Abstract
Background:
PIM serine/threonine kinases are often highly expressed in haematological malignancies. We have shown that PIM inhibitors reduced the survival and migration of leukaemic cells. Here, we investigated PIM kinases in diffuse large B-cell lymphoma (DLBCL) biopsy samples and DLBCL cell lines.
Methods:
Immunohistochemical staining for PIM kinases and CXCR4 was performed on tissue microarrays from a cohort of 101 DLBCL cases, and the effects of PIM inhibitors on the survival and migration of DLBCL cell lines were determined.
Results:
PIM1 expression significantly correlated with the activation of signal transducer and activator of transcription (STAT) 3 and 5, P-glycoprotein expression, CXCR4-S339 phosphorylation, and cell proliferation. Whereas most cases exhibited cytoplasmic or cytoplasmic and nuclear PIM1 and PIM2 expression, 12 cases (10 of the non-germinal centre DLBCL type) expressed PIM1 predominately in the nucleus. Interestingly, nuclear expression of PIM1 significantly correlated with disease stage. Exposure of DLBCL cell lines to PIM inhibitors modestly impaired cellular proliferation and CXCR4-mediated migration.
Conclusion:
This work demonstrates that PIM expression in DLBCL is associated with activation of the JAK/STAT signalling pathway and with the proliferative activity. The correlation of nuclear PIM1 expression with disease stage and the modest response to small-molecule inhibitors suggests that PIM kinases are progression markers rather than primary therapeutic targets in DLBCL.
Keywords: DLBCL, PIM kinases, CXCR4
The proto-oncogene serine/threonine-protein kinase (PIM) family comprises PIM1, PIM2 and PIM3. In the last decade, several PIM kinase-regulated signalling pathways and downstream targets have been identified (Brault et al, 2010; Nawijn et al, 2011). PIM1 was found to be an important effector of the signal transducer and activator of transcription (STAT) 3 and 5 transcription factors (Yip-Schneider et al, 1995). Interestingly, both tumour suppressors and oncogenes have been identified as potential PIM kinase substrates. However, all PIM kinases can act as survival factors, preventing cells from undergoing apoptosis by activating BCL2 (Lilly et al, 1999) and inactivating the pro-apoptotic proteins BAD (Aho et al, 2004) and ASK1 (Gu et al, 2009). PIM1 was also shown to phosphorylate the tumour suppressor p27KIP1, leading to its proteasomal degradation (Morishita et al, 2008). Although PIM kinases were first discovered in murine B- and T-cell lymphomas (Cuypers et al, 1984), their role in human malignancies has been best investigated in prostate cancer (Valdman et al, 2004) and different forms of leukaemias (Brault et al, 2010; Nawijn et al, 2011). In lymphomas, Epstein–Barr virus infection resulted in the upregulation of PIM kinases, enhancing the activity of EBNA2 (Rainio et al, 2005). PIM1 was found to be upregulated and to interact with STAT3 in mantle-cell lymphomas that are associated with poor prognosis (Zhu et al, 2002; Hsi et al, 2008). Interestingly, a functional collaboration between PIM1 and the c-MYC oncogene seems to be essential for promoting STAT3-mediated cell cycle progression in vitro (Shirogane et al, 1999).
In diffuse large B-cell lymphoma (DLBCL), PIM1 has been shown to be a target for aberrant hypermutation, particularly in extra-nodal cases (Deutsch et al, 2007), and it has recently been reported to be highly expressed in up to 50% of all studied cases at the protein level (Gomez-Abad et al, 2011; Schatz et al, 2011). A subgroup of the activated B-cell (ABC) type of DLBCL demonstrated increased PIM1 mRNA levels, which correlated with poor prognosis (Alizadeh et al, 2000; Sivertsen et al, 2006). In addition, PIM2 was reported to be upregulated during the progression of several B-cell malignancies, including chronic lymphocytic leukaemia, DLBCL, mantle-cell lymphoma and plasma-cell myeloma (Cohen et al, 2004). PIM2 seems to promote lymphoid cell survival, probably through activation of the NF-κB signalling pathway (Hammerman et al, 2004).
Resolution of the structures of the PIM kinases has allowed for the development of a number of small-molecule inhibitors (Bullock et al, 2005; Anizon et al, 2010; Morwick, 2010). We have previously demonstrated the anti-leukaemic activity of several imidazo-pyridazine small-molecule inhibitors (Fedorov et al, 2007; Pogacic et al, 2007). Interestingly, blocking PIM kinases with small-molecule inhibitors was able to impair the migration of acute myeloid leukaemia cells by downregulating the CXCR4 chemokine receptor at the cell surface (Grundler et al, 2009).
In this study, we aimed to explore the role of PIM kinases in DLBCL by immunohistochemical analysis of PIM1-3 and CXCR4 expression in tissue microarrays representing a set of 101 well-characterised cases. We frequently found elevated PIM kinase expression in DLBCL that was associated with activation of the JAK/STAT signalling pathway. Interestingly, nuclear PIM1 expression significantly correlated with disease stage and proliferative activity of the tumour cells. Two small-molecule inhibitors developed by our laboratories only modestly impaired cell survival and CXCR4-mediated migration. These observations suggest that PIM kinases represent molecular progression markers rather than primary therapeutic targets in DLBCL.
Materials and methods
Patients and tissue microarrays
The characteristics of the patients included in this study are summarised in Supplementary Table S1, and the construction details of the tissue microarrays have been previously published (Tzankov et al, 2006). All patients had given general informed consent for studies to be performed using tissue materials that remained after diagnostic procedures. Two cores from every sample were included in the arrays. All cases were classified as germinal centre (GC) B-cell like or non-GC cases according to Choi’s algorithm (Choi et al, 2009). If marker information from Choi’s decision tree was unavailable (i.e., for technical reasons, such as missing cores on the tissue microarray or poor immunostaining of the archival material), the cases could not be properly classified as DLBCL of GC or non-GC origin and were thus designated as ‘unclassifiable’.
Immunohistochemical staining
For PIM3, CXCR4 and pS339-CXCR4 staining, tissue microarrays were processed using an automated immunostainer (Nexes, Ventana, Tucson, AZ, USA), while PIM1 and PIM2 were stained manually. The streptavidin–biotin peroxidase detection technique using diaminobenzidine as the chromogen was applied. The primary antibodies were diluted in a 1% solution of bovine serum albumin in PBS (pH 7.4) and incubated for 30 min at room temperature in the automated immunostainer, and either for 2 h at room temperature (PIM1) or overnight at 4 °C (PIM2) in cases of manual staining. The dilutions and retrieval conditions for the primary antibodies used are listed in Supplementary Table S2. The immunohistochemical staining of CXCR4 and pS339-CXCR4 was established using paraffin-embedded Jurkat cell pellets known to express significant levels of CXCR4, which becomes phosphorylated on S339 upon phorbol-ester treatment (Grundler et al, 2009). Twenty percent of all cases were re-evaluated by a second observer to assess inter-observer agreement. At least 200 cells were assessed in each tumour core biopsy, and the percentage of positive cells (i.e., cells with distinct staining) was calculated.
