitly validated the use of the Fbn1Lox/− mice as a model to study tissue- and stage-specific roles of fibrillin-1 microfibrils.
FIG. 2.
Characterization of Fbn1CMV−/− mice. (a) Survival curves showing the percent survival of various mice in days after birth with indicated the number (n) of animals for each genotype. Fbn1+/+and Fbn1CMV−/− curves are not significantly different from each other (p>0.1). (b) Bar graphs summarizing qPCR analyses of Fbn1 transcripts in neonatal lungs (white, Fbn1+/+ mice; black, Fbn1CMV−/− mice). Data are from 3 independent samples of each genotype assayed in triplicate and including ± SE. (c) Anatomy of aortas collected from P16 Fbn1+/+ and Fbn1CMV−/−mice showing a large dilation (arrowhead) in the latter sample. (c) Illustrative cross-section image of a dilated ascending aorta of Fbn1CMV−/− mice demonstrating a tear through the aortic wall with a large thrombus (between arrows); image taken at 4X.
The above assertion was further explored in conditional Fbn1 inactivation experiments aimed at refining the characterization of reduced bone mass (osteopenia) in MFS mice (Nistala et al., 2010a). Peak bone mass in these earlier studies was examined in Fbn1mgR/mgR rather than Fbn1−/− mice because absence of fibrillin-1 microfibrils leads to neonatal death from ruptured aortic aneurysm (Carta et al., 2006). The analyses have demonstrated that osteopenia in 3 month-old Fbn1mgR/mgR mice is mostly accounted for by increased bone resorption, which cell culture experiments have related to enhanced osteoblast-supported osteoclast activity (Nistala et al., 2010a). Micro-computer tomography (μCT) was employed here to compare bone mass in the femurs of 3 month-old Fbn1mgR/mgR mice and of mice in which Fbn1 was conditionally inactivated in skeletal progenitor cells of the forming limbs by the action of the Prx1-Cre transgene (Fbn1Prx−/−mice; Logan et al., 2002) or in early pre-osteoblasts by the action of the Osx-Cre transgene (Fbn1Osx−/− mice; Rodda and McMahon, 2006). Measurements of bone mineral density (BMD) and bone volume over total volume (BV/TV) as well as average trabecular number (Tb.N.), thickness (Tb.Th.) and separation (Tb. Sp.) revealed that both Fbn1Prx−/− and Fbn1Osx−/− femurs have comparable lower than normal bone mass and worse trabecular architecture than the Fbn1mgR/mgR counterparts (Fig. 3). The latter finding may conceivably reflect the greater negative impact of Fbn1 inactivation than underexpression in bone remodeling. This point notwithstanding, the results imply that normal osteoblast activity depends in part on the continued production of fibrillin-1 microfibrils throughout osteoblastogenesis, from the specification of osteochondral progenitors to the differentiation of osteoblast precursors.
FIG. 3.
Osteopenia in Fbn1 mutant mice. Representative μCT images of distal epiphyses of femurs isolated from 3 month-old mice of the indicated genotypes with bar graphs below summarizing the measurements of average BMD, BV/TV, Tb.Th., Tb.N. and Tb.Sp. values (± SE); sample numbers (n) are also included along with asterisks signifying statistical significance of mutant compared to wild type values. Data from Fbn1mgR/mgR and Fbn1 conditional null mice and their Fbn1+/+ littermates are shown separately because the two sets of animals were respectively examined on pure C57Bl/6 or mixed C57Bl/6;129SvEv genetic backgrounds.
