Abstract
MicroRNAs (miRNAs) are short RNAs with essential roles in gene regulation in various organisms including higher plants. In contrast to the vast information on miRNAs from many economically important plants, almost nothing has been reported on the identification or analysis of miRNAs from rubber tree (Hevea brasiliensis L.), the most important natural rubber-producing crop. To identify miRNAs and their target genes in rubber tree, high throughput sequencing combined with a computational approach was performed. Four small RNA libraries were constructed for deep sequencing from mature and young leaves of two rubber tree clones, PB 260 and PB 217, which provide high and low latex yield, respectively. 115 miRNAs belonging to 56 known miRNA families were identified, and northern hybridization validated miRNA expression and revealed developmental stage-dependent and clone-specific expression for some miRNAs. We took advantage of the newly released rubber tree genome assembly and predicted 20 novel miRNAs. Further computational analysis uncovered potential targets of the known and novel miRNAs. Predicted target genes included not only transcription factors but also genes involved in various biological processes including stress responses, primary and secondary metabolism, and signal transduction. In particular, genes with roles in rubber biosynthesis are predicted targets of miRNAs. This study provides a basic catalog of miRNAs and their targets in rubber tree to facilitate future improvement and exploitation of rubber tree.
Keywords: miRNAs, Rubber tree, miR396, miR172, Rubber elongation factor, Hevamine A
Introduction
microRNAs (miRNAs) are a class of short, single-stranded, non-coding RNAs in plants and animals that are derived from primary miRNAs (pri-miRNAs) transcribed from specific MIR genes (see Chen 2009, Jones-Rhoades et al. 2006, and Voinnet 2009 for reviews on plant miRNAs). In plants, pri-miRNAs are processed by DICER-LIKE1 (DCL1) to produce 60–300 nucleotide (nt) hairpin RNAs known as pre-miRNAs, which are further processed into miRNA:miRNA* duplexes with 2 nt 3′ overhangs. A mature miRNA is derived from one strand of the duplex and bound by an argonaute protein, guiding the complex to target messages to effect posttranscriptional silencing. In plants, miRNAs guide the precise cleavage of their target RNAs or lead to their translational inhibition. Within the repertoire of miRNA species in any given organism, some miRNAs are conserved across large evolutionary distances while others are restricted to a particular species or lineage. In plants, roughly 20 miRNA families are conserved in angiosperms. Functional analyses of conserved miRNAs revealed their involvement in multiple developmental processes including leaf development and polarity, hormone signaling, meristem boundary formation and organ separation, lateral root formation, and floral organ identity specification (Chen 2009; Zhang et al. 2006). They also regulate plants’ responses to environmental stimuli such as biotic and abiotic stresses (Shukla et al. 2008; Sunkar 2010; Sunkar et al. 2007). The evolutionary conservation of some miRNAs across diverse plant lineages enabled a powerful bioinformatics approach that predicts miRNAs from publicly available expressed sequence tag (EST) databases (Zhang et al. 2005). With this method, the number of known plant miRNAs has been dramatically increased in diverse plant species. However, this method is limited by the amount of genome sequence data available, especially for plants in the economically important Euphorbiaceae family.
Rubber tree (Hevea brasiliensis L.), belonging to the family Euphorbiaceae, is of major economic importance because its sap-like extract (known as latex) is the primary source of natural rubber. Natural rubber consists mainly of cis-polyisoprene, which is the most widely used natural plant polymer. Natural rubber has superior properties compared to synthetic rubber such as resistance to abrasion and impact, elasticity, efficient heat dispersal, and resilience and malleability at low temperature. These properties are lacking in synthetic rubber probably because they result from the unique secondary compounds (lipids, carbohydrates, and minerals) in rubber tree (Mooney 2009).
Identification of miRNAs and their target genes is essential for a full understanding of gene expression in plants during development and stress responses. Direct cloning followed by high throughput sequencing is considered the most effective way for miRNA discovery in plants (Lu et al. 2005), especially in plants without completely sequenced genomes. This powerful technology enables the discovery of conserved miRNAs as well as species-specific miRNAs. Moreover, the read frequencies can reflect quantities such that the technology can be utilized for expression profiling.
Recently, Zeng et al. (2010) performed a genome-scale systematic study of miRNAs in Euphorbiaceae, by combining computational prediction and experimental analysis. They predicted 85 conserved miRNAs in castor bean (Ricinus communis), and experimentally verified and characterized 58 of the 85 miRNAs in at least 1 of 4 Euphorbiaceous plants: castor bean, cassava (Manihot esculenta Crantz), jatropha (Jatropha curcas), and rubber tree during seedling development.
To obtain a better knowledge of miRNAs and their target genes in rubber tree, we combined high throughput sequencing and bioinformatics analyses to identify miRNAs in rubber tree. In this study, we identify 115 known miRNAs that belong to 56 miRNA families and 20 novel miRNAs that do not match any known miRNAs in miRBase (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008). Targets of the known and novel miRNAs have been predicted, and genes acting in rubber biosynthesis are putative miRNA targets. Some miRNAs exhibit developmental stage-dependent or clone-specific accumulation patterns.
