Abstract
Developmental endothelial cell locus-1 (Del-1) glycoprotein is secreted by endothelial cells and a subset of macrophages. Del-1 plays a regulatory role in vascular remodeling and functions in innate immunity through interaction with integrin αVβ3. Del-1 contains 3 epidermal growth factor (EGF)-like repeats and 2 discoidin-like domains. An Arg-Gly-Asp (RGD) motif in the second EGF domain (EGF2) mediates adhesion by endothelial cells and phagocytes. We report the crystal structure of its 3 EGF domains. The RGD motif of EGF2 forms a type II′ β turn at the tip of a long protruding loop, dubbed the RGD finger. Whereas EGF2 and EGF3 constitute a rigid rod via an interdomain calcium ion binding site, the long linker between EGF1 and EGF2 lends considerable flexibility to EGF1. Two unique O-linked glycans and 1 N-linked glycan locate to the opposite side of EGF2 from the RGD motif. These structural features favor integrin binding of the RGD finger. Mutagenesis data confirm the importance of having the RGD motif at the tip of the RGD finger. A database search for EGF domain sequences shows that this RGD finger is likely an evolutionary insertion and unique to the EGF domain of Del-1 and its homologue milk fat globule-EGF 8.—Schürpf, T., Chen, Q., Liu, J., Wang, R., Springer, T. A., Wang, J. The RGD finger of Del-1 is a unique structural feature critical for integrin binding.
Keywords: crystal structure, cell adhesion, innate immunity, extracellular matrix protein
Developmental endothelial cell locus-1 (Del-1) was originally identified as an extracellular matrix protein expressed by endothelial cells during embryonic vascular development. Del-1 was shown to promote adhesion of endothelial cells through its interaction with integrin receptors. It acts as a negative factor in vascular remodeling in the breaking-and-restructuring process during embryonic development (1). The 52-kDa Del-1 molecule has 3 N-terminal EGF-like domains, followed by 2 discoidin I-like or factor V C domains (2), C1 and C2. Interestingly, the second epidermal growth factor (EGF) domain (EGF2) contains an Arg-Gly-Asp (RGD) motif (1). The 3-residue RGD peptide motif was originally identified in fibronectin as responsible for mediating cell attachment. RGD motifs that function in integrin binding were later found in many other extracellular matrix proteins (3). All known RGD receptors on cell surfaces belong to the integrin family. Eight vertebrate integrins recognize an RGD motif in ligands (4, 5). In Del-1, this RGD motif is responsible for mediating the attachment of endothelial cells through binding to integrin αVβ3 (1). An Arg-Ala-Asp (RAD) mutant Del-1 was inactive in αVβ3-binding (6). Furthermore, Del-1 expression is low in normal brain tissue and is enhanced after cerebral ischemia, suggesting that this protein may participate in ischemia-induced angiogenesis (7, 8). More recently, Del-1 was found to bind leukocyte integrins (9).
Del-1 has been further implicated as a bridge between apoptotic cells and phagocytes. Del-1 specifically binds phosphatidylserine (PS) on apoptotic cells in a high-affinity interaction with its C1 and C2 lipid-binding domains (10). This leaves the RGD-bearing EGF domain of Del-1 available for binding to integrins on phagocytes. A closely related structural and functional homologue of Del-1 is milk fat globule-EGF 8 (MFG-E8; ref. 11). When cells are triggered to undergo apoptosis, PS will be exposed on the cell surface as an “eat me” signal. Del-1 and MFG-E8 function as secreted PS opsonins to facilitate the “tethering” of the apoptotic cells to phagocytes, for further “tickling” and clearance of the apoptotic cells. Del-1 and MFG-E8 provide molecular diversity to innate immunity to eliminate these dead cells. These two proteins are produced by distinct populations of macrophages (10). Moreover, whereas Del-1 is secreted from endothelial cells, MFG-E8 is secreted from epithelial cells (12). The bridging function of Del-1 and MFG-E8 is reminiscent of the mechanism with which innate immunity eliminates pathogens. That process involves complement C3 as an opsonin, bridging pathogens and phagocytes for pathogen clearance (13). As ligands for integrins αVβ3 and αVβ5, Del-1 and MFG-E8 also participate in angiogenesis and may represent relevant targets in cancer and other related diseases (14).
We report here the crystal structure of the 3 EGF domains of Del-1. We show that the RGD motif is located at the tip of the RGD finger, a long protruding loop at the N-terminal end of the EGF2 domain. We describe the calcium-binding site and O-linked as well as N-linked glycosylation sites in the EGF domains. Through cell adhesion assays, we demonstrate the critical importance of having the RGD motif at the tip of the RGD finger. The functional significance of these structural features and the comparison with other RGD motif-containing structures are discussed.
MATERIALS AND METHODS
Protein expression and purification
The 3 EGF-like domains of human Del-1 (D24-K157; EGF123) were PCR-amplified from Del-1 cDNA (MGC: 26287; Open Biosystems, Lafayatte, CO) and ligated into the pLEXm vector (15), together with an N-terminal mouse immunoglobulin κ chain signal peptide and a C-terminal hexahistidine tag. The Del-1-Fc fusion construct was made by subcloning a DNA fragment containing Kozak sequence, signal peptide, and EGF123 into the ET-5 vector, which fuses the C terminus of EGF123 in frame to human Fcγ1. Point mutations and deletions were introduced by overlap-extension PCR and confirmed by DNA sequencing.
His-tagged Del-1 was expressed in human embryonic kidney (HEK) 293S GnTI− cells (16) by transient transfection with pLEXm-Del-1, as described previously (17). Del-1-Fc fusion proteins were obtained by transfecting HEK293T cells with ET-5-Del-1 following the same procedure. In functional assays, supernatant from mock-transfected HEK293T cells was used as a control. His-tagged Del-1 was purified from cell culture supernatants using NiNTA beads (Qiagen, Valencia, CA) followed by gel filtration over a Superdex 200 10/300 GL column (GE Healthcare, Piscataway, NJ) equilibrated with 20 mM HEPES (pH 7.5) and 100 mM NaCl. Del-1-containing fractions were pooled and concentrated to 15 mg/ml. Serum-free transfection supernatants containing Del-1-Fc proteins were cleared by centrifugation, 0.2-μm filtered, and used directly for experiments.