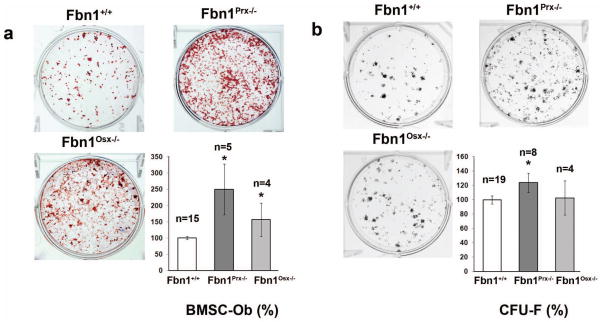
Although unable to assess peak bone mass in adult Fbn1−/− mice, we have nonetheless shown that calvarial osteoblast (cOb) cultures from newborn Fbn1−/− mice differentiate and support osteoclast activity more than those from Fbn1+/+ mice (Nistala et al., 2010b,c). We extended these earlier observations by evaluating osteoblast differentiation of primary cultures of bone marrow stromal cells (BMSCs) derived from the long bones of 3 month-old Fbn1+/+, Fbn1Prx−/− and Fbn1Osx−/− mice. The finding that both mutant cell cultures produce more mineral nodules than wild type BMSC cultures confirmed the negative impact of loss fibrillin-1 synthesis on normal osteoblastogenesis (Fig. 4a). Next, we performed a colony-forming unit fibroblast (CFU-F) efficiency assay to compare the number of functional mesenchymal stem cells in BMSCs isolated from the same wild type and mutant mice (Kuznetsov and Robey, 1996). The experiments documented the ability of BMSCs from 3 month-old Fbn1Prx−/− mice to yield statistically more CFU-Fs than those derived from Fbn1Osx−/− or Fbn1+/+ mice (Fig. 4b). We interpret these results to implicate fibrillin-1 synthesis in modulating the performance of mesenchymal stem cells.
FIG. 4.
Ex vivo cell culture experiments. (a) BSMC differentiation into osteoblasts (BSMC-Ob) showing representative images of alizarin red-stained cultures (top) and bar graphs quantification (± SE) of mineral nodules (below) with indicated sample numbers (n). (b) CFU-F efficiency assays with representative images of crystal violet-stained cell clones generated by BMSCs of the indicated genotypes (top) and bar graphs summarizing the quantification (± SE) of CFU-F data (below) with indicated sample numbers (n). Asterisks in both panels signify statistical significance of mutant data in comparison with wild type values.
Based on the above findings and our previous in vivo and ex vivo studies of Fbn1 deficient mice (Nistala et al., 2010a-c), we conclude that fibrillin-1 microfibrils normally restrict recruitment of osteoprogenitor cells, pre-osteoblast differentiation and osteoblast-supported osteoclast activity. We further speculate that enhanced osteoblast-dependent osteoclast activity together with premature depletion of osteoprogenitor cells may account for progressive bone loss in MFS. The availability of Fbn1Lox/− mice will enable us to test this and related hypotheses and ultimately delineate the full contribution of extracellular fibrillin-1 assemblies to skeletogenesis and bone physiology.
METHODS
Generation of Fbn1 conditional mutant mice
A bacterial artificial chromosome (BAC) clone containing the mouse Fbn1 gene was obtained by screening a 129SvEv genomic library. Gene Bridges engineered the targeting vector by inserting the 5’ loxP site into the BAC clone, excising an appropriate 18 kb BAC fragment and sub-cloned it into a minimal vector system in which a neo-expressing cassette with a 3’ loxP site was inserted immediately downstream of exon 1. The targeting vector was sequenced, linearized and electroporated into R1 mES cells by InGenious Targeting Laboratory; positively targeted ES clones were identified and characterized as previously described (Pereira et al., 1997 and 1999). Likewise, generation of chimeric mice and assessment of germ line transmission of the targeted Fbn1 allele were performed in our Mouse Genetics Core Facility (Director Dr. K. Kelley) according to our published protocols (Pereira et al., 1997 and 1999).