Materials and Methods
RNA extraction
Total RNA was extracted from young and mature leaves of the PB260 and PB217 clones of rubber tree using plant RNA isolation reagent (PRIR) (Invitrogen, USA). In brief, 1 g of tissue was ground in a frozen mortar until it became a fine powder. The frozen powder was transferred into a tube and resuspended in 5 ml of extraction buffer. The mixture was incubated for 5 min at room temperature and centrifuged for 10 min at 4 °C at 12,000g to precipitate insoluble material and the clarified supernatant was transferred to a clean tube. The supernatant was mixed with 2 ml of 5 M NaCl and 6 ml of chloroform by vortexing. The mixture was centrifuged for 10 min at 4 °C at 12,000g to separate the 2 phases; the top phase was then transferred to a new tube. The RNA was precipitated by adding 0.9 volumes of isopropanol followed by a 10 min incubation at room temperature. The mixture was centrifuged for 30 min at 4 °C at 12,000g and the pellet was washed with 5 ml of cold 75 % ethanol. The RNA pellets were dried and dissolved in RNase-free water. The total RNA was further assessed for quantity and quality using Nanodrop ND-1000 (Nanodrop technologies, USA) and denaturing gel electrophoresis and then stored at −80 °C.
Construction of small RNA libraries
500 μg of total RNA was precipitated with ethanol. The pellet was washed with 70 % ethanol, dried, and dissolved in RNase-free water. The quality and quantity of total RNA were subsequently determined by Nanodrop and denaturing agarose gel electrophoresis.
500 μg of RNA was resolved in a 15 % polyacrylamide/8 M urea/0.5x TBE gel. A gel slice corresponding to small RNAs between 18 and 30 nt was excised and the RNA was eluted and dissolved in 20-μl RNase-free water. Small RNAs were then ligated to a 3′ adaptor using T4 RNA ligase and the ligated species were recovered after size fractionation by denaturing polyacrylamide gel electrophoresis. The 5′ adaptor was then ligated and the ligated products were similarly recovered. The ligated small RNAs were reverse-transcribed to cDNA and amplified by PCR. The amplified products were purified by gel electrophoresis and sequenced in the Hiseq2000 (Illumina). In this study, libraries from young and mature leaves of PB217 and PB260 were separately constructed with 5′ adaptors containing different bar codes, mixed and sequenced together in one lane. The unique barcodes allowed the identification of reads from the different sources.
Identification of known and novel miRNAs in rubber tree
The raw sequences were filtered to remove low quality reads and the ones that passed the quality filter were trimmed to remove the adaptor sequences. Next, sequences of 20–24 nt that are represented by at least ten reads in all four libraries combined were selected as the raw small RNA sequences. The raw small RNA sequences were mapped to the miRNA database (http://www.mirbase.org/) (Griffiths-Jones 2004; Griffiths-Jones et al. 2006, 2008; Kozomara and Griffiths-Jones 2011) using the ssearch36 software in FASTA36 (Pearson 1991; Smith and Waterman 1981). Sequences containing not more than 2 nt mismatches were considered as known miRNAs in rubber tree.
The prediction of novel miRNAs was performed with a bioinformatics pipeline as described (Barrera-Figueroa et al. 2011). In brief, the raw small RNA sequences were mapped to the newly assembled rubber tree genome (http://www4a.biotec.or.th/rubber/Search) as well as to publicly available expressed sequence tags (ESTs) from rubber tree. The rubber tree sequences (ESTs or genomic sequences) with small RNA matches were interrogated in sliding windows for the potential to form secondary structures using UNAFold (Markham and Zuker 2008). The structures were further examined for free energy, mismatches between the miRNA and miRNA*, the number of asymmetric bulges in the stem region, dominance of the miRNA relative to other small RNAs in abundance, and precise cleavage as revealed by the presence of miRNA and miRNA* sequences, to satisfy the criteria defined for plant miRNA annotation (Meyers et al. 2008).
Target prediction
The potential targets of rubber tree miRNAs were predicted using the psRNATarget program (http://plantgrn.noble.org/psRNATarget/) (Dai and Zhao 2011) with default parameters. The program uses a scale of 0–5 to indicate the stringency of miRNA-target pairing with the smaller numbers representing higher stringency. A score of 3 or 3.5 was used in our target prediction. Since the EST database from rubber tree is much limited in size, the EST databases for leafy spurge (Euphorbia esula DFCI gene index release 1) and cassava (Manihot esculenta DFCI gene index release 1) were also used in the target searches.