Crystallization and structure determination
Crystals were grown at room temperature by the hanging-drop vapor-diffusion method. Equal volumes of protein solution (15 mg/ml in 20 mM HEPES, pH 7.5, and 100 mM sodium chloride) and reservoir solution (100 mM sodium acetate, pH 4.6; 30% PEG 4000; and 200 mM ammonium acetate) were mixed, and crystals appeared in 1-2 d. The reservoir solution supplemented with 15% (w/v) glycerol was used as a cryoprotectant. Diffraction data were collected at beamlines 24-ID-C and 19-ID at the Argonne National Laboratories (Argonne, IL, USA). The structure of Del-1 EGF repeats was determined by molecular replacement using the structure of human Notch-1 EGF domains 12-13 (Protein Data Bank [PDB] code 2VJ3) as the search model. Structure refinement and model building were performed with PHENIX (18, 19) and Coot (20). The model was validated with MolProbity (21). The data statistics and refinement results are listed in Supplemental Table S1. The coordinates and structure factors of the Del-1 structure have been deposited in the PDB (code 4D90).
Molecular weight measurement
Size-exclusion chromatography/multiangle light scattering (SEC/MALS; Wyatt Technology, Santa Barbara, CA, USA) analysis was used to measure the molecular weight of the Del-1 EGF123 fragment in solution. Protein (6.2 mg/ml, 200 μl) was loaded on a Superdex 200 column equilibrated with 20 mM HEPES (pH 7.5) and 100 mM sodium chloride. The molecular weight was measured according to the user manual.
HEK293-αVβ3 cells
HEK293 cells were cotransfected with full-length αV and β3 cDNAs in pEF1-puro (22) and pcDNA3.1/myc-His (23), respectively. Cells were selected in medium containing puromycin (1 μg/ml) and G418 (400 μg/ml) 40 h post-transfection. Live cells were further sorted based on αVβ3 expression to obtain single clones. Control HEK293 cells were transfected with vector cDNA only.
Cell adhesion assay
Del-1 substrates were prepared by coating wells of a microtiter plate with protein A (10 μg/ml in 100 mM carbonate, pH 9.6; Sigma, St. Louis, MO, USA) and blocking with heat-treated BSA. Transfection supernatants containing Del-1-Fc fusion proteins were diluted 2-fold with HBS/BSA (20 mM HEPES, pH 7.4; 135 mM NaCl; 5 mM KCl; 5.5 mM glucose; and 1% BSA) and directly added to protein A-coated wells (50 μl) for 1 h at 37°C. Remaining protein A binding sites were blocked with 1 mg/ml human γ-globulin (Sigma). HEK293-αVβ3 cells were fluorescently labeled as described previously (17) and resuspended at 2 × 106 cells/ml in HBS/BSA containing 2 mM Mn2+/0.2 mM Ca2+ and 25 μg/ml of mouse IgG1 (Sigma) or blocking anti-β3 monoclonal antibody 7E3 (24). Cell adhesion to Del-1 substrates was measured as described previously (17). Adhesion to wells coated with mock transfection supernatant was subtracted from adhesion to Del-1-coated wells.
Multimeric Del-1 binding assay
Multimeric Del-1-Fc immune complexes were formed by adding FITC-labeled goat anti-human Fcγ (100 μg/ml; Sigma) to Del-1-Fc transfection supernatants, which were diluted 2-fold with HBS and incubated for 30 min at room temperature in the dark. As a control, supernatant from mock-transfected cells or human myeloma IgG1,κ (2.5 μg/ml; Sigma) were complexed with anti-human Fcγ-FITC as described above. HEK293-αVβ3 transfectants were washed once with HBS/10 mM EDTA and twice with HBS. Aliquots of 25 μl (2×105 cells) were incubated with 25 μl of fluorescent immune complexes in the presence of either 1 mM Ca2+ and 1 mM Mg2+ or 2 mM Mn2+ and 0.2 mM Ca2+and 25 μg/ml of either mouse IgG1 or 7E3 antibody for 45 min at 37°C in the dark. Cells were washed, and cell-associated fluorescence was measured by flow cytometry on a FACScan (Becton Dickinson, Franklin Lakes, NJ, USA). Background mean fluorescence intensity (MFI) obtained with mock transfection control complexes was subtracted from Del-1 complex MFI values.
RESULTS
Structure of EGF domains of Del-1
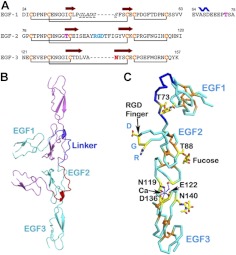
The Del-1 EGF123 protein expressed in the HEK293S GnTI− cell line yielded diffraction quality crystals in the P32 space group with 62% solvent. The structure has been determined to 2.6 Å resolution with molecular replacement using Notch EGF domains 12-13 (PDB code 2VJ3) as a search model. Figure 1A shows the sequence of EGF123 with secondary structures and disulfide bonds assigned, and Fig. 1B is the ribbon diagram of the 2 EGF123 molecules in the asymmetric unit. The EGF3 domain abuts EGF2. By contrast, a 12-residue, highly acidic linker separates the EGF1 and EGF2 domains (Fig. 1A). The linker has a short 310 helix in the middle. This long linker should render the EGF1 domain mobile relative to EGF2-EGF3 in solution. In the crystal, most parts of the EGF1 domain do not have any contacts with neighboring molecules. However, the linker between EGF1 and EGF2 curves around a protruding loop of the other molecule's EGF2 domain, such that the 2 molecules pack in a head-to-head fashion in the EGF2 regions, with the EGF1 domains bending ∼90° away (Fig. 1B). The very C-terminal segment of the EGF1 domain, Ser60-Ser61-Val62, actually forms a short antiparallel β structure with main-chain atoms of Tyr95 on the other molecule's protruding loop of EGF2 domain (Supplemental Fig. S1). The segment of Ser60-Ser61-Val62 is just next to the disulfide bond of Cys59-Cys50. These hydrogen bonds allow for the small EGF1 domain to be positioned in the crystal, but with a significantly higher average temperature factor of 130 as opposed to 52 and 54 for EGF2 and EGF3 (Supplemental Table S1). Accordingly, electron densities of the EGF1 domain are less well defined than in the other 2 domains, and densities are missing for residues 40–44 of molecule A and residues 40–45 of molecule B, which are at the very tip of their EGF1 domains (Fig. 1B and Supplemental Fig. S1). MALS analysis of a solution containing 6.2 mg/ml Del-1 EGF123 shows that the molecular weight ranges from ∼15 to 20 kDa, with an average of 17.2 kDa for the glycosylated protein. This agrees well with the calculated size of 15.3 kDa for the nonglycosylated monomeric protein and suggests that Del-1 EGF123 is primarily a monomer in solution, if glycosylation is taken into account (Supplemental Fig. S2). Figure 1C depicts the α-carbon backbone of a single EGF123 molecule in a view rotated ∼90° around a vertical axis from that in Fig. 1B.
