Abstract
This article describes predictive, preventive value of genetic tests and the implication of the use of testing for personalized treatment. This year marks the 10th anniversity of publishing of the sequence of the human genome. One important area of application of this mega project is a development of genetic tests for mutation detection in single gene disorders that has impact for pediatric age group patients and analyzing susceptibility genes as risk factors in common disorders. Types of genetic tests, new emerging technologies will enable developments of high-throughput approaches by microarrays of great application capacity as described here. As it is usual for all technologies used in health care, bioethical concerns has to be delt with. The ethical, social and governance issues associated with genetic testing are discussed.
Keywords: Genotyping, Predictive value, SNP, Arrays, Personalized treatment, Ethics
The Human Genome Project (HGP), which was initiated in the 1990s, is usually labeled as the mega project of 20th century that has revolutionized biomedical sciences [1]. With the sequencing of the human genome completed in the last decade medical science is looking forward to advances in the diagnosis, prevention and treatment of Mendelian as well as common disorders. Consequently, we are moving to the area characterized as personalized medicine in health care [2–4]. It is also expected that genomics will play a major role in new drug design for disease related pathways and also be used as a predictive tool for individual responses to drugs [5]. The research dedicated to genetics/genomics is expected to have a high impact on pediatric practice where one area of application relevant to information gained from the HGP is genetic testing of germline DNA variations or acquired changes in the somatic tissues such as cancer [6]. There are over 5000 inherited monogenic diseases affecting children and prevelance at birth of this diseases is around 10/1000 [7]. These rare diseases are very severe and usually can not be cured. Therefore, genetic testing in diagnosis of monogenic disorders is a relevant area of research and health care practice in pediatric age group of patients [8].
There are different types of genetic testing defined as [9, 10]:
Newborn screening is used immediately following birth to identify treatable conditions to ensure that treatment begins as early as possible before irreversible damage takes hold [11]. One of the diseases first introduced in the newborn screening programs was phenylketonuria (PKU), the first example of a treatable genetic condition. Today, there are more than 30 diseases that are included in the screening programs.
The following criteria proposed by James M.G. Wilson and Gunnar Jungner [12] as given below, are widely used to assess the validity of screening for a given condition:
“The condition being screened for should be an important health problem;
The natural history of the condition should be well understood;
There should be a detectable early stage;
Treatment at an early stage should be of more benefit than at a later stage;
A suitable test should be devised for the early stage;
The test should be acceptable;
Intervals for repeating the test should be determined;
Adequate health service provision should be made for the extra clinical workload resulting from screening;
The risks, both physical and psychological, should be less than the benefits.”
Diagnostic testing is done to confirm or rule out a diagnosis made by physical examination. It can be done at the post-natal or pre-natal stage during pregnancy. There are more than 2,000 clinical and research tests available today (www.ncbi.nlm.nih.gov/sites/GeneTests).
In prenatal testing, fetal cells can be obtained by invasive procedures such as amniocentesis at the 13th-15th of pregnancy by the aspiration of amniotic fluid sample [13]. Another invasive procedure is chorionic vilus sampling where chorionic tissue of fetal trophoblast origin can be obtained at the 12th week of gestation [14].
For genetic testing new non-invasive procedures are coming into use. Fetal cells from maternal circulation and cell free fetal nucleic acid (cffNA) from pregnant women are beginning to be used for diagnosis [15]. Analysis of fetal aneupleudies is a recent application of the use of cffNA [16]. Prenatal diagnosis can allow for prevention or very early intervention for management of monogenic diseases.
Pre-implantation testing is a form of prenatal testing where embryos created through in vitro fertilization are tested for a particular disease, and only the healthy embryos are implanted in the uterus for the pregnancy [17].
Carrier screening is done to test if any person with a positive family history carries a mutant copy of the gene. In some ethnic groups where a particular gene mutation is high, couples can request premarital testing to obtain information about the risk of their having a child with the disease. One well known population screening program was established for the Ashkenazi Jewish population where the frequency of Tay-Sachs Disease (GM2 Gangliosidosis type B) is quite high (1/3600) [18].