Northern blot analysis
Total RNA isolation was performed using Plant RNA Isolation Reagent (Invitrogen) according to the manufacturer’s instructions. For northern blot analysis, 10 μg of total RNA was resolved by electrophoresis in 15 % polyacrylamide/8 M urea/0.5x TBE gels. The RNA was transferred to Hybond N+ membrane and probed with labeled 32P DNA oligonucleotides complementary to the miRNA sequence. Hybridizations were performed at 50 °C overnight. After hybridization, membranes were washed twice in 2x SSC/0.1 % SDS buffer and analyzed using a PhosphorImager (Amersham). Where indicated, membranes were stripped, re-exposed to a PhosphorImager screen to ensure complete signal removal and reused for a second hybridization. A DNA oligonucleotide complementary to 5S rRNA was used as a probe to detect 5S rRNA as an internal loading control. Sequences of the DNA oligonucleotide probes used for northern blotting are shown in Supplementary Table 6.
miRNA detection by stem–loop RT-PCR
miRNA detection by stem–loop RT-PCR was performed as described (Varkonyi-Gasic et al. 2007). DNase-treated total RNA from mature and young leaves from PB260 and PB217 clones were reverse transcribed to cDNAs using a stem–loop RT primer. The stem–loop RT, forward, and reverse primers were designed according to criteria previously described (Varkonyi-Gasic et al. 2007) and their sequences are shown in Supplementary Table 6. To detect novel miRNAs, the PCR reactions were conducted with the following schemes: 94 °C for 2 min, followed by 28–35 cycles of 94 °C for 15 s and 60–64 °C for 1 min. The expected size of the PCR product obtained from this method should be ~60 bp.
Results
Construction and sequencing of rubber tree small RNA libraries
In general, two major approaches, computational and experimental, are used separately or in combination to identify miRNAs in plants. For rubber tree, pure computational approaches are limited by the lack of a completely sequenced and assembled genome and the small number of available ESTs such that the identification of all known conserved miRNA families or the prediction of novel miRNAs in rubber tree is not possible. Here we employed experimental cloning followed by high throughput sequencing to interrogate the small RNA repertoire of rubber tree. miRNAs were then identified from the small RNA reads based on their sequence similarity with known miRNAs. De novo miRNA prediction was also applied to the small RNA reads using the newly available, partial rubber tree genomic sequence assemblies (GSS) and the limited rubber tree EST database to identify novel miRNAs.
We constructed small RNA libraries using total RNAs extracted from young and mature leaves from two rubber tree clones, PB260 with a high latex yield and PB217 with a low latex yield. The four libraries were barcoded, mixed and sequenced on an Illumina HiSeq2000. A total of 203,588,835 sequences were recovered from the four libraries. The raw sequences were computationally analyzed to remove low quality sequences, adaptors, and reads shorter than 18 nt after adaptor removal to yield 69,205,862 small RNA sequences. As shown in Fig. 1, approximately 70 % of the small RNAs ranged 20–24 nt in length, with the 24 nt class being the highest in total abundance (21.5 %) followed by the 21 nt (18.6 %) and 18 nt classes (12.8 %). The size distribution of small RNAs from rubber tree mimics that of Arabidopsis (Kasschau et al. 2007).
Fig. 1.
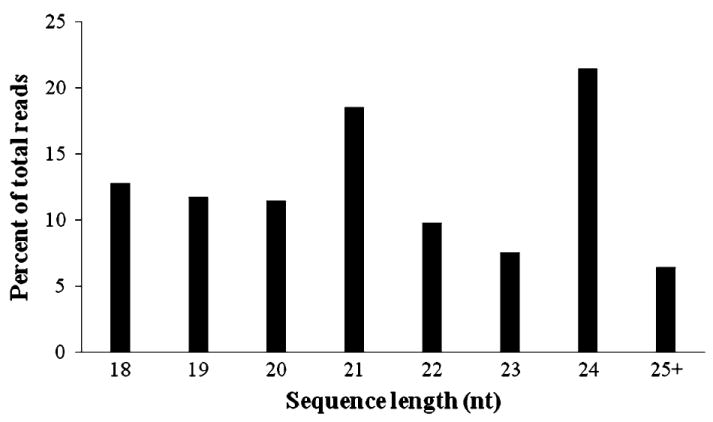
Size distribution of small RNAs in rubber tree leaves. The percentage of small RNAs of a particular length in the total small RNA population is shown
Identification of known miRNAs in rubber tree
To identify miRNAs in rubber tree that are already known in other species, we searched the small RNA sequences of 20–24 nt in length for matches to mature miRNA sequences deposited in miRBase. We obtained a total of 115 potential miRNAs representing 56 miRNA families in rubber tree (Supplemental Table 1). The identified miRNA families are conserved in dicotyledonous plants only, both dicotyledonous and monocotyledonous plants, or in most land plants (Supplemental Table 1). Rubber tree miRNA families showed obvious differences in abundance between each other, as reflected by the read frequencies in the libraries (Supplemental Table 1; Fig. 2). For instance, reads for hbr-miR396, hbr-miR159, and hbr-miR167 were at 48,807, 43,127, and 18,330 transcripts per million (TPM), respectively; whereas the hbr-miR169 family was of relatively low abundance (113 TPM). The read frequencies indicated that miR396 (48,807 TPM) had the highest expression level in rubber tree leaves followed by miR159 (43,127 TPM), miR167 (18,330 TPM), miR166 (7,248 TPM), and miR858 (5,005 TPM) (Fig. 2).
Fig. 2.