Figure 1.
Structure of 3 EGF domains of Del-1. A) Sequences of 3 EGF domains with secondary structures and disulfide bonds assigned. The linker between EGF1 and EGF2 is shown separately after EGF1. Brown arrow denotes β strands, and the blue wave sign indicates a short helix. Glycosylation sites are shown in red, and the RGD site in EGF2 is in cyan. The missing part of EGF1 in the current model is in italic and underlined with a broken line. B) The 2 molecules in ribbon drawing pack head-to-head in the asymmetric unit. The 3 EGF domains in the cyan molecule are labeled. The linker between EGF1 and EGF2 is in dark blue. C) α-Carbon skeleton drawing of the cyan molecule. View is rotated roughly 90° around a vertical axis from that in panel B. The RGD finger, the cation Ca, the metal-binding site, and 3 glycosylation sites are depicted. The 3 disulfide bonds in each of the EGF domains are shown in orange. The linker is in dark blue as in panel B.
EGF is a small module, with the size of only 35–50 amino acid residues. It is stabilized by 6 conserved cysteines forming 3 disulfide bonds in the pattern of 1–3, 2–4, and 5–6 (25). EGF domains 1, 2, and 3 of Del-1 have 40, 43 and 37 residues, respectively. In addition to the 3 disulfide bonds, there is a small segment of β ribbon, as shown in Fig. 1A, B. More than a dozen intradomain hydrogen bonds seen in each EGF domain should help to keep the small domain in a defined shape.
Calcium-stabilized rodlike EGF2-EGF3 entity
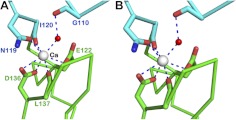
A large subgroup of EGF domains bind calcium (26). In the difference electron density map, we observed a peak above 6σ level at the interface between EGF2 and EGF3 in each molecule in the asymmetric unit. We assigned the observed density as Ca2+ (Fig. 1C). The cation has 7 ligands forming a pentagonal bipyramid, as seen in EGF domains from human clotting factor IX (27). In the EGF123 structure, among the 7 Ca2+ ligands, 3 are contributed by EGF2 and 4 by EGF3 (Figs. 1C and 2A). Two negatively charged residues from EGF3 (Glu122 and Asp136) directly bind and neutralize the cation. From EGF2, 1 side-chain amide oxygen of Asn119 coordinates the cation. In addition, 3 main-chain carbonyl oxygens from Ile120 and Gly110 (indirectly through a water molecule) of EGF2 and Leu137 of EGF3 also participate in coordination. A proposed calcium-binding consensus sequence of Asp-Leu/Ile-Asp-Gln-Cys (26) is similar to the sequence observed here, Asn119-Ile-Asn-Glu-Cys, the junction of the EGF2 and EGF3 domains. The coordination geometry is similar to that in Notch EGF 12-13 (Fig. 2B), the search model used to determine our structure. Indeed, the two abutting Notch EGF 12-13 domains can be superimposed onto EGF2-3 of Del-1 with an RMSD value of 1.9 Å for 83 residues. Supplemental Fig. S3 depicts the overlay of Notch EGF12-13 onto EGF2-EGF3 of Del-1 and shows that the interdomain orientation between EGF12 and EGF13 in Notch is essentially identical to that between EGF2 and EGF3 in Del-1. This finding agrees with the notion that a Ca2+ rigidifies the interdomain junction, leading to a near-linear, rodlike arrangement of 2 adjacent EGF domains (28).
Figure 2.
Calcium coordination geometry. A) Calcium coordination in Del-1. The metal is in silver, and the smaller water molecule in red. The calcium coordination has a typical pentagonal bipyramid shape. Two negatively charged D136 and E122 directly interact with the cation. Three carbonyl oxygen atoms from G110 (via a water molecule), I120, L137, and an oxygen atom from the amide group of N119, participate in coordination. B) A similar Ca-coordination observed in Notch EGF domains 12-13 (PDB 2VJ3).
Ca2+ binding appears important for orienting neighboring modules relative to each other in a manner that is required for biological function (26). The neighboring module can be another EGF domain, as seen here in Del-1, or can also be a different small disulfide-rich module, as in fibrillin (29, 30). Therefore, like many other EGF-containing proteins, Del-1 has its EGF2 and EGF3 domains lined up in a stiff rodlike structure, with a flexible EGF1 domain attached N-terminally to it.
Unusual glycosylation sites
EGF domains are known to have some unusual post-translational modifications, as manifested in Notch. Among others, O-linked fucose has been found attached to Thr or Ser in a consensus sequence motif of C2X4–5(S/T)C3 (where X4–5 are any 4–5 residues and C2 and C3 are the second and third Cys in an EGF domain, respectively; ref. 31). The addition of O-linked fucose directly to proteins is a highly specialized type of glycosylation. It is present in cysteine-rich sequences in EGF domains (32) and structurally observed in the type I repeats of thrombospondin-1 (33). The O-fucose of Notch extracellular EGF domains can be further elongated by a fucose-specific enzyme Fringe (reviewed in ref. 34).