Predictive testing can be done in cases with positive family history to test for mutations that cause disease only later in life, such as Huntington’s disease. Since there are no signs of disease at the time of testing, this type of testing can also be called presymptomatic testing. With increased information available to medical practitioners, it is expected that predictive testing will have a broad applications for common disorders such as diabetes, hypertension, coronary heart disease and Alzheimer’s, where genetic variations that cause susceptibility to a particular disease and, thus, become a risk factor can be detected very early in life. This type of testing holds the promise of prevention through life style improvements or personalized medication, where each genotype will direct the course of specific therapy. However, pediatric applications of predictive testing is rife with ethical issues and controversies that need to be addressed properly, so that testing results can be used for the patient benefits, rather than harming the patients [19].
Pharmacogenetic testing is used to assess responsiveness to drug therapy. Taking into account individual factors as contributing elements to the outcome of therapy, before assigning the use of a drug, paves the way to “personalized medication.” So far, this type of testing has found applications mostly in cancer drugs. There is still a long way for methodologies to become available in a hospital setting, so that personal drug prescriptions according to genotypic findings can have a real positive impact on clinical practices [20, 21].
Modifications on the predicted effects of genetic variations
The basic outcome of a genetic test is the physician’s ability to predict the phenotype of the patient and take preventive measures or design the management of the disease. Hopefully, by further advances and appliactions of genotypic predictions personalized therapies based on the individual genome information will come into practice.
However, investigations into the molecular pathophysiology of Mendelian diseases have revealed various mechanisms that make it difficult to prediction the phenotypic outcome [22].
Genetic heterogeneity and genetic modifiers can play a role, so that a determination of the predictive variation on the single gene level may not reflect the phenotype directly.
These mechanisms need to be taken into consideration during the interpretation of test results as well as the counseling of patients and their families.
Deafness is a well known example with high degree of heterogeneity. With over one hundred genes identified, a testing is overwhelming. The variability of the phenotype, further, contributes to the complexity of the prediction of the outcome based on an individual’s test results [23].
Another cause of variability is the presence of modifying affects to the phenotype. Cystic Fibrosis (CF) is an example of an AR monogenic disease where modifiers have been identified. The severity of the lung disease in CF has been shown to be affected by modifier genes and environmental factors. Polymorphisms identified in genes for transforming growth factor β1, Mannose binding lectin 2, tumor necrosis factor α and mutations in alpha1 antitrypsin gene, all lead to a variability in the lung function of CF patients [24, 25]. The further source of variability are nucleotide repeats in intronic regions of the CF transmembrane conductance regulator (CFTR) gene [26].
Another example can be given for more than 20 neuromuscular and neurodegenerative diseases, where a disease phenotype is due to expanded poly-amino acid tracts. These trinucleotide repeats can take place in coding or noncoding regions of genes involved in diseases such as Huntington’s disease, myotonic dystrophy, fragile X mental retardation syndrome (FMR1). The repeats become unstable, if expansion increases through transmission to next generations with the concomitant increase of severity of the phenotype, the phenomenon called anticipation [27, 28]. That makes it difficult to predict the outcome of disease solely according to the detected DNA variation.
Predictive value of genetic testing for personalized therapy
Individualized drug therapy based on findings of DNA variations in pediatrics is exemplified in monogenic diseases such as CF and Duchenne Muscular Dystrophy (DMD), and also in childhood cancers such as Acute Lenfoblastic Leukemia (ALL).
In CF, mutations in the CFTR gene cause abnormal salt and water transport across epithelial surfaces in systems such as the hepato-bilary gastrointestinal, respiratory and reproductive systems, as well as in the sweat glands. The identification of the CFTR gene confirmed the findings that chloride secretion was defective in CF [29].