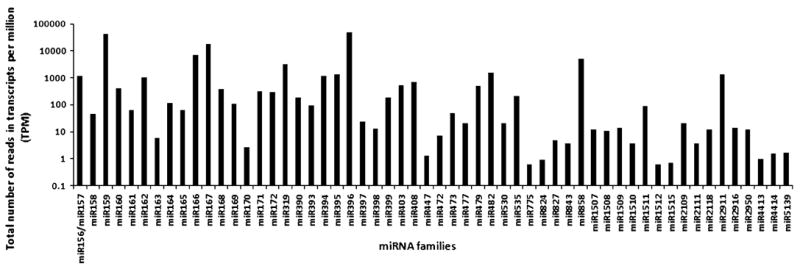
The read frequencies of conserved miRNAs in rubber tree leaves. The total number of reads corresponding to a particular miRNA family was normalized against the number of total small RNA reads and expressed in transcripts per million (TPM)
The sensitivity of the high throughput sequencing technology also enabled the detection of miRNA*, the strand that pairs with the miRNA in the DCL1 product. 35 distinct miRNA* sequences representing 23 miRNA* families were found in the four libraries (Supplemental Table 2). The determination of which strand of the miRNA:miRNA* duplex is incorporated into the RNA-induced silencing complex (RISC), is largely based on the identity of the first nucleotide in plants. Plant miRNAs tend to begin with a U, a preferred 5′-nucleotide for AGO1, the major miRNA effector (Baumberger and Baulcombe 2005; Mi et al. 2008). The miRNA* is usually not incorporated into RISC and is typically degraded and often not detectable in vivo. Most of the detected miRNA*s did not begin with a 5′ U, suggesting that they may not be bound by AGO1. Surprisingly, miR170*/171* (the two have the same sequence and are thus grouped together) was represented by more reads (1,748 TPM) than miR170 and miR171 (316 TPM as the sum of the two) in the four libraries combined. This raises the possibility that both strands of miR170/miR170* and miR171/miR171* have biological functions. Intriguingly, miR170*/171* reads were extremely enriched in young leaves of the rubber tree clone 260 (Supplemental Table 2), suggesting that the retention of star strands can occur in a developmental stage- and clone-specific manner.
Target prediction for known miRNAs in rubber tree
For most plant miRNAs, targets with nearly perfect complementarity can be identified with various algorithms. We employed psRNATarget (Dai and Zhao 2011) to predict the potential targets for conserved miRNAs in rubber tree. We used an expectation score of 3 or 3.5 (in a scale of 0–5 with the smaller numbers representing higher stringency) as the cutoff in the prediction. We first used the EST database of rubber tree (ESTTIK) for target finding because a complete genome sequence with annotated transcripts is not available. Due to the limited size of the EST database, potential targets were only found for 18 and 20 of the 56 miRNAs at the expectation scores of 3 and 3.5, respectively (Supplemental Table 3).
To better understand the targeting potential of the rubber tree miRNAs, we resorted to target prediction using the larger EST databases from two related plants in the same Euphorbiaceae family, Manihot esculenta (cassava) and Euphorbia esula (leafy spurge) with the assumption that the conserved miRNAs should have conserved targets in these related species. This effort identified potential target genes for 51 of the 56 miRNA families at the expectation score of 3.0 (Supplemental Table 3). When the expectation score was raised to 3.5, potential targets were also identified for the remaining five miRNA families (Supplemental Table 3). Of these predicted targets, a large category consists of transcription factor genes. Genes encoding Squamosa Promoter Binding protein, type III HD-Zip protein, Nuclear Transcription Factor Y subunit A (NFYA), PHAP2B and LIPLESS (AP2-domain containing proteins), a homeodomain protein, a Myb transcription factor, and AUX/IAA proteins were predicted as targets of miR156/157, miR165/166, miR169, miR172, miR396, miR858, and miR1510, respectively (Supplemental Table 3). A second large group consisted of genes related to plant defense and stress responses. For example, genes encoding glutathione-S-transferase, superoxide dismutase, and a hypoxia-responsive family protein were predicted targets of miR162, miR398, and miR535, respectively (Supplemental Table 3). Moreover, disease resistance genes are among the predicted targets of miR399, miR1507, and miR1510 (Supplemental Table 3). A third group of miRNA targets consisted of genes involved in signal transduction, especially in hormone signaling pathways. For instance, the auxin receptor TIR1 was a predicted target of miR393. A gene annotated as “abscisic acid responsive element-binding protein 2” was a potential target of miR482. A gene encoding a mitogen-activated protein kinase kinase kinase was predicted to be a target of miR472. We also found some miRNAs with potential roles in nutrient assimilation: miR395, miR399, and miR1508 could target ATP-sulfurylase, phosphate transporter, and nitrate reductase genes, respectively. Interestingly, rubber elongation factor and Hevamine A, genes involved in rubber biosynthesis, were predicted targets of miR172 and miR160, respectively (Supplemental Table 3; Fig. 3).
Fig. 3.