Genotype and analysis of Fbn1 conditional null mice
Fbn1Lox/+ mice were genotyped using forward primer 1F (AGAAGAGGCGGCGGAGGACACGAT) and reverse primer 3R (TGCCAAGCCTTAGACAGCCAACCT). Amplifications conditions included denaturation at 95°C per 5 min, annealing and amplification at 95°C per 1 min, 61°C per 1 min, 72°C per 1 min for 34 cycles and 72°C per 10 min extension. Forward primer 3F (CAGGACAGCCGGACAGTGAGACAG) was used in combination with reverse primer 3R to monitor deletion of exon 1 using the following amplification conditions: denaturation at 95°C per 5 min, annealing and amplification at 95°C per 1 min, 60°C per 1 min, 72°C per 1 min for 34 cycles and at 72°C per 10 min extension. CMV-Cre and Prx-Cre transgenic mice were purchased from Jackson Laboratories and FLPeR mice from InGenious Targeting Laboratory; transgenic mice were genotyped using PCR amplification primers according to instructions provided by the companies. Osx-Cre mice, a generous gift of Dr. A. McMahon, were genotyped as described (Rodda and McMahon, 2006). Survival curves were generated using GraphPad (Prism); aortas were isolated and imaged under a dissecting microscope and sectioned and stained (Carta et al., 2006); and RNA isolated from neonatal lungs was subject to qPCR analysis (Nistala et al., 2010b). Comparative RNA quantification was performed using three independent samples per genotype each run in triplicate; statistical significance was evaluated by an unpaired t test, assuming significance with p < 0.05.
BMSC differentiation and CFU-F efficiency ex vivo assays
A modification of the technique originally described by Kuznetsov and Robey (1996) was used for the CFU-F efficiency assays. Briefly, the femurs and tibias of wild type and Fbn1 mutant mice were surgically removed and marrow cells were flushed from the bone shafts. Nucleate marrow cells were plated at 4x104 per cm2 in presence of marrow non nucleated cells. The cells were maintained at 37°C in a sterile and humidified atmosphere of 5% CO2 and cultured in α-MEM containing 20% not heat-inactivated FBS supplemented with streptomycin, penicillin, and fungizone (Invitrogen/Gibco). After 10 days cells were rinsed with PBS to remove non-adherent cells, fixed with paraformaldehyde and stained with an aqueous solution of saturated crystal violet (Sigma). Colony-forming ability was evaluated by enumeration of colonies containing more than 50 cells. For primary BMSC differentiation, cells were cultured in α-MEM containing 10% FBS containing 1% penicillin/streptomycin (Invitrogen/Gibco). Once confluent, cells were induced to differentiate by supplementing the culture medium with 50 μg/ml of ascorbic acid and 10 mM of β-glycerophosphate (Nistala et al., 2010a,b). Resulting mineral deposits were visualized by alizarin red staining (Sigma) and quantified with MetaMorph imaging software (MDS Analytical Technologies). Statistical significance was evaluated by an unpaired t test, assuming significance with p < 0.05.
Bone μCT analyses
Formalin-fixed femurs of wild type and mutant mice were isolated and scanned using Skyscan 1172 μCT system with an isotropic voxel resolution of 6.7 μm. Scans were normalized using a density calibration phantom containing air, water and a hydroxyapatite standard rods of 0.25g/cm3 and 0.75 g/cm3 (Skyscan) to determine BMD and BV/TV. Images were analyzed using Skyscan data acquisition software (version 1.5), NRecon image reconstruction software (version 1.6.4.1) and μCT analyzer 1.11 visualization and analysis software (Skyscan). Analyses of reconstructed bone μCT images were carried out as previously described (Nistala et al., 2010a,b); statistical significance was evaluated by an unpaired t test, assuming significance with p < 0.05.
Acknowledgments
Contract grant sponsors: NIH/NIAMS to F.R. (contract grant number: R01AR42044), NIH/ NIGMS-NHLBI to J.R.C. (contract grant number: T32GM007280) and the National Marfan Foundation to F.R.
We are indebted to Drs. A. McMahon and F. Laub for providing Osx-Cre mice and designing the targeting strategy, respectively, and to Dr. M. Schaffler for invaluable assistance with the μCT analyses. We also thank Ms. C. Liu and J. Basta-Pljakic for excellent technical support and Ms. K. Johnson for organizing the manuscript.