Cell lines
The human GC-type (SU-DHL-4, SU-DHL-6, SU-DHL-7, and SU-DHL-10) and non-GC-type (HBL-1, U2932, OCI-Ly3, and OCI-Ly10) DLBCL cell lines were kindly provided by M Thome of the University of Lausanne (Hailfinger et al, 2009) and were kept in RPMI-1640 containing glutamine (Invitrogen, Carlsbad, CA, USA) plus penicillin/streptomycin and 15–20% foetal bovine serum at 37 °C in a humidified atmosphere containing 5% CO2, with the exception of OCI-Ly10 cells, which were kept in IMDM containing glutamine (Invitrogen) plus penicillin/streptomycin and 20% human plasma. K-562 (Ph+, blast crisis CML) cells, which are known to express significant amounts of PIM1 and PIM2, served as positive controls for protein and mRNA expression levels (Adam et al, 2006). HEK-293T cells, which are known to express low amounts of PIM1 and PIM2, were used as negative controls.
Quantitative RT–PCR
Messenger RNA expression was determined by quantitative real-time PCR (RT–PCR) using SYBR Green. One microgram of total mRNA, which was isolated using the RNeasy kit (Qiagen, Hilden, Germany), was used for cDNA synthesis using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA), and 50 ng of cDNA was used for RT–PCR using the Power SYBR Green PCR Mastermix (Applied Biosystems) on an ABI Prism 7700 sequence detection system (Applied Biosystems). Expression levels were normalised to glyceraldehyde-3-phosphate dehydrogenase mRNA using the 2ΔΔCt-method. See Supplementary Table S3 for the oligonucleotides PCR primers used.
Immunoblotting
Harvested cells were disrupted in lysis buffer (10 mmol l−1 Tris-HCl (pH 7.4), 150 mmol l−1 NaCl, 1% Triton X-100, 0.5 mmol l−1 EDTA, 10% glycerol, 10 mmol l−1 NaF, 1 mmol l−1 Na3VO4), supplemented with protease inhibitor cocktail (Calbiochem, La Jolla, CA, USA) for 20 min on ice followed by centrifugation at 12 000 g for 15 min. The cleared lysates were assayed for protein concentration using the Bradford protein assay system (Bio-Rad, Hercules, CA, USA). A total of 50 μg of protein was separated in a 15% SDS–PAGE gel and transferred onto a PVDF membrane (Macherey-Nagel, Dueren, Germany). Immunoblotting was performed using the following antibodies: anti-PIM1 (1 : 500) (12H8; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-PIM2 (1 : 1000) (HPA000285; Sigma, St Louis, MO, USA), anti-PIM3 (1 : 500) (AP7171a; Abgent, San Diego, CA, USA), anti-CXCR4 (1 : 1000) (Ab2074; Abcam, Cambridge, MA, USA) and anti-β-Actin (1 : 5000) (AC-74; Sigma). The bound primary antibodies were detected using either horseradish peroxidase-goat anti-rabbit antibodies or horseradish peroxidase-goat anti-mouse antibodies (Thermo Scientific, Ilkirch, France). Enhanced chemiluminescence western blotting detection was performed using the West Femto SuperSignal reagent (Thermo Scientific).
Cytotoxicity assay
To investigate cell viability effects, either the small-molecule PIM kinase inhibitor K00486 or Compound 20 (0 to 250 μℳ in 0.1% DMSO) was added to the culture medium, and the cultures were incubated for 48 h (Pogacic et al, 2007; Huber et al, 2012). Cell viability was assessed using the cell proliferation reagent WST-1 (Roche Diagnostics, Mannheim, Germany) according to the manufacturer’s instructions. Cell survival was calculated as a percentage normalised to control cultures, and the IC50 values were calculated using Graph Pad Prism (Graph Pad Software, La Jolla, CA, USA).
Cell death measurement
Cells were treated with the PIM inhibitor for 48 h and were analysed by flow cytometry using both the violet fluorescent dye PO-PRO-1 (a sensitive indicator of apoptotic cells) and 7-amino-actinomycin D (an indicator of dead cells) from the Vybrant Apoptosis Assay Kit #13 (Molecular Probes, Eugene, OR, USA) according to the manufacturer’s instructions.
Determination of CXCR4 surface expression
Cell surface expression of CXCR4 was analysed by staining with an APC-conjugated anti-human CD184 (1 : 50) (12G5) antibody (Pharmingen, Becton-Dickinson, San Jose, CA, USA), and dead cells were excluded by DAPI staining. Nonspecific binding was assessed using an APC-conjugated rat IgG2b antibody as an isotype control. Two-colour flow cytometric analysis was performed on a CyAn flow cytometer (Beckman Coulter, Fullerton, CA, USA) using the 405- and 633-nm lasers to acquire DAPI (BP 450/50) and APC (LP 750), respectively.
Migration assay
A total of 5 × 105 cells were allowed to migrate for 4 h toward a CXCL12 gradient (100 ng ml−1) in Transwell chambers (5.0 μm pore size; Costar, Cambridge, MA, USA). The migration index was calculated as a percentage of input cells. Where indicated, the cells were pre-treated for 2 h with 10 μℳ of K00486.
Statistical analysis
Statistical analyses, including data description, were performed using the Statistical Package of Social Sciences version 19.0 for Windows (SPSS, Chicago, IL, USA). The degree of agreement between observers was evaluated by interclass correlation coefficients using Cronbach’s Alpha reliability analysis; an alpha >0.75 indicated very good reproducibility. The χ2 test was used to assess distribution differences. The Spearman test was used to analyse relationships between the markers, and only correlations with a ρ⩾0.2 or ⩽−0.2 were further considered. The ANOVA, Mann–Whitney and Kruskal–Wallis tests were applied where appropriate to assess mean differences between groups. The prognostic performance of the variables and the determination of optimal cutoff values for continuous variables were established as described elsewhere (Tzankov et al, 2010). After these cutoff values had been applied, survival was analysed using the Kaplan–Meier method and compared using the log-rank test. Statistical significance was defined as P<0.05, and two-sided tests were used throughout.