In Del-1, clear densities can be seen for assigning a fucose moiety attached to Thr88 within the sequence Cys83-His-Asn-Gly-Gly-Thr-Cys89 of EGF2 (Supplemental Fig. S4), which fits the consensus sequence mentioned above. No densities for further carbohydrate residues can be assigned. In the literature, the function of this O-fucose alone and its glycan elongation have received extensive discussion, as reviewed for the Notch receptors (31, 32, 34). There is evidence that Escherichia coli-expressed human Notch1 ligand-binding domains without glycosylation appear to be well folded (35). A recent elegant study comparing structures of a chemically synthesized nonglycosylated EGF domain with a glycosylated version reveals a correctly formed structure without glycosylation. The NMR data also show that the methyl group of the fucose in the O-fucosylated form may stabilize the conformation of the domain (35, 36). The fucose in our structure has a B factor of 53, similar to the average B factors of EGF2, and much lower than the B factors of other glycans described below [N-linked N-acetyl-d-glucosamine (N-GlcNAc): 108; O-linked N-acetyl-d-galacosamine (O-GalNAc): 116], implying that this fucose may indeed help stabilize the EGF domain. The O-fucosylation site on Del-1 is on EGF2, on the opposite side of the integrin-binding RGD loop. As will be discussed later, it is therefore unlikely that the sugar moiety is directly involved in integrin binding.
The difference maps also show significant density extending from the side chain of Thr73 in the linker between EGF1 and EGF2. Both GlcNAc and GalNAc can be O-linked to Thr, and the density does not allow for distinguishing between these two sugars. We have built O-β-GalNAc at this site, which is the most common O-linked carbohydrate residue. In addition to the 2 O-linked glycosylation sites, there are 2 potential N-linked glycosylation sites in EGF123. Clear density demonstrates a sugar residue attached to Asn140 of EGF3 (Fig. 1C). The other potential glycosylation site is at the sequon Asn58-Cys-Ser within EGF1. However, we could not see any density for a glycan attached to Asn58, even though partial Endo H digestion confirms at least partial glycosylation of Asn58 (data not shown). Like the O-fucosylation site, the 3 residues (Asn58, Thr73, and Asn140) and their glycosylation modifications are located at the opposite side of the RGD loop (Fig. 1C). Therefore, these glycans are unlikely to directly engage in integrin binding.
Projection of an RGD finger as a long loop
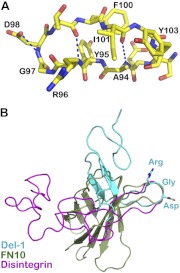
A biologically most interesting feature of the structure is the 13-residue loop (Ile91 to Tyr103) projecting out of the β ribbon in the EGF2 domain. An RGD motif is located at the very tip of this loop that we term the RGD finger (Fig. 1C). Figure 3A illustrates the local conformation of the loop. One remarkable feature of the finger is to have hydrophobic side-chain contacts on either side. A cluster of Ala94-Ile101-Tyr103 is located on one side of the finger, whereas on the other side, Tyr95 packs against Phe100 at a right angle, a commonly observed, energetically favorable interaction between aromatic rings in protein structures (37). There are hydrogen bonds elsewhere along the loop (Fig. 3A). In addition, as discussed above, when the two molecules pack into the crystal, the linker from one molecule wraps around the RGD finger of the other molecule, making important contacts, as seen in Fig. 1B and Supplemental Fig. S1. These structural features all help define the long and otherwise floppy loop.
Figure 3.
An RGD finger is located at the tip of a protruding loop. A) The RGD motif on a 13-residue loop of the Del-1 EGF2 domain. Note that Arg96-Gly-Asp-Thr form a β turn of the loop. One carboxyl oxygen atom of Asp98 forms a hydrogen bond with the main-chain nitrogen of the same Asp98 to stabilize the turn. Two main-chain hydrogen bonds and a few hydrophobic side-chain contacts help maintain the long loop. B) Superposition of RGD-containing loops from 3 proteins (superposition is based on Cα atoms of RGDX, X representing any other residues). EGF2 of Del-1 is in cyan, fibronectin type III domain 10 (PDB 1FNF) is in olive, and disintegrin (PDB 1J2L) is in magenta. The 3 proteins are conformationally very different, yet they all have their RGD positioned at the tip of a protruding loop.
Note that in fibronectin, fibronectin type III domain 10 (FN10) also has an RGD motif positioned at the tip of the FG loop, significantly projecting out of the domain as well (38). Another similar example of having an RGD motif at the tip of a long protruding loop is found in trimestatin, a member of the disintegrin family (39). Despite having very different overall domain structures, these three representative molecules share one feature in common: A loop projection presents the RGD motif out of the molecular body at about the same distance for integrin binding. Figure 3B is a superposition of FN10 and disintegrin onto EGF2 of Del-1, based on the loop tips, which depicts the similar protrusion of the RGD-containing loop in these structures.
The binding of RGD-containing peptides to integrins αVβ3 and αIIbβ3 have been structurally documented (5, 40, 41). In all cases, the RGD motif binds into a depression formed by the two integrin subunits. The Asp binds the metal ion-dependent adhesion site (MIDAS) on the β I domain, whereas the Arg side chain reaches into a binding cavity in the β-propeller domain of the integrin α subunit. These two residues constitute the major binding parts for interaction with the RGD-binding subset of integrins that do not have an I domain in their α chain. We envision that an RGD finger in integrin ligands like Del-1 and others is well suited for such a “pointing-in” binding.
RGD finger is an evolutionary insertion unique to the EGF domain of Del-1 and MFG-E8
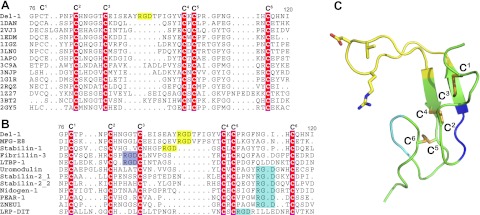
The 13-residue RGD finger in a small EGF2 domain of only 43 residues is an extraordinary structural feature. It connects the two antiparallel β strands and is located between cysteines C3 and C4 (Fig. 1A, C). We set out to search whether this is unique to Del-1. Using the Protein Structure Searching software by DaliLite (http://ekhidna.biocenter.helsinki.fi/dali_server/start), we hit many known EGF domain structures that aligned with Del-1's EGF2 domain. It is striking that the C3-C4 loop in Del-1 is longer than in any other known EGF domain structure. Figure 4A lists just the top 13 hits with a Z score above 4.