Diagrams of miRNA-target pairing between two conserved miRNAs (miR160 and miR172) and 20 novel miRNAs and their selected predicted targets. A–U and G–C hydrogen bonds are represented by two dots and the G–U hydrogen bonds are represented by a single dot. The numbers in the target genes represent the positions of the target sites within the indicated EST sequences. EC606382 is a GenBank accession number for a rubber tree EST. EC606382 encodes a protein identical to rubber elongation factor (GenBank accession P15252 from Hevea brasiliensis). The accession numbers for the other targets are from either the Euphorbia esula (leafy spurge) DFCI gene index release 1 or the Manihot esculenta (cassava) DFCI gene index release 1. TC12051 is a cassava EST that encodes a protein homologous to Hevamine A from rubber tree (GenBank accession P23472 from Hevea brasiliensis)
We also attempted to predict targets for the detected miRNA* species in rubber tree small RNA libraries. Surprisingly, potential targets could be predicted for all 23 miRNA* families including miR170*/171* (Supplemental Table 4).
Discovery of novel rubber tree miRNAs
In the initial phase of our studies, our ability to predict miRNAs de novo was limited by the lack of a sequenced and assembled rubber tree genome. But we still attempted to predict novel miRNAs with the rubber tree EST database. Small RNA reads matching to the ESTs were interrogated with a bioinformatics pipeline (Barrera-Figueroa et al. 2011) that follows the updated plant miRNA annotation criteria (Meyers et al. 2008) for miRNA prediction. In total, 8,15,937 reads representing 5,471 unique sequences matched to the EST database and only 2 emerged from the pipeline as potential miRNAs. One was miR166 and the other did not match to any known miRNAs in miRBase, and is therefore a potentially novel miRNA (hbr-candidate 1; Table 1; Supplemental Figure 1). We did not find any targets for this miRNA from the rubber tree ESTs, but two potential targets were predicted from the leafy spurge and cassava ESTs (Supplemental Table 5; Fig. 3).
Table 1.
Predicted novel miRNAs from rubber tree
| miRNA_ID | Mature miRNA | Length | Databasea | Read frequency (in TPM)
|
|||
|---|---|---|---|---|---|---|---|
| 217_m | 217_y | 260_m | 260_y | ||||
| Hbr-cand01 | AUUGGAUAACGGGAUUACAUGGU | 23 | Rubber tree EST | 0 | 0.2 | 0.3 | 0.3 |
| Hbr-cand02 | GAAAUGGGUGGAUGGGAGUGA | 21 | Rubber tree genome | 0.3 | 1.3 | 0.2 | 45.6 |
| Hbr-cand03 | UACCCAGGUGGAAGCUUUGA | 20 | Rubber tree genome | 0.2 | 0.2 | 0.3 | 0.1 |
| Hbr-cand04 | UCUUGGUGCGGUGGACUGCUG | 21 | Rubber tree genome | 0 | 0.2 | 0.2 | 0.9 |
| Hbr-cand05 | AGAUGGGUGGCUGGGCAAGAAG | 22 | Rubber tree genome | 108.2 | 4,322.5 | 108.3 | 4,590.1 |
| Hbr-cand06 | UAGUAUUUCUCUUUUCUCUCU | 21 | Rubber tree genome | 0.9 | 0 | 1.1 | 4.7 |
| Hbr-cand07 | UCAAUAUACUGGUGAUGUGAU | 21 | Rubber tree genome | 0.3 | 0 | 0.2 | 0.1 |
| Hbr-cand08 | CAAGAAACAGAAGAGAGGGAU | 21 | Rubber tree genome | 0 | 0 | 0 | 0.9 |
| Hbr-cand09 | AUGUGGAUUGCUGAAGGCUUU | 21 | Rubber tree genome | 0.1 | 0 | 0 | 0.5 |
| Hbr-cand10 | CACAGCUUUCUUGAACUUUCU | 21 | Rubber tree genome | 8.7 | 9.5 | 7 | 20.8 |
| Hbr-cand11 | UGGACCGUGUUUAAGUAAGAA | 21 | Rubber tree genome | 0.3 | 0 | 0.9 | 0.6 |
| Hbr-cand12 | AGCCGUAAACGAUGGAUACU | 20 | Rubber tree genome | 21.6 | 21.4 | 10.2 | 0.8 |
| Hbr-cand13 | GGAGCGACCUGAGAUCACAUG | 21 | Rubber tree genome | 6.1 | 4.2 | 3.3 | 1.8 |
| Hbr-cand14 | AUGCUAAGCUGUCAGACUUUG | 21 | Rubber tree genome | 0.1 | 0.2 | 0.2 | 0.8 |
| Hbr-cand15 | UGGUGGCUUUUAGGGCUCAAG | 21 | Rubber tree genome | 0.1 | 0 | 0 | 0.7 |
| Hbr-cand16 | UUUGUGUGGACUUUGUAGGUG | 21 | Rubber tree genome | 0.4 | 0.4 | 0 | 3.4 |
| Hbr-cand17 | UUUCCGAAACCUCCAAUUCCAA | 22 | Rubber tree genome | 14.8 | 9.4 | 32.3 | 22.1 |
| Hbr-cand18 | CGGAGGCUACGAGGAAUUCGG | 21 | Rubber tree genome | 1.8 | 0.2 | 0.5 | 0 |
| Hbr-cand19 | UUUGGUGUUGUACUUUUAGAA | 21 | Rubber tree genome | 48.7 | 44.9 | 85 | 622.7 |
| Hbr-cand20 | UGUAGUCGGGUAAGGGGUGUCACA | 24 | Rubber tree genome | 0 | 0.5 | 0.2 | 1.8 |
TPM transcripts per million, m mature leaves, y young leaves, 217 and 260 two rubber tree clones
The databases from which the miRNAs were predicted
A partially sequenced and assembled rubber tree genome became available recently. We took advantage of this genomic resource to predict novel miRNAs from the high-throughput sequencing data. 11,708,729 reads representing 63,513 unique reads were mapped to the partial rubber tree genome. 37 potential miRNAs were predicted from these reads, among which 16 were known miRNAs. Among the 21 that did not match any known miRNAs in miRbase, 2 were removed due to suboptimal precursor structures to result in 19 candidate novel miRNAs (hbr-candidates 2–20; Table 1). Altogether, 20 novel miRNAs were predicted from rubber tree ESTs and genomic sequences (Table 1). The predicted targets of the 20 novel miRNAs included metabolic enzymes, transcription factors, and protein kinases (Supplemental Table 5; Fig. 3).