Results
Expression of PIM 1–3 kinases in DLBCL and clinico-pathological correlations
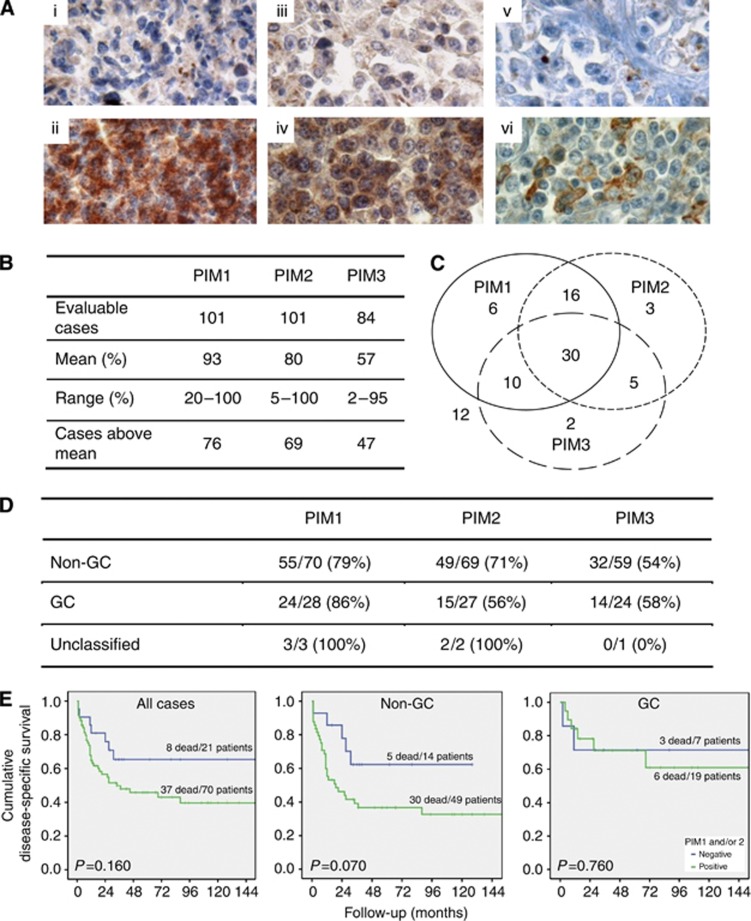
We first established immunohistochemical staining for the PIM kinases using formalin-fixed, paraffin-embedded pellets of cell lines expressing high (K-562) or very low (HEK-293T) PIM levels, as well as cell pellets of all examined DLBCL cell lines (see ‘cell lines’) and human tissues (prostate cancer, tonsils, and lymph nodes) (Adam et al, 2006) (Supplementary Figure S1). Using this protocol, all 101 cases of DLBCL were evaluable for PIM1 and PIM2 staining, while only 84 were evaluable for PIM3. As shown in Figure 1A, the staining for PIM1 and PIM2 was generally more intensive than that for PIM3. The kappa values for the overall agreement between observers were 0.8 for PIM1, 0.8 for PIM2, and 0.95 for PIM3. Diffuse large B-cell lymphoma cases were considered PIM positive when the percentage of stained cells per case was above the mean (Figure 1B). Of the 84 cases with complete data available for all three PIM kinases, 30 cases expressed all PIM kinases. Except for these 30 cases, 32 cases were positive for PIM1 either alone or co-expressed with PIM2 or PIM3, 24 cases were positive for PIM2 alone or in combination with PIM1 or PIM3 and 17 cases were positive for PIM3 alone or together with PIM1 or PIM2 (Figure 1C). On the basis of the cell-of-origin classification (Choi et al, 2009), 79% of the non-GC cases demonstrated PIM1 positivity compared with 86% GC and all unclassifiable cases (P=0.942); 71% of the non-GC cases expressed PIM2 compared with 56% of the GC cases (P=0.115); and finally, 54% of the non-GC cases expressed PIM3 compared with 58% of the CG cases and none of the unclassifiable cases (P=0.810) (Figure 1D). Although co-expression of all three PIM kinases was more prevalent in the non-GC-derived cases, the distribution differences between GC- and non-GC cases, as defined by either the Choi- or the Hans-algorithm (Hans et al, 2004; Choi et al, 2009), did not reach statistical significance. Similarly to what has recently been reported (Schatz et al, 2011), we observed a trend towards lower overall survival in DLBCL cases expressing high levels of PIM1 and/or PIM2; however, these differences did not reach statistical significance within our cohort (Figure 1E).
Figure 1.
PIM kinase expression in DLBCL cases. (A) Representative cases for PIM1, PIM2 and PIM3: (i) PIM1 negative, (ii) PIM1 positive, (iii) PIM2 negative, (iv) PIM2 positive, (v) PIM3 negative and (vi) PIM3 positive. (B) Quantitative data on PIM expression. (C) Distribution of PIM1-3 in DLBCL cases (n=84, included are only cases with available data on the expression of all three PIM). (D) Expression of PIM1-3 with respect to DLBCL subtype. (E) Disease-specific survival of DLBCL cases with respect to PIM1 and/or PIM2 expression shown for all cases (left) and non-GC (centre) and GC cases (right) according to the Choi algorithm. The survival curves were plotted using the Kaplan–Meier method and compared by the log-rank test.
We next determined whether PIM kinase expression levels might correlate with other molecular markers that we have previously assessed using the same DLBCL cohort (Tzankov et al, 2006, 2008; Meier et al, 2009; Nagel et al, 2010). As shown in Table 1, PIM1 significantly correlated with pSTAT3, pSTAT5 and with the fraction of actively proliferating cells as detected by high levels of MIB-1 labelling. Expression of PIM1 but not PIM2 or PIM3 also correlated with the expression of CD44 (ρ=0.320, P=0.002). PIM1 and PIM2 both significantly correlated with the expression of the permeability glycoprotein (Pgp; ρ=0.422–0.430, P⩽0.001). By contrast, no significant correlations were found between PIM expression and the status of the c-MYC, BCL2 and BCL6 genes, which were previously studied in the same cohort (Obermann et al, 2009a, 2009b), nor was an association with the presence of the Epstein–Barr virus in lymphoma cells, as suggested by EBER staining, observed (data not shown).
Table 1. Correlation between markers and stage.
| PIM1 | PIM2 | PIM3 | |
|---|---|---|---|
| PIM2 | 0.439 (<0.001) | ||
| PIM3 | 0.313 (0.004) | ||
| pSTAT3 | 0.318 (0.002) | 0.285 (0.006) | 0.292 (0.009) |
| pSTAT5 | 0.305 (0.002) | 0.348 (0.001) | |
| CD44 | 0.320 (0.002) | ||
| Pgp | 0.430 (<0.001) | 0.422 (>0.001) | 0.343 (0.003) |
| MIB-1 | 0.265 (0.013) | ||
| Stage | 0.386a (<0.001) |
Abbreviations: Pgp=permeability glycoprotein; PIM=proto-oncogene serine/threonine-protein kinase; STAT=signal transducer and activator of transcription.
Numbers indicate correlation coefficients, P-values are indicated in parentheses.
This correlation applies only to nuclear PIM1.
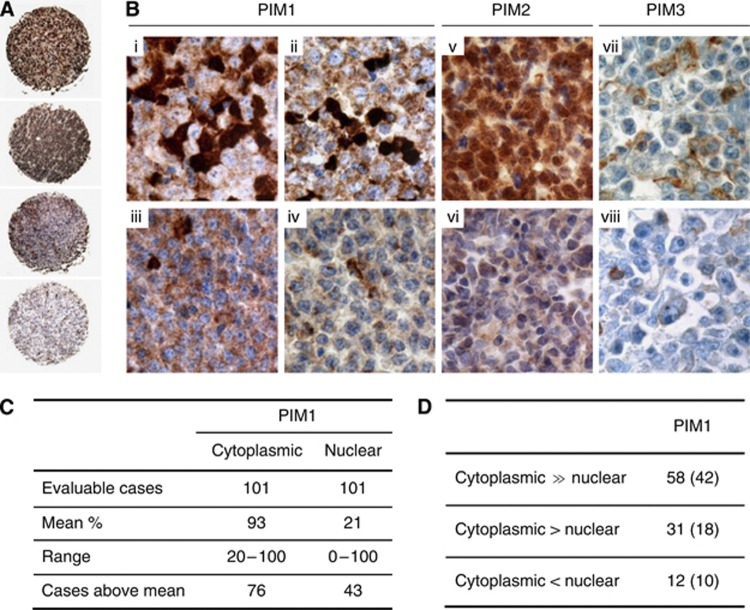
In general, signals for PIM1 and PIM2 staining were found in the cytoplasm and the nucleus of the tumour cells, whereas PIM3 appeared only in the cytoplasm (Figure 2). Interestingly, a distinct distribution of nuclear and cytoplasmic PIM1 staining was observed; in most of the cases, PIM1 cytoplasmic staining exceeded the nuclear signals. Importantly, 10 of 12 DLBCL cases with predominantly nuclear PIM1 staining were of the non-GC type. One additional case with nuclear PIM1 staining could not be categorised (Figure 2). Considering the clinical parameters, nuclear expression of PIM1 significantly correlated with Ann Arbor disease stage (ρ=0.386; P<0.001) but not with any other prognostic parameters, such as IPI, age, LDH level, B-symptoms, or extra-nodal disease origin.
Figure 2.