Figure 4.
The RGD finger is an evolutionary insertion unique to the EGF domains of Del-1 and MFG-E8. A) The top 13 hits of DALI search for Del-1 EGF2 domain. Compared to these known EGF structures, the Del-1 EGF2 domain appears to have an RGD motif inserted between cysteines C3 and C4. B) List of known EGF domains containing an RGD motif resulting from a genome-wide search. C) Locations of an RGD finger shown in panel B are displayed on the Del-1 EGF2 structure. Six cysteines in orange are labeled. Yellow, cyan, and dark blue regions indicate where an RGD motif positions in panel B.
Since Dali-search only looks for known structures, we further performed a human genome-wide search of EGF-like domains using the Simple Modular Architecture Research Tool (SMART; ref. 42; http://smart.embl-heidelberg.de/). There are 268 genes encoding proteins that contain ≥1 EGF domain, slightly more than the earlier estimation of 222 genes, which makes the EGF-like domain the 13th most populated module in human proteins (43). Within these EGF-like-domain-containing proteins, 12 EGF domains are found to have an RGD motif. These EGF-like domains can be divided into 3 categories with respect to where the RGD motif is located (Fig. 4B). The RGD-bearing C3-C4 loop in Del-1 is 15 residues long (Fig. 4B, C), so RGD motifs in shorter loop C2-C3 (Fig. 4B) are unlikely to serve as a pointing-in integrin-binding ligand (Fig. 4C). The third class of proteins has the RGD motif between C5 and C6 (Fig. 4B) and seems to have a protruding loop (Fig. 4C). Whether this group of proteins uses its RGD-containing EGF domains as integrin ligands remains an intriguing subject for exploration.
Only 3 EGF domains in human proteins have an RGD motif between C3 and C4 (Fig. 4B). As mentioned above, Del-1 and MFG-E8 belong to the same family. They are structurally and functionally homologous. MFG-E8 of most species is composed of 2 EGF domains at the N terminus followed by 2 discoidin-like domains. The first 2 of 3 EGF domains of Del-1 are homologous to the 2 EGF domains of MFG-E8. Del-1 and MFG-E8 both have an RGD motif in the loop between C3 and C4 of their EGF2 domain for integrin binding (44). Interestingly human MFG-E8 has only one EGF domain, corresponding to the EGF2 in Del-1 and MFG-E8 of most other species (45). From Fig. 4B, it is apparent that the RGD motif in the integrin-binding EGF domain of Del-1 and MFG-E8 is indeed centered in an unusually long loop, which makes the RGD reaching far out as a finger, suitable for contacting a depression formed between 2 integrin subunits. The long linker between EGF1 and EGF2 is unique to Del-1. The mouse MFG-E8 does not have such a linker between the two EGF domains, and human MFG-E8 has only one EGF domain (ref. 45 and Supplemental Fig. S5). The third protein in the category discussed is stabilin-1 (Fig. 4B), which was reported to be responsible for lymphocyte trafficking both in vascular and lymphatic vessels (46) and also plays a role as scavenger receptor (47). Since the RGD-carrying loop is substantially shorter than that of Del-1, stabilin-1 seems less likely to be an integrin ligand.
Centered position of the RGD sequence on the finger is crucial for integrin binding
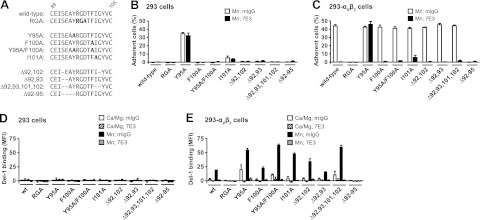
To address whether the position of RGD at the tip and the length of the loop are critical for functions of Del-1, we carried out adhesion experiments. We tested the functional importance of 3 features of the RGD finger: the hydrophobic residues surrounding the RGD motif, the length of the finger, and the relative position of the RGD on the fingertip (Fig. 5A). Mutants containing the 3 EGF-like domains of Del-1 were fused to a C-terminal human Fcγ1 tag. All mutants expressed at levels similar to wild-type Del-1, as confirmed by nonreducing SDS-PAGE (not shown). Adhesion of HEK293 transfectants expressing αVβ3 to wild-type Del-1 was stimulated by Mn2+ and completely RGD and αVβ3-dependent, because the Del-1 RGA mutant failed to support cell adhesion, adhesion was blocked by the anti-β3 antibody 7E3, and nontransfected HEK293 control cells did not adhere (Fig. 5B, C).
Figure 5.
Position of the Del-1 RGD sequence at the RGD fingertip is critical for interaction with integrin αVβ3. A) Point mutations or deletions in the RGD loop between C89 and C105 of Del-1. B, C) Adhesion of fluorescently labeled HEK293 control cells (B) or HEK293-αVβ3 transfectants (C) to Del-1-Fc proteins immobilized on protein A substrates. Adhesion was measured in buffer containing 2 mM Mn2+/0.2 mM Ca2+ and 25 μg/ml of either mouse IgG (open bars) or anti-β3 antibody 7E3 (solid bars). Adhesion to control substrates was <1.5% and subtracted from the data. D, E) Del-1-Fc proteins were complexed in solution with an FITC-labeled goat anti-human Fcγ antibody. Binding of multimeric Del-1 to HEK293 control cells (D) or HEK293-αVβ3 transfectants (E) in buffer with either 1 mM Ca2+/1 mM Mg2+ (open and striped bars), 2 mM Mn2+/0.2 mM Ca2+ (solid and shaded bars), in the presence of 25 μg/ml of either mouse IgG (open and solid bars) or anti-β3 antibody 7E3 (striped and shaded bars) was measured by flow cytometry. As a control, supernatant from mock-transfected cells was used for complex formation and gave mean fluorescence intensity (MFI) values of less than 8, which were subtracted from the Del-1 data to get specific binding. Human IgG1,κ control complexes bound with MFI values between 3.0 and 6.6. Data are averages ± sd from 2 independent experiments done either in triplicate (B, C) or in single determinations (D, E).