LITERATURE CITED
- Carta L, Pereira L, Arteaga-Solis E, Lee-Arteaga SY, Lenart B, Starcher B, Merkel CA, Sukoyan M, Kerkis A, Hazeki N, Keene DR, Sakai LY, Ramirez F. Fibrillins 1 and 2 perform partially overlapping functions during aortic development. J Biol Chem. 2006;281:8016–8023. doi: 10.1074/jbc.M511599200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Habashi JP, Judge DP, Holm TM, Cohn RD, Loeys BL, Cooper TK, Myers L, Klein EC, Liu G, Calvi C, Podowski M, Nepturne ER, Halushka MK, Bedja D, Gabrielson K, Rifkin DB, Carta L, Ramirez F, Huso DL, Dietz HC. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312:117–121. doi: 10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm TM, Habashi JP, Doyle JJ, Bedja D, Chen Y, van Erp C, Lyndsay ME, Kim D, Schoenhoff F, Cohn RD, Loeys BL, Thomas CJ, Marugan JJ, Judge DP, Dietz HC. Noncanonical TGFβ signaling contributes to aortic aneurysm progression in Marfan syndrome mice. Science. 2011;332:358–361. doi: 10.1126/science.1192149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov S, Robey PG. Species differences in growth requirements for bone marrow stromal fibroblast colony formation In Vitro. Calcif Tissue Int. 1996;59:265–270. doi: 10.1007/s002239900121. [DOI] [PubMed] [Google Scholar]
- Lima BL, Santos EJ, Fernandes GR, Merkel C, Mello MR, Gomes JP, Soukoyan M, Kerkis A, Massironi SM, Visintin JA, Pereira LV. A new mouse model for Marfan syndrome presents phenotypic variability associated with the genetic background and overall levels of Fbn1 expression. PLosOne. 2010;5:e14136. doi: 10.1371/journal.pone.0014136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- Nistala H, Lee-Arteaga S, Carta L, Cook JR, Smaldone S, Siciliano G, Rifkin AN, Dietz HC, Rifkin DB, Ramirez F. Differential effects of alendronate and losartan therapy on osteopenia and aortic aneurysm in mice with severe Marfan syndrome. Hum Mol Genet. 2010a;19:4790–4798. doi: 10.1093/hmg/ddq409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistala H, Lee-Arteaga S, Smaldone S, Siciliano G, Carta L, Ono RN, Sengle G, Arteaga-Solis E, Levasseur R, Ducy P, Sakai LY, Karsenty G, Ramirez F. Fibrillin-1 and -2 differentially modulate endogenous TGF-β and BMP bioavailability during bone formation. J Cell Biol. 2010b;190:1107–1121. doi: 10.1083/jcb.201003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nistala H, Lee-Arteaga S, Smaldone S, Siciliano G, Ramirez F. Extracellular microfibrils control osteoblast-supported osteoclastogenesis by restricting TGFβ stimulation of RANKL production. J Biol Chem. 2010c;285:34126–34133. doi: 10.1074/jbc.M110.125328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Andrikopoulos K, Tian J, Lee SY, Keene DR, Ono R, Reinhardt DP, Sakai LY, Jensen-Biery N, Bunton T, Dietz HC, Ramirez F. Targeting of the gene coding fibrillin-1 recapitulates the vascular phenotype of Marfan syndrome in the mouse. Nat Genet. 1997;17:218–222. doi: 10.1038/ng1097-218. [DOI] [PubMed] [Google Scholar]
- Pereira L, Lee SY, Gayraud B, Andrikopoulos K, Shapiro SD, Bunton T, Biery NJ, Dietz HC, Sakai LY, Ramirez F. Pathogenetic sequence for aneurysm revealed in mice underexpressing fibrillin-1. Proc Natl Acad Sci, USA. 1999;96:3819–3823. doi: 10.1073/pnas.96.7.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Dietz HC. Marfan syndrome: from molecular pathogenesis to clinical treatment. Curr Opin Genet Dev. 2007;17:252–258. doi: 10.1016/j.gde.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Ramirez F, Rifkin DB. Extracellular microfibrils: contextual platforms for TGFβ and BMP signaling. Curr Opin Cell Biol. 2009;21:616–622. doi: 10.1016/j.ceb.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]