The nuclear and cytoplasmic PIM1 and PIM2 expression pattern in DLBCL. (A) Tissue microarray data showing (first case) significant nuclear expression of PIM1, (second case) combined nuclear and cytoplasmic expression of PIM1, (third case case) cytoplasmic expression of PIM1 and (fourth case) a negative case ( × 4). (B) DLBCL case with (i) nuclear expression of PIM1 in 50% of the tumour cells, (ii) nuclear expression of PIM1 in 15% of the tumour cells, (iii) exclusive cytoplasmic expression of PIM1 in 100% of the tumour cells, and (iv) without expression of PIM1 (note internal positive control). DLBCL case (v) with nuclear expression of PIM2 in almost all of the tumour cells and (vi) without expression of PIM2. PIM3 in DLBCL cases (vii) with expression in 10% of the tumour cells and (viii) without expression of PIM3 (note internal positive control) ( × 400). (C) Quantitative data on PIM expression. (D) Intracellular microtopographic localisation of PIM1 in DLBCL cases; the non-GC case numbers are indicated in parentheses.
Taken together, our observations suggest that elevated expression of PIM1 and PIM2 is frequent in DLBCL, that it correlates with proliferation and JAK/STAT pathway activation, and that their nuclear expression is more prevalent in non-GC DLBCL and advanced (high stage) cases.
Effect of small-molecule PIM kinase inhibitors on proliferation of DLBCL cell lines
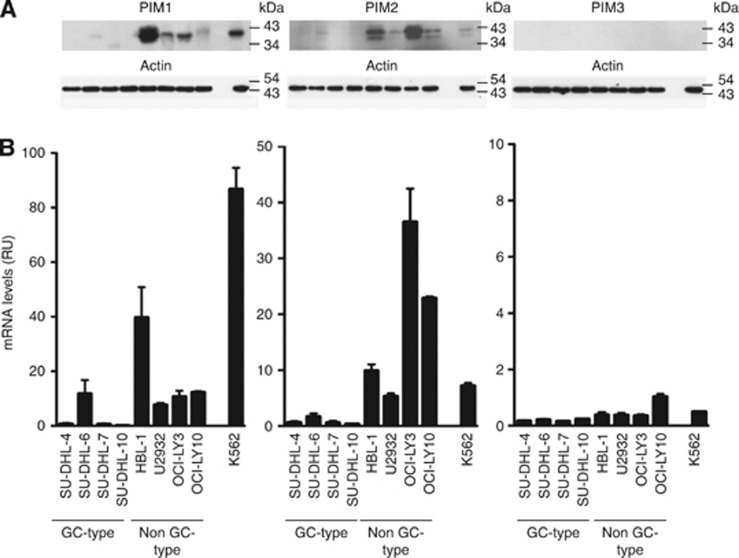
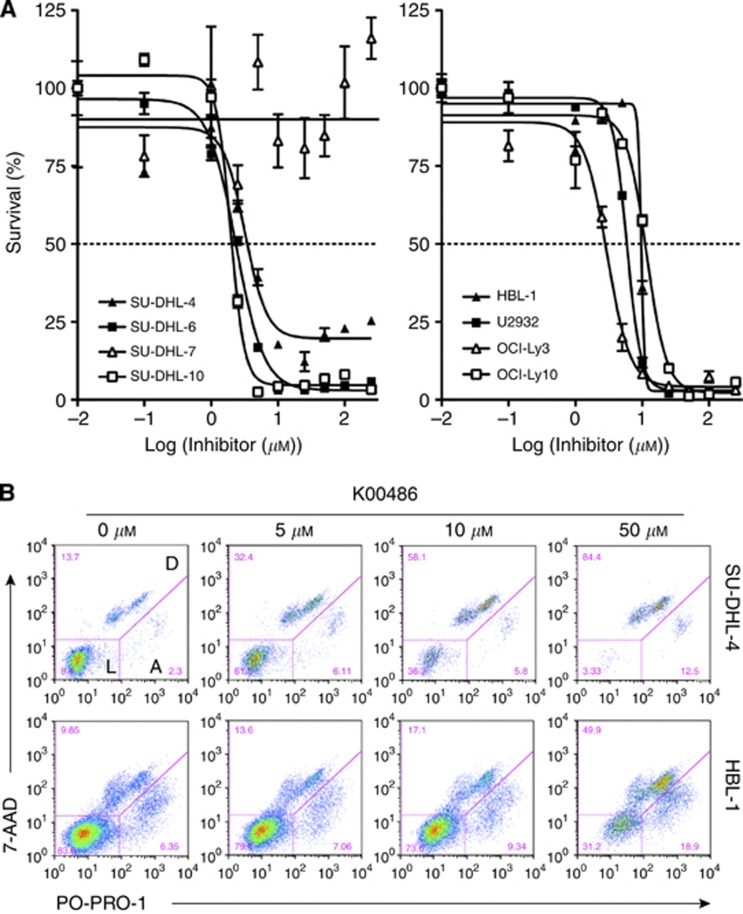
To explore the role of PIM kinases as potential therapeutic targets in DLBCL, we evaluated the effects of two structurally different small-molecule PIM inhibitors (K00486, Compound 20) in a panel of well-established human DLBCL cell lines. We first measured PIM mRNA and protein expression levels in DLBCL cell lines. As shown in Figure 3, the non-GC-derived DLBCL cell lines HBL-1, U2932, OCI-Ly3, and OCI-Ly10 expressed higher mRNA and protein levels of PIM1 and PIM2 than the GC-derived cell lines SU-DHL-4, SU-DHL-6, SU-DHL-7, and SU-DHL-10. A statistically significant difference in PIM1 and PIM2 expression (P<0.001) in GC vs non-GC cell lines confirmed recent observations (Gomez-Abad et al, 2011). No significant PIM3 expression was detected by immunoblotting or quantitative RT–PCR. Exposure of the cells to PIM kinase inhibitors (K00486 and Compound 20) impaired cell survival in a dose-dependent manner in the low micromolar range for most of the DLBCL cell lines (Table 2 and Figure 4A). Flow cytometric assessment of cell death confirmed the impact of the PIM inhibitors on cell viability as shown by a dose-dependent decrease in the live cell percentage (Figure 4B and data not shown). It is worth noting that SU-DHL-7 cells were highly resistant to PIM kinase inhibitors.
Figure 3.
PIM expression levels in GC and non-GC-derived DLBCL cell lines. (A) Protein levels determined by immunoblots with antibodies directed against the indicated PIM and Actin as a loading control (one of two independent experiments is represented in the figure). (B) Messenger RNA levels of the indicated PIM as determined by qPCR and standardised to Jurkat T-cell expression levels; the leukaemic cell line K-562 was used as a control. The data represent the mean±s.e.m. of three independent experiments. Note the higher expression of PIM1 and PIM2 in non-GC-type cells.
Table 2. PIM inhibitor potentials, determined as the half maximal inhibitory concentration (IC50) for two structurally independent PIM inhibitors in GC and non-GC-derived DLBCL cell lines.
|
IC50
24 h (μℳ) |
IC50
48 h (μℳ) |
|||
|---|---|---|---|---|
| Cell line | K00486 | Compound 20 | K00486 | Compound 20 |
| SU-DHL-4 | 14.4 | 5.27 | 9.71 | 3.21 |
| SU-DHL-6 | 12.1 | 3.29 | 8.40 | 2.35 |
| SU-DHL-7 | >100 | >100 | >100 | >100 |
| SU-DHL-10 | 6.2 | 2.65 | 3.77 | 1.95 |
| HBL-1 | 33.8 | 10.6 | 18.34 | 9.70 |
| U2932 | 24.5 | 8.79 | 8.62 | 5.88 |
| OCI-Ly3 | 10.36 | 4.80 | 6.042 | 2.98 |
| OCI-Ly10 | >100 | 13.2 | 51.15 | 11.63 |
Abbreviations: DLBCL=diffuse large B-cell lymphoma; GC=germinal centre; IC50=half maximal inhibitory concentration; PIM=proto-oncogene serine/threonine-protein kinase.