Therapies for CF have so far aimed at alleviating the end organ damage in different systems, such as pancreatic enzyme replacement, and in therapies for lung disease, such as mucus retention, inflammation and infection. However, none of these therapies can correct the underlying molecular defects of the mutant CFTR gene. Compounds that are classified as potentiators and correctors are being investigated as gene targeting chemicals. Potentiators play a role in allowing the CFTR channel to stay open longer, thus allowing more chloride ions to pass through and are candidates for corrections of Class I mutations such as G551D and delta F508 [30]. VX-770 is a potentiator (Vertex Pharmaceuticals) that is being used in Phase III clinical trials for patients with at least one allele carrying the G551D mutation, and also in Phase II trials in homozygous patients for delta F508 mutation (www.clinicaltrials.gov).
As for correctors, these are compounds that facilitate the movement of delta F508 CFTR from the ER to the apical membranes. Quinazoline VRT-325, aminoarylthiazoles and bisaminomethylbithiazoles are therapeutics under investigation [31].
Table 1: In CF therapies [31, www.cff.org, www.clinicaltrials.gov] are directed to the consequences of the non-functional CFTR which are related to abnormal ion transport. New therapies are emerging where the novel compounds are targeted specifically against the defect of the gene variant which holds promise for personalized medicine.
Table 1.
Therapies for cystic fibrosis
| Treatment of Clinical Consequences | |
| 1. Pancreatic Disease | • Pancreatic enzyme replacement (PERT) |
| 2. Lung Disease | |
| a) Mucus clearance | • DNAse - Pulmozyme (Genentech) |
| • UTP analog - Donufosol (Inspire Pharmaceuticals) | |
| • Lantibiotic - Duramycin (Moli 1901 - Lantibio) | |
| • Hypertonic saline | |
| b) Anti-inflammation | • Nonsteroidal anti-inflammatory drugs - Ibuprofen |
| • Corticosteroids - Prednisone | |
| • Macrolide antibiotics - Zithromax | |
| c) Anti-microbial | • TOBI (Novartis) - for P.aeruginosa |
| Gene Variant Targeted Pharmaceutical Compounds (Personalized Treatment) | |
| Protein truncation | • Aminoglycoside antibiotics - Gentamicin |
| • Non-aminoglycoside compounds - PTC124 (Ataluren) (PTC Therapeutics) | |
| Channel regulation (Potentiators) | • Vx-770 (Vertex pharmaceuticals) - G551D Mutation |
| Protein folding (Correctors) | • Vx-809 (Vertex pharmaceuticals) - ΔF508 Mutation |
Another example of potential therapeutic compounds, that are being investigated, are chemicals that aim at stop codon mutations, where a premature stop in translation results in a truncated protein product with loss of function [32]. In DMD, Ataluren (PTC124-3-[5-(2-fluorophenyl)-1,2,4-oxadiazol-3-yl]benzoic acid) is an experimental drug that has been shown to suppress the premature termination and restoration of dystrophin protein production in vitro and in mdx mice [33].
In cancer patients, pharmacogenetic testing can identify those who may run the risk of developing toxicity to chemotherapeutic agents, thus allowing for an individualized regiment for therapy [34]. For example, decreased enzyme activity has been determined for variant forms of the enzyme thiopurine methyl transferase (TPMT). G238C, G460A, and A719G are the prevalent mutations in Caucasians, and A719G is the prevalent variant in Asian, African and African-American populations that lead to 80–95% decreased activity resulting in thiopurine drug toxicity in childhood ALL treatment with 6-mercaptopurine, 6-thioguanine and azathiopurine. Testing prior to therapy will help to stratify patients that will benefit from tailored therapeutics [35].
Emerging technologies
Array systems
Molecular genetic testing owes its analytical validity much to the use of Polymerase Chain Reaction (PCR) where a segment of DNA can be amplified in vitro, and the presence of a disease causing mutation can be checked by various methods, such as Restriction Fragment Length Polymorphism (RFLP) utilizing restriction enzymes or sequencing technology to identify novel mutations within the amplified segments [36]. Although these methods are now within standard operation procedures for mutation detection at many genetic testing labs, the capacity is limited for testing of a very few samples simultaneously.