Accumulation of miRNAs in rubber tree
To confirm the in vivo accumulation and to examine the accumulation patterns of potential miRNAs, 19 miRNAs identified through sequence homology to known miRNAs, including 18 widely conserved miRNAs and a legume-specific miRNA (miR1511), were analyzed in leaves at different developmental stages and in the two rubber tree clones. Northern blot analysis was performed using antisense probe sequences from Arabidopsis miRNAs (Fig. 4). The expression was normalized to the abundance of 5S rRNA present in each RNA sample and the relative levels are shown in Supplemental Figure 2.
Fig. 4.
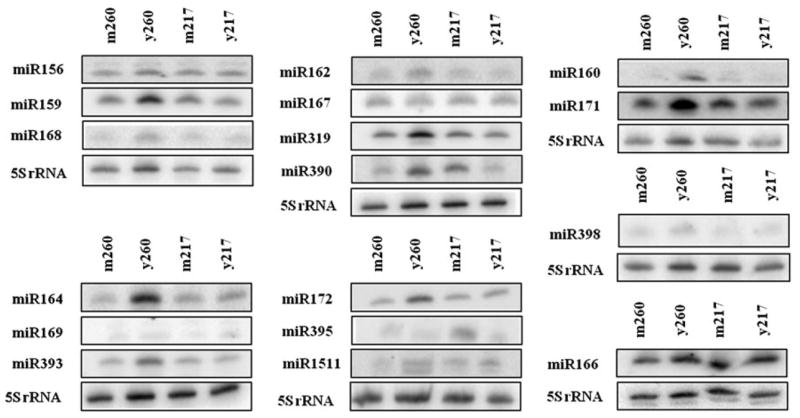
Northern blot analysis of rubber tree miRNA accumulation in leaves. Total RNA (10 μg) from mature (m) and young (y) leaves from two rubber tree clones PB260 (260) and PB217 (217) was resolved in 15 % polyacrylamide/8 M urea/0.5x TBE gels and subsequently transferred to membranes for northern blot analysis. Membranes were hybridized with an oligonucleotide antisense to each miRNA. 5S rRNA served as a loading control. In some cases, membranes were stripped and used for several hybridizations such that one 5S blot was the loading control for all the miRNAs shown above
For all miRNAs examined, a signal was detected in at least one of the four samples, which confirmed their accumulation in vivo. Most of the miRNAs displayed clone-specific and/or developmental stage-specific expression patterns. For example, miR160, miR164, and miR319 were particularly enriched in young leaves of the clone PB260 (Fig. 4; Supplemental Figure 2). miR395 accumulated to higher levels in mature leaves than in young leaves in the clone PB217 (Fig. 4; Supplemental Figure 2).
We also examined the accumulation of four randomly chosen novel rubber tree miRNAs using a sensitive stem–loop RT-PCR approach (Varkonyi-Gasic et al. 2007). All four miRNAs accumulated in vivo (Fig. 5). Hbr-cand02, hbr-cand05 and hbr-cand12 were present in all four rubber tree samples, but hbr-cand13 was only present in the two mature leaf samples (Fig. 5).
Fig. 5.