The data represent the mean of three independent experiments performed in quadruplicate.
Figure 4.
Impact of PIM inhibitors on cell viability. (A) Dose–response curves of K00486 treatment in GC (left) and non-GC-derived DLBCL cell lines (right). Cell viability was determined in the presence of increasing doses of the inhibitor after 48 h. The data represent the mean±s.d. of four individual wells. Note the increased sensitivity of GC-type cells in the graph on the left. (B) Analysis of cell death induction upon K00486 treatment in GC (SU-DHL-4) and non-GC-derived (HBL-1) DLBCL cell lines. Abbreviations: L=living cells; A=apoptotic cells; D=dead cells.
We also evaluated the potential of these PIM inhibitors in combination with two chemotherapeutic agents: adriamycin and vinblastine. SU-DHL-6 (a GC-type) and U2932 (a non-GC-type) cells, which are similar in their sensitivity to the PIM inhibitors, were selected for combination treatment (Table 2). The cells were sequentially exposed to the chemotherapeutic agents and PIM inhibitors. Upon pre-treatment with the PIM1 inhibitors for 2 h, we observed a minor sensitisation of the GC type but not the non-GC-type cell line to adriamycin; a marginal sensitivity to vinblastine was also observed (Supplementary Figure S2). When applied simultaneously or after the chemotherapeutic drugs, no potentiating effects were observed.
CXCR4 and PIM kinases in DLBCL
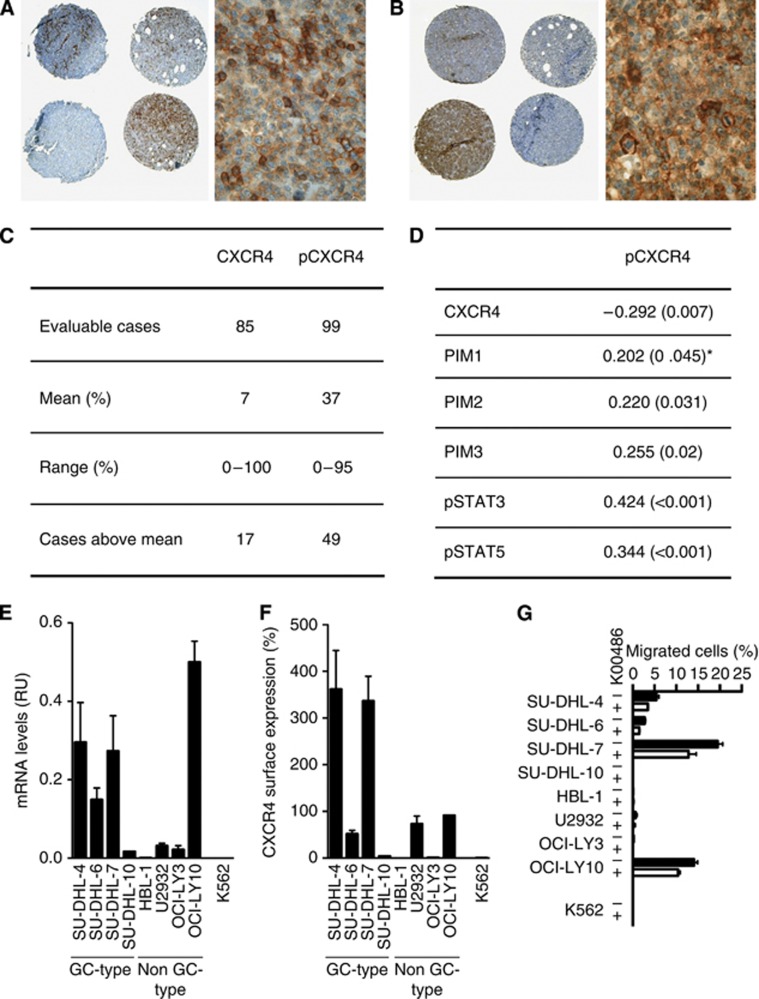
In addition to its pro-survival function, PIM1 has been reported to affect the migration of leukaemic cells by regulating CXCR4 surface expression through the phosphorylation of Serine 339 in the intracellular C-terminal tail domain (Grundler et al, 2009). We therefore analysed CXCR4/pS339-CXCR4 expression in our DLBCL cohort by immunohistochemical staining. In total, 85 cases were evaluable for CXCR4 and 99 for pS339-CXCR4 staining. It is important to note that among the available CXCR4 antibodies, only the UMB-2 clone (Epitomics, Burlingame, CA, USA) provided a specific signal when tested on formalin-fixed, paraffin-embedded biopsy samples. UMB-2 recognises an epitope in the C-terminal tail (KGKRGGHSSVSTESESSSFHSS) that includes S339 (Fischer et al, 2008) (Figures 5A and B). CXCR4 and pS339-CXCR4 staining demonstrated an inverse correlation (ρ=−0.297, P=0.007) with 29 of 85 cases being positive for pS339-CXCR4 only, 13 of 85 cases showing positive staining for non-phosphorylated CXCR4, and 3 cases exhibiting signals for both CXCR4 and pS339-CXCR4, suggesting a concurrent recognition of the same epitope. All other cases (n=40) expressed either no CXCR4 or pS339-CXCR4, or they expressed these molecules below the cutoff levels (means) (Figure 5C). Interestingly, we found that pS339-CXCR4 staining correlated with pSTAT3 (ρ=0.427, P<0.001), pSTAT5 (ρ=0.385, P<0.001) and with cytoplasmic PIM1 (ρ=0.202, P=0.048). pS339-CXCR4 expression levels also correlated with PIM2 (ρ=0.220, P=0.031) and PIM3 (ρ=0.255, P=0.02) (Figure 5D).
Figure 5.
Expression of (A) CXCR4 and (B) pCXCR4 in DLBCL (overview and detailed view of the respective positive cases; × 400). Note the inverse distribution of positivity for CXCR4 and pCXCR4 among the same cases shown in the overview magnification, which were stained for the respective marker on consecutive sections, the first core on the second line is negative for CXCR4, while being strongly positive for pCXCR4, while the second core stains for CXCR4, but not for pCXCR4 (for explanation, see ‘Expression of PIM 1–3 kinases in DLBCL and clinico-pathological correlations’ in the Results section). (C) Quantitative data on CXCR4 and pCXCR4 expression. (D) Correlation between pCXCR4 and other markers. The numbers indicate correlation coefficients, and P-values are indicated in parentheses; *correlation applies only to cytoplasmic PIM1. (E) Messenger RNA levels in DLBCL cell lines measured by qPCR and standardised to Jurkat T-cell expression levels; the leukaemic cell line K-562 was used as a control. The data represent the mean±s.e.m. of three independent experiments. (F) CXCR4 surface expression levels as determined by flow cytometry. The data represent the mean±s.e.m. of three independent experiments. (G) Migration capacity towards CXCL12 in the presence or absence of a PIM inhibitor as investigated in a Transwell migration assay. The data represent the mean±s.e.m. of three independent experiments.