Arrays may be described as chips depicted as in Fig. 1, carrying multiples of assays that allow for testing of tens of thousands of different variants simultaneously, and they are beginning to find wide clinical use [37]. Single nucleotide polymorphism (SNP) genotyping arrays and resequencing are two high throughput examples used to identify genetic variants. HGP has led to the discovery of many SNPs as a source of genetic variation, and genotyping for SNPs has played a major role in international efforts to map for disease susceptibility genes [38].
Fig. 1.
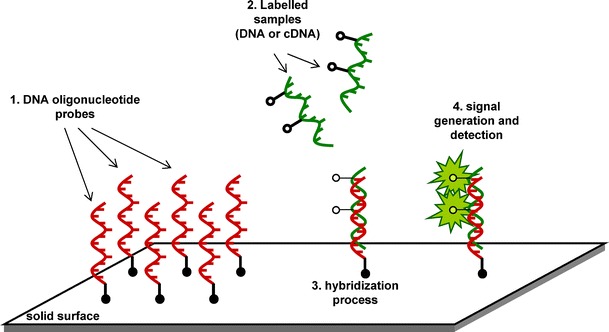
Basic steps of microarray analysis: The synthetic oligonucleotides are either plotted or directly synthesized on a solid surface that can be described as a “chip” (1). Fluorescent labelled DNA samples can be used for techniques such as SNP analysis, re-sequencing applications or cDNA samples utilized in transcriptomics for gene expression analysis (2). Sequence complementarity between the probe and the sample target allows for hybridization (3). Bioinformatics tools are used to process the generated signals followed by hybridization and washing steps (4)
In some monogenic diseases, multigenic inheritance and genetic heterogeneity make diagnosis quite taxing. In such cases, SNP microarray applications for homozygosity mapping to identify disease locus in children from consanguineous parents facilitates the diagnosis [39].
Microarrays are further employed in studies, such as those analyzing cancer tissues, to identify acquired somatic mutations or gene expresson patterns that have prognostic and therapeutic value in childhood cancers [40–42].
Array technologies can also increase the capacity of newborn screening programs. Guthrie cards, which are used to store blood spots from newborns are a valuable source of genetic material. However, the yield of extracted DNA is usually very low. With improved methods developed to obtain higher yields of DNA of better quality, genome scale microarray analysis is becoming commercially available [43]. This will increase the capacity of newborn screening programs to detect multitudes of variations that may have predictive implications for the neonate. As seen from the examples above, microarrays have a wide range of applications. Technical advances are increasingly enabling the marketing of higher density arrays with decreasing costs, resulting in the wider use of microarrays for research and clinical purposes, thus, aiding the identification of new disease genes, diagnosis of disease causing mutations and susceptibility variants with predictive value [44].
Copy Number Variation (CNV) detection: CNV can be described as a segment of DNA that carries a different copy number as compared to a reference genome sequence. Submicroscopic alterations such as insertions/duplications or deletions constitute the structural genetic variation that can be detected by tecniques such as comparative genome hybridization (CGH) or chromosomal microarray (CMA) [45].
CNVs are beginning to be found as associated with diseases such as asthma, autoimmune diseases, autism and schizophrenia. Genetic experts recommend CMA for screening children with congenital abnormalities, autism spectrum disorders and developmental delays [46, 47].
Whole genome sequencing (WGS)
The identification of any sequence of DNA with a disease causing variation has the potential for accurate diagnosis and proper therapy. The automated Sanger method is the classical sequencing technology currently used worldwide to identify genomic variations. However, this technology is not effective in sequencing an entire genome within a reasonable amount of time at a reasonable cost. Technology is taking long strides towards ‘next generation sequencing’ technologies where an entire genome can be sequenced in a short period of time with high resolution and efficiency [48, 49]. The realization of the ultimate vision of “personalized medicine” depends on the future use of WGS for genotyping of individual genomes. Although the cost is prohibitive today and WGS is used only in research settings, it is hoped that falling costs (ultimately assumed as 1,000 USD/Genome) will enable genetic testing laboratories to employ WGS in clinical settings [50].