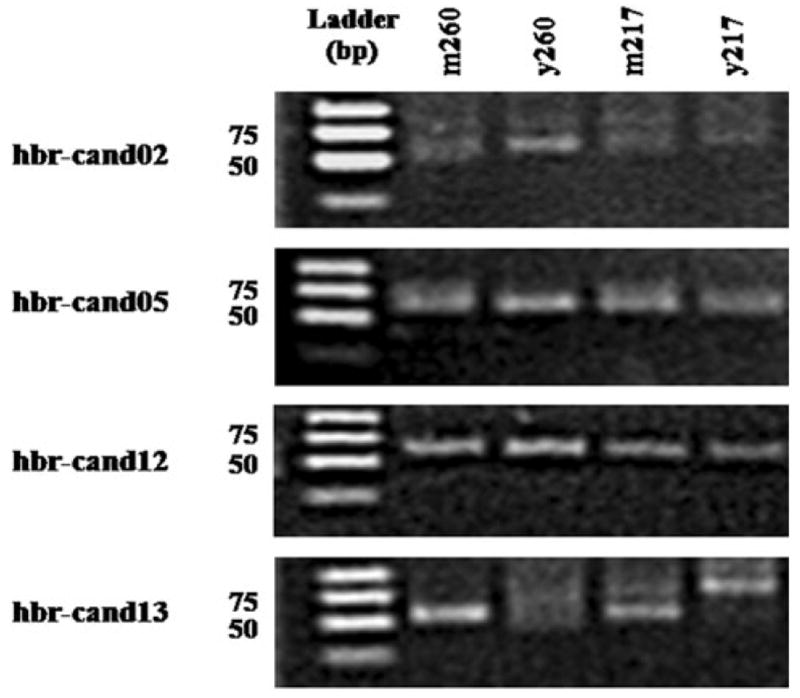
Stem–loop RT-PCR analysis of rubber tree miRNA accumulation in leaves. The expected size of the RT-PCR products is 60 bp. For hbr-cand02 and hbr-cand13, PCR were conducted for 35 cycles with annealing at 64 °C. For hbr-cand05 and hbr-cand12, PCR were conducted for 28 and 35 cycles, respectively, with annealing at 60 °C
Discussion
Using high-throughput sequencing, we generated millions of small RNA reads from young and mature leaves in two different rubber tree clones with differing latex yields. Of the millions of high-quality small RNA reads from the libraries, the 24 nt class exhibited the highest abundance followed by the 21 and 18 nt classes. Such size distribution is similar to that observed for Arabidopsis thaliana (Kasschau et al. 2007), Oryza sativa (Morin et al. 2008), Medicago truncatula (Szittya et al. 2008) and Arachis hypogaea (Zhao et al. 2010). In Arabidopsis, the 24 nt in class is composed almost exclusively of DCL3-dependent siRNAs (Kasschau et al. 2007; Xie et al. 2004). Conversely, 21 nt is the common length of miRNAs (Kasschau et al. 2007). We identified 115 conserved miRNAs representing 56 families and 20 novel miRNAs. miR396 was found to be the most abundant miRNA, accounting for close to 50 % of the total sequence reads. miR396 is conserved among dicot and monocot plants, and targets six growth-regulating factor (GRF) genes encoding putative transcription factors with roles in growth and cell proliferation in leaves in Arabidopsis (Wang et al. 2011). In Arabidopsis, miR396 is present at low levels in young leaves and its abundance gradually increases as leaves mature (Wang et al. 2011). This pattern of accumulation of miR396 was also seen in rubber tree, as reflected by the read frequencies in young and old leaves (Supplemental Table 1). Apart from the regulation of cell proliferation in plants, miR396 was also reported to be involved in responses to environmental stimuli such as drought stress (Gao et al. 2010). In addition to this miRNA, miR159/319, miR167, and miR166 families, which are believed to regulate MYB transcription factor, auxin responsive factor (ARF), and type III HD-Zip transcription factors, were also abundantly represented in our libraries. Several miRNA* species were found to accumulate to the levels comparable with their respective miRNAs, implicating a functional importance for these miRNA* species.
In this study, rubber elongation factor (REF) was predicted to be a target of miR172. REF is a major protein located on the surface of large rubber particles in latex and is involved in rubber biosynthesis (Priya et al. 2008). Hevamine A, a predicted target of miR160, is one of several genes encoding Hevamine, a chitinase/lysozyme activity found in the lutoid (vacuolar) fraction of rubber latex (Bokma et al. 2001, 1997; Subroto et al. 1996). Hevamine is important for plant defense against various bacterial and fungal pathogens. Further investigation into the regulatory relationship between the two miRNAs and their targets would potentially provide a means to enhance latex production in rubber tree.
Supplementary Material
Acknowledgments
We thank Lijuan Ji and Theresa Dinh for technical advice. This work was partially supported by National Science and Technology Development Agency (grant No. BT-B-01-PG-14-5202), University of California, Riverside, and Mahidol University. Lertpanyasampatha M. is supported by the Royal Golden Jubilee Ph.D. program (no. PHD/0216/2550) and Fulbright-TRF Thai Junior Research Scholarship Program.
Footnotes
Note added in proof While our manuscript was under revision, a paper describing the identification of rubber tree miRNAs appeared in BMC Plant Biology (2012, 12:18).
Electronic supplementary material The online version of this article (doi:10.1007/s00425-012-1622-1) contains supplementary material, which is available to authorized users.
Contributor Information
Manassawe Lertpanyasampatha, Department of Biotechnology, Faculty of Science, Mahidol University, Rama 6 Rd., Rajthewee, Bangkok 10400, Thailand.
Lei Gao, Department of Botany and Plant Sciences, Institute of Integrative Genome Biology, University of California, Riverside, CA 92521, USA.