Previously, we showed that chemical inhibition of PIM1 kinase decreased surface CXCR4 expression and impaired the migration of leukaemic cells (Grundler et al, 2009). We therefore also addressed the impact of PIM kinase inhibitors on the migration of DLBCL cells. First, we determined CXCR4 expression at the mRNA and corresponding surface protein levels. As shown in Figure 5E, steady-state CXCR4 mRNA levels varied between cell lines with SU-DHL-4, SU-DHL-7 and OCI-Ly10 expressing the highest levels. The mRNA levels correlated with the surface expression of the receptor with the exception of OCI-Ly10, which expressed moderate surface protein but significant mRNA levels (Figure 5F). As shown in Figure 5G, we found that not only SU-DHL-4, SU-DHL-6, SU-DHL-7 cells, but also OCI-Ly10 cells, despite their low-surface receptor expression, were able to migrate towards the CXCL12 ligand in Transwell migration assays. Interestingly, a short-term pre-treatment of the cells with a non-cytotoxic K00486 dose (10 μℳ) was able to affect migration of these cells.
Discussion
We explored the expression of PIM kinases in a well-characterised cohort of DLBCL cases by immunostaining using a panel of commercially available isoform-specific antibodies. Taken together, our observations suggest that upregulation of PIM1 and PIM2 is a frequent event in advanced stage and highly proliferating DLBCL and significantly correlates with the activation of STAT3 and STAT5. In addition, we found a high proportion of nuclear PIM1 in non-GC-type DLBCL cases. We are aware that our findings might have been influenced by the potential presence of somatic hypermutations targeting PIM1 (Pasqualucci et al, 2001). However, the lack of appropriate material did not allow us to determine the mutational status. Nevertheless, the detectable PIM protein expression in DLBCL-derived cell lines suggested that even in the presence of monoallelic mutations, most of the cells express a normal-sized and presumably wild-type protein (Figure 3).
During the course of this work, Schatz et al (2011) reported converging PIM kinase signalling pathways in malignant lymphoma. By immunohistochemical staining, they reported PIM1 or PIM2 expression in roughly similar proportions of DLBCL (48% of their cases expressed PIM1, compared with 43% in our cohort; 42% of their cases expressed PIM2, compared with 69% in our cohort). Unfortunately, little has been reported on the specificity and sensitivity of the establishment of their detection assay and of the PIM subcellular distribution. Another recently published study indicated that only 23% of DLBCL cases displayed strong PIM2 expression (Gomez-Abad et al, 2011). Both of these studies proposed that positive PIM staining correlated with disease outcome. Although we observed trends towards lower overall survival for DLBCL cases expressing high levels of PIM1 and/or PIM2, these differences did not reach statistical significance (Figure 1E). In addition to differences in detection methods and cutoff level definitions, the differences between these studies may also be explained by the lower proportion of anthracyclin-treated patients and limited follow-up data in one of the studies (Schatz et al, 2011). As the details of the case composition in the other study are lacking (Gomez-Abad et al, 2011), we cannot address the prognostic differences between our cohorts.
In contrast to previous studies, our work revealed for the first time that significant fractions of DLBCL cases express nuclear and/or cytoplasmic PIM1 in a mean of 93% (cytoplasmic) and 21% (nuclear) of tumour cells. Interestingly, PIM1 nuclear localisation was almost exclusively observed in non-GC DLBCL cells (Figure 2), and despite a small subset of 12/101 cases, this was highly correlated with increased proliferation and disease stage (Table 1). Conversely, only PIM1 cytoplasmic localisation correlated with pS339-CXCR4 (Figure 5). When compared with nuclear localisation, only cytoplasmic localisation correlated with Pgp. Several in vitro studies suggested that nuclear PIM1 seems to regulate cell cycle progression by direct modification of cell cycle-dependent kinase inhibitors such as p21WAF1 and p27KIP1 (Zhang et al, 2007; Morishita et al, 2008). However, we did not observe any significant correlation between nuclear PIM1 and the subcellular localisation of p21WAF1 or p27KIP1 (data not shown). Nuclear PIM1 has been proposed to be more potent in mediating cellular transformation, at least in vitro, in solid cancer cell lines (Chen et al, 2009; Peltola et al, 2009). Very little is known about the mechanisms that regulate cellular localisation of PIM1. Some in vitro experiments suggested that nuclear localisation of PIM1 may be dependent on the carboxy-terminal portion of the protein (Ishibashi et al, 2001). There are indeed several potential target sequences in that region that could be recognised by one or several upstream protein kinases, and this possibility is currently under investigation (Brault et al, 2010).
We observed a significant correlation between cytoplasmic PIM1 and PIM2 and the expression of the ABC-transporter Pgp. Previous studies have proposed that PIM1 might directly regulate drug resistance through modification of the ATP-binding cassette transporter breast cancer resistance protein (Xie et al, 2008) and Pgp-mediated drug resistance by phosphorylation and protection from degradation (Xie et al, 2010). In addition, PIM1 staining also correlated with CD44 expression. Previous studies found elevated CD44 expression in DLBCL (Tzankov et al, 2003; Nagel et al, 2010), thus underlining the association of PIM1 expression and disease progression. Whether PIM1 is able to directly modulate CD44 expression or signalling remains to be elucidated.
The strong association with known upstream regulators, such as pSTAT3 and pSTAT5, confirmed PIM1 and/or PIM2 as downstream effectors of the aberrantly activated JAK/STAT signalling in DLBCL and is potentially relevant for maintenance of the transformed phenotype. Therefore, we explored the activity of small-molecule PIM kinase inhibitors in a panel of well-characterised human DLBCL cell lines (Brault et al, 2010; Morwick, 2010). We used two structurally different PIM kinase inhibitors: K00486, an imidazo(1,2-b)pyridazine compound with low nanomolar in vitro potency (against PIM1 and PIM3) that significantly impaired growth and survival and surface expression of the CXCR4 chemokine receptor on myeloid leukaemia cell lines (Pogacic et al, 2007; Grundler et al, 2009), and Compound 20, a carboline-derivate that has been identified as a potent PIM kinase inhibitor (Huber et al, 2012). Both compounds impaired the proliferation of DLBCL cells (Figure 4). The higher cellular activity of Compound 20 is presumably the consequence of a lower selectivity and a higher number of ‘off-targets’ that are inherently associated with all currently available small-molecule PIM kinase inhibitors (Huber et al, 2012). For both PIM inhibitors, the modest potentiation of chemotherapeutic drug activity confirmed their moderate impact on DLBCL cell survival (Supplementary Figure S2). These findings suggest that elevated PIM kinase may not be essential for maintenance of the transformed state of DLBCL cells. Indeed, transgenic overexpression of PIM1 or PIM2 in the lymphoid compartment leads to formation of lymphomas after very long latency periods, suggesting that PIM kinases are oncogenic but not sufficient to drive disease (Berns et al, 1999). Additionally, PIM kinases expression levels did not predict the sensitivity of DLBCL cell lines to small-molecule inhibitors and the most sensitive cell lines expressed low levels of the kinases. Similarly, DLBCL cell lines expressing low level of PIM have been shown to be the most sensitive to another PIM kinase inhibitor (ETP-39010) (Gomez-Abad et al, 2011). These findings indicate that the sensitivity to PIM inhibitors is not directly correlated with the expression level of the kinases but might be driven by more complex drug-resistance associated mechanisms. Indeed, when compared with myeloid leukaemia cells that are very sensitive to PIM inhibitors with sub-micromolar IC50 values, we observed K00486 and Compound 20 activities in the micromolar IC50 range in most DLBCL cell lines (Table 2). It is likely that DLBCL cell lines express high levels of drug-resistance mediating pumps and/or proteins such as Pgp that could antagonise the effects of these PIM inhibitors. In agreement with this hypothesis, Pgp expression levels significantly correlated with elevated PIM1 and PIM2 expression in our DLBCL cohort (Table 1).