Genome Wide Association Studies (GWAS)
GWAS is a powerful tool for prediction of risk in common diseases. When compared to monogenic diseases where only mutations in one single gene are the causative factor, in complex diseases multiple genes interact with the environment to lead to the phenotype. Since they are more prevalent in the populations, it is obvious that trying to predict the genetic risk factors from an early age and developing appropriate genetic tests for these multifactorial diseases will be an important advance in health care. Prevention of disease and personalized treatment modalities based on the genetic make up of individuals is a priority area of biomedical research. Towards this goal, new high resolution technologies are being developed. GWAS relies on the new microarray technologies to study simultaneously the associative role of thousands of SNPs as genetic risk factors for common traits. So far, these studies are in preclinical phase to use GWAS results in health care and management of disease, that needs to definite association of one single variant as the susceptibility factor for the disease phenotype. GWAS for complex diseases such as bipolar disorder, coronary artery disease, Crohn’s disease, hypertension, rheumatoid arthritis, type 1 and 2 diabetes was carried out by the Welcome Trust Case Control Consortium [51]. Since GWAS studies rely on control populations it is very important that for different population SNP haplotypes will be investigated prior to undertaking such studies.
The integration of data sets from genomics and transcriptomics platforms will have an impact in the future for disease stratification and testing for individual genomic variations leading to the final goal of HGP: the prevention of disease and the introduction of therapies based on individual genomics information [52].
Ethical issues
Genetic testing has some aspects that differentiates it from other medical tests, requiring the use of an entirely different ethical framework [53–55]. This type of testing has implications for the individual, family and the society [56]. For the individual, the decision to test should take into account the benefits versus the risks, such as the psychological effects, and stigmatization. For the members of the family, the result of the testing will have the potential of stigmatization, parental guilt and anxiety in individual members of the family knowing they may also run the risk of a certain disease. Societies need to be able to debate and reflect on basic ethical questions such as the right to access health care, non-marginalization and discrimination [57]. In some diseases such as X-linked monogenic diseases, the determination of the gender of the fetus may have predictive value. However, in some societies, sex determination may also be used as a method of selecting babies with a desired gender. This is, of course, against basic ethical values such as human dignity and human rights.
There are different phases during a genetic test that need to be governed. An ideal flow of the procedures would begin with pre-testing counseling and end with post-testing counseling. Furthermore, basic ethical guidelines should be followed: a written informed consent must be obtained from the parents or legal guardians of pediatric patients; the privacy of the data and confidentiality during testing must be secured and the results must not be released to third parties such as school officials or insurance agencies without authorization [58].
There are presently no treatments for most genetic disorders, so testing does not alter the course of the disease or its outcome. Thus, the question of testing will become an ethical dilemma that needs careful reflection on the part of medical practitioner and the family of the patient. In particular, predictive testing in children for adult onset disorders calls for special attention to the psychological risks such as anxiety attached to testing [59].
Considering the ethical, social and psychological burden of genetic testing, counseling becomes an integral part of services both before and after testing as previously mentioned.
Quality assurance and governance
The quality of genetic testing must be assured in order to minimize risk due to erroneous results [60]. This is possible through licensing, accreditation and certification as the methods of oversight for laboratories that offer genetic testing. To gain a license, a laboratory has to have authorization from a relevant government agency. Through accreditation, the competence of the laboratory is benchmarked against an external standard. Certification is a method to indicate the assessment of the quality management of the laboratory and both procedures are dependent on external audits [61].
The analytical validity of the tests from a laboratory performing genetic testing is usually benchmarked through an external quality assurance scheme (EQA) or proficiency testing. Besides laboratory procedures used for the tests, other procedures such as the interpretation of results and issuing of the patients’ reports are very important indicators of quality assurance for genetic testing laboratories.
Europe wide efforts are being implemented to increase the quality of genetic testing. One example is the EC Project EuroGentest, which is an NoE (network of excellence) project supported through the Framework Programs (FP6) with the mission to harmonize the genetic testing. The major outputs of the project are the organization of external quality assurance schemes, advice on accreditation and certification of genetic testing services, and the development of guidelines for genetic testing (www.eurogentest.org). Another European effort to promote quality in genetic testing is European Molecular Genetics Quality Assurance Network (EMQN) (www.emqn.org) which works closely with EuroGentest and the CF Network to provide external quality assessment schemes to many labs in Europe and worldwide.