Panida Kongsawadworakul, Department of Plant Science, Faculty of Science, Mahidol University, Bangkok, Thailand.
Unchera Viboonjun, Department of Plant Science, Faculty of Science, Mahidol University, Bangkok, Thailand.
Hervé Chrestin, Institut de Recherche pour le Développement (IRD), Montpellier, France.
Renyi Liu, Department of Botany and Plant Sciences, Institute of Integrative Genome Biology, University of California, Riverside, CA 92521, USA.
Xuemei Chen, Department of Botany and Plant Sciences, Institute of Integrative Genome Biology, University of California, Riverside, CA 92521, USA xuemei.chen@ucr.edu.
Jarunya Narangajavana, Department of Biotechnology, Faculty of Science, Mahidol University, Rama 6 Rd., Rajthewee, Bangkok 10400, Thailand scjnr@mahidol.ac.th.
References
- Barrera-Figueroa BE, Gao L, Diop NN, Wu Z, Ehler JD, Roberts PA, Close TJ, Zhu J, Liu R. Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol. 2011 doi: 10.1186/1471-2229-11-127. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci U S A. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokma E, van Koningsveld GA, Jeronimus-Stratingh M, Beintema JJ. Hevamine, a chitinase from the rubber tree Hevea brasiliensis, cleaves peptidoglycan between the C-1 of N-acetyl-glucosamine and C-4 of N-acetylmuramic acid and therefore is not a lysozyme. FEBS Lett. 1997;411:161–163. doi: 10.1016/s0014-5793(97)00682-0. [DOI] [PubMed] [Google Scholar]
- Bokma E, Spiering M, Chow K-S, Mulder PPMFA, Subroto T, Beintema JJ. Determination of cDNA and genomic DNA sequences of hevamine, a chitinase from rubber tree Hevea brasiliensis. Plant Physiol Biochem. 2001;39:367–376. [Google Scholar]
- Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155–W159. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Bai X, Yang L, Lv D, Li Y, Cai H, Ji W, Guo D, Zhu Y. Over-expression of osa-MIR396c decreases salt and alkali stress tolerance. Planta. 2010;231:991–1001. doi: 10.1007/s00425-010-1104-2. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 2007;5:e57. doi: 10.1371/journal.pbio.0050057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating micro-RNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK. Criteria for annotation of plant microRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, Chen S, Hannon GJ, Qi Y. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney BP. The second green revolution? Production of plant-based biodegradable plastics. Biochem J. 2009;418:219–232. doi: 10.1042/BJ20081769. [DOI] [PubMed] [Google Scholar]
- Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res. 2008;18:571–584. doi: 10.1101/gr.6897308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson WR. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith-Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- Priya P, Venkatachalam P, Thulaseedharan A. Molecular cloning and characterization of the rubber elongation factor gene and its promoter sequence from rubber tree (Hevea brasiliensis): A gene involved in rubber biosynthesis. Plant Sci. 2008;171:470–480. doi: 10.1016/j.plantsci.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Shukla LI, Chinnusamy V, Sunkar R. The role of microRNAs and other endogenous small RNAs in plant stress responses. Biochim Biophys Acta. 2008;1779:743–748. doi: 10.1016/j.bbagrm.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Smith TF, Waterman MS. Identification of common molecular subsequences. J Mol Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Subroto T, van Koningsveld GA, Schreuder HA, Soedjanaatmadja UM, Beintema JJ. Chitinase and beta-1,3-glucanase in the lutoid-body fraction of Hevea latex. Phytochemistry. 1996;43:29–37. doi: 10.1016/0031-9422(96)00196-3. [DOI] [PubMed] [Google Scholar]
- Sunkar R. MicroRNAs with macro-effects on plant stress responses. Semin Cell Dev Biol. 2010;21:805–811. doi: 10.1016/j.semcdb.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu JK. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Szittya G, Moxon S, Santos DM, Jing R, Fevereiro MP, Moulton V, Dalmay T. High-throughput sequencing of Medicago truncatula short RNAs identifies eight new miRNA families. BMC Genomics. 2008;9:593. doi: 10.1186/1471-2164-9-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Wang L, Gu X, Xu D, Wang W, Wang H, Zeng M, Chang Z, Huang H, Cui X. miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J Exp Bot. 2011;62:761–773. doi: 10.1093/jxb/erq307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, Wang W, Zheng Y, Chen X, Bo W, Song S, Zhang W, Peng M. Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucleic Acids Res. 2010;38:981–995. doi: 10.1093/nar/gkp1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang BH, Pan XP, Wang QL, Cobb GP, Anderson TA. Identification and characterization of new plant microRNAs using EST analysis. Cell Res. 2005;15:336–360. doi: 10.1038/sj.cr.7290302. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- Zhao CZ, Xia H, Frazier TP, Yao YY, Bi YP, Li AQ, Li MJ, Li CS, Zhang BH, Wang XJ. Deep sequencing identifies novel and conserved microRNAs in peanuts (Arachis hypogaea L.) BMC Plant Biol. 2010;10:3. doi: 10.1186/1471-2229-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