The hydrophobic residues Y95, F100, and I101 on the RGD-bearing loop were individually mutated to Ala. Furthermore, since Y95 and F100 pack against each other (Fig. 3A), they were mutated to Ala simultaneously, as well (Fig. 5A). All Del-1 mutants, in which a hydrophobic side chain was mutated, supported cell adhesion under activating conditions (Fig. 5C), indicating that RGD-mediated adhesion is not affected by these loop residues. Unexpectedly the Y95A but not the Y95A/F100A mutant gained adhesiveness for a HEK293 cell surface protein (Fig. 5B, C). This protein was different from αVβ3 and endogenously expressed on HEK293 cells, because cell adhesion was not blocked by 7E3 and nontransfected HEK293 control cells adhered to the Y95A mutant similar to the HEK293-αVβ3 transfectants (Fig. 5B, C). This activity gain appears to require Phe at position 100, because the Y95A/F100A double mutant showed wild-type behavior. Thus, the observed activity gain was not induced by disrupting the packing of Y95 and F100. The I101A mutant showed a similar non-αVβ3 activity, although to a much lesser extent (Fig. 5B, C). Adhesion to the F100A and Y95A/F100A mutants was completely αVβ3 dependent.
Either 1 or 2 residues were deleted from each side of the RGD finger in mutants Δ92,102 and Δ92,93,101,102, respectively, thereby shortening finger length while keeping the RGD motif still centered at the fingertip (Fig. 5A). The deleted residues were selected because they do not interact with residues outside the finger. Cell adhesion to Del-1 mutants with RGD centered on shorter fingers was not diminished, compared to wild-type Del-1, and completely αVβ3 dependent (Fig. 5B, C). To test for the importance of having the RGD motif centered at the fingertip, Del-1 mutants Δ92,93 and Δ92–95 were compared. Those constructs have the RGD motif offcentered on similarly shortened loops (Fig. 5A). Del-1 Δ92,93 supported levels of cell adhesion similar to wild-type. Deleting 4 residues on the same side of the finger (mutant Δ92–95), however, completely abolished αVβ3-mediated cell adhesion (Fig. 5C). This contrasts with deletion of the same number of residues on either side of the finger in mutant Δ92,93,101,102, highlighting the importance of having the RGD motif located at the tip of the loop, as an RGD finger.
Soluble Del-1-Fc multimer binding to αVβ3 transfectants or HEK293 control cells was also tested by flow cytometry (Fig. 5D, E). Wild-type Del-1 binding was inducible by manganese and was αVβ3 and RGD-dependent (Fig. 5D, E). Binding of the hydrophobic residue mutants Y95A, F100A, I101A, and Y95A/F100A to activated αVβ3 was equal or higher compared to wild-type protein binding (Fig. 5E). Unlike cell adhesion, binding in solution of all hydrophobic residue mutants was completely αVβ3-dependent (Fig. 5D, E). Shortening of the RGD finger by 2 and 4 residues with a centered RGD in Δ92,102 and Δ92,93,101,102, respectively, progressively enhanced integrin binding compared to wild-type. In contrast, offcentering RGD by 2 residues (Δ92–93) slightly diminished binding, and offcentering by 4 residues (Δ92–95) completely abolished binding to activated integrin (Fig. 5E). All soluble ligand-binding results, except the Y95A mutant, were therefore consistent with what was found in cell adhesion assays.
Taken together, our data show that the hydrophobic residues surrounding the RGD sequence are not required to maintain the loop structure for integrin binding or to occupy hydrophobic binding pockets. We also demonstrate that Del-1 with RGD fingers shortened by 1 or 2 residues on either side retain their integrin binding activity, as long as the RGD sequence remains on the fingertip, whereas offcentering the RGD sequence by 4 residues completely blocks the integrin binding capability of Del-1.
DISCUSSION
Del-1 was originally cloned and characterized as a matrix protein that promotes adhesion of endothelial cells through interaction with the integrin receptor αVβ3. An RGD motif on the EGF2 domain was thought to be responsible for integrin binding (1). The RGD-associated binding was later confirmed, because an RAD mutant was inactive (6). The work we report here provides the structural basis for how Del-1 presents an RGD finger for integrin binding.
The 3 EGF domains of Del-1 are configured such that the abutting EGF2-EGF3 domains form a rigidified rodlike entity through an interdomain calcium coordination. In contrast, EGF1 at the N terminus is flexibly linked through 12 residues to EGF2. Crystal packing constrains the EGF1 domain to bend away from the EGF2-EGF3 rod, so that the RGD finger on a long protruding loop of EGF2 is fully exposed. The substantially higher average temperature factor of EGF1 compared to the rest of the molecule reflects the mobility of this domain. Notably, soluble Del-1-Fc multimer binding to αVβ3 transfectants has confirmed that in solution EGF1 does not interfere with the integrin-binding ability of the RGD motif. The 2 glycans (on Thr88 and Asn140) are located on the side of the EGF2-EGF3 rod opposite from the RGD motif, and the other glycan on Thr73 is in the linker and probably mobile. These are all favorably outside the integrin-binding interface.
One key observation of our Del-1 EGF domains structure is that the integrin-binding RGD motif resides at the very tip of a remarkable 13-residue protruding C3-C4 loop, the RGD finger, of EGF2. Our cell adhesion assays clearly demonstrate that neither neighboring hydrophobic residues in the loop nor slight shortening of the loop affects the RGD-mediated adhesion. Rather, it is vital to have the RGD motif centered at the tip of a protruding finger. The ligand binding site of integrins is known to be located in a shallow depression formed between its 2 subunits (48). On binding to integrin, an RGD-bearing ligand presumably has to insert its RGD-containing loop into this depression for recognition. Our structural and functional findings on Del-1 show that an RGD finger protruding out from a domain fits this requirement well. It is intriguing to observe that an RGD-containing loop in FN10 and disintegrin also have their RGD at the tip of a long protruding loop as discussed above (Fig. 3B).