Taking these findings together, we found that the levels of expression of the PIM kinases in DLBCL correlated with active STAT signalling, higher lymphoma proliferative activity, and more advanced disease stage, indicating that PIM kinases may represent valuable markers for DLBCL progression. The studied small-molecule PIM kinase inhibitors moderately impaired proliferation and CXCR4-mediated migration of DLBCL cells. Their rather modest activity suggests that such compounds could find a place in the therapeutic arsenal, although probably only in combination with compounds blocking functionally cooperative signalling pathways.
Acknowledgments
This study was partially supported by Stiftung zur Krebsbekämpfung grant 269 to AT and a grant from the Swiss Cancer League (OCS 2357-02-2009) to JS. JS is a research professor supported by the Gertrude von Meissner Foundation. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from Abbott, the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Eli Lilly and Company, Genome Canada, GlaxoSmithKline, the Ontario Ministry of Economic Development and Innovation, the Novartis Research Foundation, Pfizer and the Wellcome Trust. We thank Prof. Franz Bracher (Munich University) for making Compound 20 available for this study.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Adam M, Pogacic V, Bendit M, Chappuis R, Nawijn MC, Duyster J, Fox CJ, Thompson CB, Cools J, Schwaller J (2006) Targeting PIM kinases impairs survival of hematopoietic cells transformed by kinase inhibitor-sensitive and kinase inhibitor-resistant forms of Fms-like tyrosine kinase 3 and BCR/ABL. Cancer Res 66(7): 3828–3835 [DOI] [PubMed] [Google Scholar]
- Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ (2004) Pim-1 kinase promotes inactivation of the pro-apoptotic bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett 571(1–3): 43–49 [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM (2000) Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403(6769): 503–511 [DOI] [PubMed] [Google Scholar]
- Anizon F, Shtil AA, Danilenko VN, Moreau P (2010) Fighting tumor cell survival: advances in the design and evaluation of pim inhibitors. Curr Med Chem 17(34): 4114–4133 [DOI] [PubMed] [Google Scholar]
- Berns A, Mikkers H, Krimpenfort P, Allen J, Scheijen B, Jonkers J (1999) Identification and characterization of collaborating oncogenes in compound mutant mice. Cancer Res 59(7 Suppl): 1773s–1777s [PubMed] [Google Scholar]
- Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J (2010) PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica 95(6): 1004–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock AN, Debreczeni JE, Fedorov OY, Nelson A, Marsden BD, Knapp S (2005) Structural basis of inhibitor specificity of the human protooncogene proviral insertion site in moloney murine leukemia virus (PIM-1) kinase. J Med Chem 48(24): 7604–7614 [DOI] [PubMed] [Google Scholar]
- Chen J, Kobayashi M, Darmanin S, Qiao Y, Gully C, Zhao R, Kondo S, Wang H, Yeung SC, Lee MH (2009) Hypoxia-mediated up-regulation of Pim-1 contributes to solid tumor formation. Am J Pathol 175(1): 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WW, Weisenburger DD, Greiner TC, Piris MA, Banham AH, Delabie J, Braziel RM, Geng H, Iqbal J, Lenz G, Vose JM, Hans CP, Fu K, Smith LM, Li M, Liu Z, Gascoyne RD, Rosenwald A, Ott G, Rimsza LM, Campo E, Jaffe ES, Jaye DL, Staudt LM, Chan WC (2009) A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res 15(17): 5494–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AM, Grinblat B, Bessler H, Kristt D, Kremer A, Schwartz A, Halperin M, Shalom S, Merkel D, Don J (2004) Increased expression of the hPim-2 gene in human chronic lymphocytic leukemia and non-Hodgkin lymphoma. Leuk Lymphoma 45(5): 951–955 [DOI] [PubMed] [Google Scholar]
- Cuypers HT, Selten G, Quint W, Zijlstra M, Maandag ER, Boelens W, Vanwezenbeek P, Melief C, Berns A (1984) Murine leukemia virus-induced T-cell lymphomagenesis - integration of proviruses in a distinct chromosomal region. Cell 37(1): 141–150 [DOI] [PubMed] [Google Scholar]
- Deutsch AJ, Aigelsreiter A, Staber PB, Beham A, Linkesch W, Guelly C, Brezinschek RI, Fruhwirth M, Emberger W, Buettner M, Beham-Schmid C, Neumeister P (2007) MALT lymphoma and extranodal diffuse large B-cell lymphoma are targeted by aberrant somatic hypermutation. Blood 109(8): 3500–3504 [DOI] [PubMed] [Google Scholar]
- Fedorov O, Marsden B, Pogacic V, Rellos P, Muller S, Bullock AN, Schwaller J, Sundstrom M, Knapp S (2007) A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci USA 104(51): 20523–20528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Nagel F, Jacobs S, Stumm R, Schulz S (2008) Reassessment of CXCR4 chemokine receptor expression in human normal and neoplastic tissues using the novel rabbit monoclonal antibody UMB-2. PLoS One 3(12): e4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Abad C, Pisonero H, Blanco-Aparicio C, Roncador G, Gonzalez-Menchen A, Martinez-Climent JA, Mata E, Rodriguez ME, Munoz-Gonzalez G, Sanchez-Beato M, Leal JF, Bischoff JR, Piris MA (2011) PIM2 inhibition as a rational therapeutic approach in B-cell lymphoma. Blood 118(20): 5517–5527 [DOI] [PubMed] [Google Scholar]
- Grundler R, Brault L, Gasser C, Bullock AN, Dechow T, Woetzel S, Pogacic V, Villa A, Ehret S, Berridge G, Spoo A, Dierks C, Biondi A, Knapp S, Duyster J, Schwaller J (2009) Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J Exp Med 206(9): 1957–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JJ, Wang Z, Reeves R, Magnuson NS (2009) PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene 28(48): 4261–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, Penas EM, Dierlamm J, Chan WC, Staudt LM, Thome M (2009) Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci USA 106(47): 19946–19951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerman PS, Fox CJ, Cinalli RM, Xu A, Wagner JD, Lindsten T, Thompson CB (2004) Lymphocyte transformation by Pim-2 is dependent on nuclear factor-kappaB activation. Cancer Res 64(22): 8341–8348 [DOI] [PubMed] [Google Scholar]
- Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC (2004) Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 103(1): 275–282 [DOI] [PubMed] [Google Scholar]
- Hsi ED, Jung SH, Lai R, Johnson JL, Cook JR, Jones D, Devos S, Cheson BD, Damon LE, Said J (2008) Ki67 and PIM1 expression predict outcome in mantle cell lymphoma treated with high dose therapy, stem cell transplantation and rituximab: a Cancer and Leukemia Group B 59909 correlative science study. Leuk Lymphoma 49(11): 2081–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber K, Brault L, Fedorov O, Gasser C, Filippakopoulos P, Bullock AN, Fabbro D, Trappe J, Schwaller J, Knapp S, Bracher F (2012) 7,8-Dichloro-1-oxo-beta-carbolines as a versatile scaffold for the development of potent and selective kinase inhibitors with unusual binding modes. J Med Chem 55(1): 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi Y, Maita H, Yano M, Koike N, Tamai K, Ariga H, Iguchi-Ariga SM (2001) Pim-1 translocates sorting nexin 6/TRAF4-associated factor 2 from cytoplasm to nucleus. FEBS Lett 506(1): 33–38 [DOI] [PubMed] [Google Scholar]