The EGF module was thought to provide a convenient structural scaffold for various functions, and in some cases to act as a spacer unit on cell-surface proteins (49). Genome-wide search for all human EGF domains shows that only Del-1 and its homologous MFG-E8 have an RGD motif located at a similar position (Fig. 4B). This conceivably reflects the evolutionary result of having an adequately long insertion between 2 cysteines, C3 and C4, in the small EGF domain of Del-1 and MFG-E8 for integrin binding. In other words, the evolutionary insertion of the RGD finger of the right size on an EGF domain may be unique to Del-1 and MFG-E8, 2 important extracellular matrix proteins that are binding integrins and participate in innate immunity.
Supplementary Material
Acknowledgments
The authors acknowledge U.S. National Institutes of Health grant HL103526 to T.A.S. and J.-H.W. The authors also thank the staff members at Advanced Photon Source beamlines 24-ID and 19-ID (Argonne National Laboratory, Argonne, IL, USA) for the help in X-ray data collection. Structural data have been deposited in the Protein Data Bank (PDB code 4D90).
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
- Del-1
- developmental endothelial cell locus-1
- EGF
- epidermal growth factor
- FN10
- fibronectin type III domain 10
- GalNAc
- N-acetyl-d-galacosamine
- GlcNAc
- N-acetyl-d-glucosamine
- HEK
- human embryonic kidney
- MFG-E8
- milk fat globule-EGF 8
- MFI
- mean fluorescence intensity
- PDB
- Protein Data Bank
- PS
- phosphatidylserine
- RAD
- Arg-Ala-Asp
- RGD
- Arg-Gly-Asp
- SEC/MALS
- size-exclusion chromatography/multiangle light scattering
REFERENCES
- 1. Hidai C., Zupancic T., Penta K., Mikhail A., Kawana M., Quertermous E. E., Aoka Y., Fukagawa M., Matsui Y., Platika D., Auerbach R., Hogan B. L., Snodgrass R., Quertermous T. (1998) Cloning and characterization of developmental endothelial locus-1: an embryonic endothelial cell protein that binds the alphavbeta3 integrin receptor. Genes Dev. 12, 21–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fuentes-Prior P., Fujikawa K., Pratt K. P. (2002) New insights into binding interfaces of coagulation factors V and VIII and their homologues lessons from high resolution crystal structures. Curr. Protein Pept. Sci. 3, 313–339 [DOI] [PubMed] [Google Scholar]
- 3. D'Souza S. E., Ginsberg M. H., Plow E. F. (1991) Arginyl-glycyl-aspartic acid (RGD): a cell adhesion motif. Trends Biochem. Sci. 16, 246–250 [DOI] [PubMed] [Google Scholar]
- 4. Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 5. Springer T. A., Zhu J., Xiao T. (2008) Structural basis for distinctive recognition of fibrinogen gammaC peptide by the platelet integrin alphaIIbbeta3. J. Cell Biol. 182, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Penta K., Varner J. A., Liaw L., Hidai C., Schatzman R., Quertermous T. (1999) Del1 induces integrin signaling and angiogenesis by ligation of alphaVbeta3. J. Biol. Chem. 274, 11101–11109 [DOI] [PubMed] [Google Scholar]
- 7. Fan Y., Zhu W., Yang M., Zhu Y., Shen F., Hao Q., Young W. L., Yang G. Y., Chen Y. (2008) Del-1 gene transfer induces cerebral angiogenesis in mice. Brain Res. 1219, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ho H. K., Jang J. J., Kaji S., Spektor G., Fong A., Yang P., Hu B. S., Schatzman R., Quertermous T., Cooke J. P. (2004) Developmental endothelial locus-1 (Del-1), a novel angiogenic protein: its role in ischemia. Circulation 109, 1314–1319 [DOI] [PubMed] [Google Scholar]
- 9. Choi E. Y., Chavakis E., Czabanka M. A., Langer H. F., Fraemohs L., Economopoulou M., Kundu R. K., Orlandi A., Zheng Y. Y., Prieto D. A., Ballantyne C. M., Constant S. L., Aird W. C., Papayannopoulou T., Gahmberg C. G., Udey M. C., Vajkoczy P., Quertermous T., Dimmeler S., Weber C., Chavakis T. (2008) Del-1, an endogenous leukocyte-endothelial adhesion inhibitor, limits inflammatory cell recruitment. Science 322, 1101–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hanayama R., Tanaka M., Miwa K., Nagata S. (2004) Expression of developmental endothelial locus-1 in a subset of macrophages for engulfment of apoptotic cells. J. Immunol. 172, 3876–3882 [DOI] [PubMed] [Google Scholar]
- 11. Hanayama R., Tanaka M., Miwa K., Shinohara A., Iwamatsu A., Nagata S. (2002) Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187 [DOI] [PubMed] [Google Scholar]
- 12. Wu Y., Tibrewal N., Birge R. B. (2006) Phosphatidylserine recognition by phagocytes: a view to a kill. Trends Cell Biol. 16, 189–197 [DOI] [PubMed] [Google Scholar]
- 13. Van Lookeren Campagne M., Wiesmann C., Brown E. J. (2007) Macrophage complement receptors and pathogen clearance. Cell. Microbiol. 9, 2095–2102 [DOI] [PubMed] [Google Scholar]
- 14. Neutzner M., Lopez T., Feng X., Bergmann-Leitner E. S., Leitner W. W., Udey M. C. (2007) MFG-E8/lactadherin promotes tumor growth in an angiogenesis-dependent transgenic mouse model of multistage carcinogenesis. Cancer Res. 67, 6777–6785 [DOI] [PubMed] [Google Scholar]
- 15. Aricescu A. R., Lu W., Jones E. Y. (2006) A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. Allogr. D Biol. Crystallogr. 62, 1243–1250 [DOI] [PubMed] [Google Scholar]
- 16. Reeves P. J., Kim J. M., Khorana H. G. (2002) Structure and function in rhodopsin: a tetracycline-inducible system in stable mammalian cell lines for high-level expression of opsin mutants. Proc. Natl. Acad. Sci. U. S. A. 99, 13413–13418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schurpf T., Springer T. A. (2011) Regulation of integrin affinity on cell surfaces. EMBO J. 30, 4712–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Adams P. D., Gopal K., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Pai R. K., Read R. J., Romo T. D., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C. (2004) Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 11, 53–55 [DOI] [PubMed] [Google Scholar]
- 19. Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Allogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. Allogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Allogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takagi J., Erickson H. P., Springer T. A. (2001) C-terminal opening mimics ‘inside-out’ activation of integrin alpha5beta1. Nat. Struct. Biol. 8, 412–416 [DOI] [PubMed] [Google Scholar]
- 23. Smagghe B. J., Huang P. S., Ban Y. E., Baker D., Springer T. A. (2010) Modulation of integrin activation by an entropic spring in the β-knee. J. Biol. Chem. 285, 32954–32966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coller B. S. (1985) A new murine monoclonal antibody reports an activation-dependent change in the conformation and/or microenvironment of the platelet glycoprotein IIb/IIIa complex. J. Clin. Invest. 76, 101–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carpenter G., Cohen S. (1979) Epidermal growth factor. Annu. Rev. Biochem. 48, 193–216 [DOI] [PubMed] [Google Scholar]
- 26. Stenflo J., Stenberg Y., Muranyi A. (2000) Calcium-binding EGF-like modules in coagulation proteinases: function of the calcium ion in module interactions. Biochim. Biophys. Acta 1477, 51–63 [DOI] [PubMed] [Google Scholar]
- 27. Rao Z., Handford P., Mayhew M., Knott V., Brownlee G. G., Stuart D. (1995) The structure of a Ca(2+)-binding epidermal growth factor-like domain: its role in protein-protein interactions. Cell 82, 131–141 [DOI] [PubMed] [Google Scholar]
- 28. Smallridge R. S., Whiteman P., Werner J. M., Campbell I. D., Handford P. A., Downing A. K. (2003) Solution structure and dynamics of a calcium binding epidermal growth factor-like domain pair from the neonatal region of human fibrillin-1. J. Biol. Chem. 278, 12199–12206 [DOI] [PubMed] [Google Scholar]
- 29. Jensen S. A., Iqbal S., Lowe E. D., Redfield C., Handford P. A. (2009) Structure and interdomain interactions of a hybrid domain: a disulphide-rich module of the fibrillin/LTBP superfamily of matrix proteins. Structure 17, 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee S. S., Knott V., Jovanovic J., Harlos K., Grimes J. M., Choulier L., Mardon H. J., Stuart D. I., Handford P. A. (2004) Structure of the integrin binding fragment from fibrillin-1 gives new insights into microfibril organization. Structure 12, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tien A. C., Rajan A., Bellen H. J. (2009) A Notch updated. J. Cell Biol. 184, 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris R. J., Spellman M. W. (1993) O-linked fucose and other post-translational modifications unique to EGF modules. Glycobiology 3, 219–224 [DOI] [PubMed] [Google Scholar]
- 33. Tan K., Duquette M., Liu J. H., Dong Y., Zhang R., Joachimiak A., Lawler J., Wang J. H. (2002) Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. J. Cell Biol. 159, 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stanley P., Okajima T. (2010) Roles of glycosylation in Notch signaling. Curr. Top. Dev. Biol. 92, 131–164 [DOI] [PubMed] [Google Scholar]
- 35. Cordle J., Redfieldz C., Stacey M., van der Merwe P. A., Willis A. C., Champion B. R., Hambleton S., Handford P. A. (2008) Localization of the delta-like-1-binding site in human Notch-1 and its modulation by calcium affinity. J. Biol. Chem. 283, 11785–11793 [DOI] [PubMed] [Google Scholar]
- 36. Hiruma-Shimizu K., Hosoguchi K., Liu Y., Fujitani N., Ohta T., Hinou H., Matsushita T., Shimizu H., Feizi T., Nishimura S. (2010) Chemical synthesis, folding, and structural insights into O-fucosylated epidermal growth factor-like repeat 12 of mouse Notch-1 receptor. J. Am. Chem. Soc. 132, 14857–14865 [DOI] [PubMed] [Google Scholar]
- 37. Burley S. K., Petsko G. A. (1985) Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science 229, 23–28 [DOI] [PubMed] [Google Scholar]
- 38. Leahy D. J., Aukhil I., Erickson H. P. (1996) 2.0Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell 84, 155–164 [DOI] [PubMed] [Google Scholar]
- 39. Fujii Y., Okuda D., Fujimoto Z., Horii K., Morita T., Mizuno H. (2003) Crystal structure of trimestatin, a disintegrin containing a cell adhesion recognition motif RGD. J. Mol. Biol. 332, 1115–1122 [DOI] [PubMed] [Google Scholar]
- 40. Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. (2002) Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 41. Xiao T., Takagi J., Coller B. S., Wang J. H., Springer T. A. (2004) Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 432, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lander E. S., et al. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 44. Raymond A., Ensslin M. A., Shur B. D. (2009) SED1/MFG-E8: a bi-motif protein that orchestrates diverse cellular interactions. J. Cell. Biochem. 106, 957–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Couto J. R., Taylor M. R., Godwin S. G., Ceriani R. L., Peterson J. A. (1996) Cloning and sequence analysis of human breast epithelial antigen BA46 reveals an RGD cell adhesion sequence presented on an epidermal growth factor-like domain. DNA Cell Biol. 15, 281–286 [DOI] [PubMed] [Google Scholar]
- 46. Irjala H., Elima K., Johansson E. L., Merinen M., Kontula K., Alanen K., Grenman R., Salmi M., Jalkanen S. (2003) The same endothelial receptor controls lymphocyte traffic both in vascular and lymphatic vessels. Eur. J. Immunol. 33, 815–824 [DOI] [PubMed] [Google Scholar]
- 47. Prevo R., Banerji S., Ni J., Jackson D. G. (2004) Rapid plasma membrane-endosomal trafficking of the lymph node sinus and high endothelial venule scavenger receptor/homing receptor stabilin-1 (FEEL-1/CLEVER-1). J. Biol. Chem. 279, 52580–52592 [DOI] [PubMed] [Google Scholar]
- 48. Luo B. H., Carman C. V., Springer T. A. (2007) Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Campbell I., Bork P. (1993) Epidermal growth factor-like modules. Curr. Opin. Struct. Biol. 3, 385–392 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.