- Lilly M, Sandholm J, Cooper JJ, Koskinen PJ, Kraft A (1999) The PIM-1 serine kinase prolongs survival and inhibits apoptosis-related mitochondrial dysfunction in part through a bcl-2-dependent pathway. Oncogene 18(27): 4022–4031 [DOI] [PubMed] [Google Scholar]
- Meier C, Hoeller S, Bourgau C, Hirschmann P, Schwaller J, Went P, Pileri SA, Reiter A, Dirnhofer S, Tzankov A (2009) Recurrent numerical aberrations of JAK2 and deregulation of the JAK2-STAT cascade in lymphomas. Mod Pathol 22(3): 476–487 [DOI] [PubMed] [Google Scholar]
- Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N (2008) Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res 68(13): 5076–5085 [DOI] [PubMed] [Google Scholar]
- Morwick T (2010) Pim kinase inhibitors: a survey of the patent literature. Expert Opin Ther Patents 20(2): 193–212 [DOI] [PubMed] [Google Scholar]
- Nagel S, Hirschmann P, Dirnhofer S, Gunthert U, Tzankov A (2010) Coexpression of CD44 variant isoforms and receptor for hyaluronic acid-mediated motility (RHAMM, CD168) is an International Prognostic Index and C-MYC gene status-independent predictor of poor outcome in diffuse large B-cell lymphomas. Exp Hematol 38(1): 38–45 [DOI] [PubMed] [Google Scholar]
- Nawijn MC, Alendar A, Berns A (2011) For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer 11(1): 23–34 [DOI] [PubMed] [Google Scholar]
- Obermann EC, Csato M, Dirnhofer S, Tzankov A (2009a) Aberrations of the MYC gene in unselected cases of diffuse large B-cell lymphoma are rare and unpredictable by morphological or immunohistochemical assessment. J Clin Pathol 62(8): 754–756 [DOI] [PubMed] [Google Scholar]
- Obermann EC, Csato M, Dirnhofer S, Tzankov A (2009b) BCL2 gene aberration as an IPI-independent marker for poor outcome in non-germinal-centre diffuse large B cell lymphoma. J Clin Pathol 62(10): 903–907 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R (2001) Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature 412(6844): 341–346 [DOI] [PubMed] [Google Scholar]
- Peltola K, Hollmen M, Maula SM, Rainio E, Ristamaki R, Luukkaa M, Sandholm J, Sundvall M, Elenius K, Koskinen PJ, Grenman R, Jalkanen S (2009) Pim-1 kinase expression predicts radiation response in squamocellular carcinoma of head and neck and is under the control of epidermal growth factor receptor. Neoplasia 11(7): 629–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogacic V, Bullock AN, Fedorov O, Filippakopoulos P, Gasser C, Biondi A, Meyer-Monard S, Knapp S, Schwaller J (2007) Structural analysis identifies imidazo[1,2-b]pyridazines as PIM kinase inhibitors with in vitro antileukemic activity. Cancer Res 67(14): 6916–6924 [DOI] [PubMed] [Google Scholar]
- Rainio EM, Ahlfors H, Carter KL, Ruuska M, Matikainen S, Kieff E, Koskinen PJ (2005) Pim kinases are upregulated during Epstein-Barr virus infection and enhance EBNA2 activity. Virology 333(2): 201–206 [DOI] [PubMed] [Google Scholar]
- Schatz JH, Oricchio E, Wolfe AL, Jiang M, Linkov I, Maragulia J, Shi W, Zhang Z, Rajasekhar VK, Pagano NC, Porco JA, Teruya-Feldstein J, Rosen N, Zelenetz AD, Pelletier J, Wendel HG (2011) Targeting cap-dependent translation blocks converging survival signals by AKT and PIM kinases in lymphoma. J Exp Med 208(9): 1799–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirogane T, Fukada T, Muller JM, Shima DT, Hibi M, Hirano T (1999) Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity 11(6): 709–719 [DOI] [PubMed] [Google Scholar]
- Sivertsen EA, Galteland E, Mu D, Holte H, Meza-Zepeda L, Myklebost O, Patzke S, Smeland EB, Stokke T (2006) Gain of chromosome 6p is an infrequent cause of increased PIM1 expression in B-cell non-Hodgkin’s lymphomas. Leukemia 20(3): 539–542 [DOI] [PubMed] [Google Scholar]
- Tzankov A, Gschwendtner A, Augustin F, Fiegl M, Obermann EC, Dirnhofer S, Went P (2006) Diffuse large B-cell lymphoma with overexpression of cyclin e substantiates poor standard treatment response and inferior outcome. Clin Cancer Res 12(7 Pt 1): 2125–2132 [DOI] [PubMed] [Google Scholar]
- Tzankov A, Meier C, Hirschmann P, Went P, Pileri SA, Dirnhofer S (2008) Correlation of high numbers of intratumoral FOXP3+ regulatory T cells with improved survival in germinal center-like diffuse large B-cell lymphoma, follicular lymphoma and classical Hodgkin’s lymphoma. Haematologica 93(2): 193–200 [DOI] [PubMed] [Google Scholar]
- Tzankov A, Pehrs AC, Zimpfer A, Ascani S, Lugli A, Pileri S, Dirnhofer S (2003) Prognostic significance of CD44 expression in diffuse large B cell lymphoma of activated and germinal centre B cell-like types: a tissue microarray analysis of 90 cases. J Clin Pathol 56(10): 747–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzankov A, Zlobec I, Went P, Robl H, Hoeller S, Dirnhofer S (2010) Prognostic immunophenotypic biomarker studies in diffuse large B cell lymphoma with special emphasis on rational determination of cut-off scores. Leuk Lymphoma 51(2): 199–212 [DOI] [PubMed] [Google Scholar]
- Valdman A, Fang X, Pang ST, Ekman P, Egevad L (2004) Pim-1 expression in prostatic intraepithelial neoplasia and human prostate cancer. Prostate 60(4): 367–371 [DOI] [PubMed] [Google Scholar]
- Xie Y, Burcu M, Linn DE, Qiu Y, Baer MR (2010) Pim-1 kinase protects P-glycoprotein from degradation and enables its glycosylation and cell surface expression. Mol Pharmacol 78(2): 310–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Xu K, Linn DE, Yang X, Guo Z, Shimelis H, Nakanishi T, Ross DD, Chen H, Fazli L, Gleave ME, Qiu Y (2008) The 44-kDa Pim-1 kinase phosphorylates BCRP/ABCG2 and thereby promotes its multimerization and drug-resistant activity in human prostate cancer cells. J Biol Chem 283(6): 3349–3356 [DOI] [PubMed] [Google Scholar]
- Yip-Schneider MT, Horie M, Broxmeyer HE (1995) Transcriptional induction of pim-1 protein kinase gene expression by interferon gamma and posttranscriptional effects on costimulation with steel factor. Blood 85(12): 3494–3502 [PubMed] [Google Scholar]
- Zhang Y, Wang Z, Magnuson NS (2007) Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol Cancer Res 59: 909–922 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Hollmen J, Raty R, Aalto Y, Nagy B, Elonen E, Kere J, Mannila H, Franssila K, Knuutila S (2002) Investigatory and analytical approaches to differential gene expression profiling in mantle cell lymphoma. Br J Haematol 119(4): 905–915 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.