Conclusion and perspectives
Genomics offers powerful tools for health care systems where early predictive value of genetic markers can be exploited for the prevention and personalized therapy of disease. These tools are being used to develop genetic tests in monogenic diseases and it is expected that multifactorial disease diagnosis and management will also benefit from genomics approaches. To be able to incorporate the use of predictive disease markers in the clinical applications, we need to wait for the reduction of cost, in order to increase their applicability in health care systems. To be able to detect the risk of disease at a very early stage, even in prenatal stages of life, more non-invasive methods of sampling such as fetal cells in the maternal circulation and cell free fetal nucleic acid needs to be developed and their sensitivity and robustness should be tested. More medical professionals need to be informed about genomics (and other *omics technologies) to be able to use them for the benefit of their patients, that needs a new approach in medical education in the new era of medicine. Furthermore, awareness in the societies is necessary for proper application of new technologies and demand for genetic testing as a part of health care delivery systems. Predictive testing is already beginning to become a large market item [62]. Especially in terms of consumer benefits; genetic testing becomes an issue for different levels of regulations [63].
Since the ultimate benefit of determining the risk at an early stage of a disease development advantages by personalized management and treatment of disease, in different societies should be covered in terms of their access to genomic technologies. This is a bioethical concern when health is considered as a common good for all human beings and access to adequate health care is a basic ethical imperative of human dignity. As a result, international collaborations and open access to information is crucial to remove the disparities in genomics medicine for many countries: when used properly, genomics holds hope for prediction, prevention and treatment of disease based on individual patients profiles.
References
- 1.The human genome. Science genome map. Science. 2001; 291:1218. [PubMed]
- 2.Ginsburg GS, Willard HF. Bottom of form genomic and personalized medicine: foundations and applications. Transl Res. 2009;154:277–87. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Hong KW, Oh B. Overview of personalized medicine in the disease genomic era. BMB Rep. 2010;43:643–8. doi: 10.5483/BMBRep.2010.43.10.643. [DOI] [PubMed] [Google Scholar]
- 4.Lander ES. Initial impact of the sequencing of the human genome. 2011;470:187–97 [DOI] [PubMed]
- 5.Weinshilboum R, Wang L. Pharmacogenomics: bench to bedside. Nat Rev Drug Discov. 2004;3:739–48. doi: 10.1038/nrd1497. [DOI] [PubMed] [Google Scholar]
- 6.Genetic Testing , Policy Issues for the New Millennium. OECD. 2000.
- 7.“Genomics and Word Health”, Report of the Advisory Committee on Health Research. WHO. 2002.
- 8.Boat TF. The future of pediatric research. J Pediatr. 2007;151:21–27. doi: 10.1016/j.jpeds.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 9.Kaariainen H, Nippert I, Nys H, Kristoffersson U, Schmidtke J, Soini S. Euro Gentest. Definitions of genetic testing in European and other legal documents (Varga O and Sequeiros J) http://www.eurogentest.org/students/info/public/unit3/DefinitionsGeneticTestingFinal.xhtml. [DOI] [PMC free article] [PubMed]
- 10.Javaher P, Kaariainen H, Kristoffersson U, Nippert I, Sequeiros J, Zimmern R, Schmidtke J. EuroGentest: DNA-based testing for heritable disorders in Europe. Community Genet. 2008;11:75–120. doi: 10.1159/000111984. [DOI] [PubMed] [Google Scholar]
- 11.Watson MS, Lloyd-Puryear MA, Mann MY, Rinaldo P, Howell RR (eds). Newborn screening: Toward a uniform screening panel and system. Genet Med. 2006; 8:12S–252S. [DOI] [PMC free article] [PubMed]
- 12.Wilson JM, Jungner G. Principles and practice of screening for disease. Public Health WHO. 1968.
- 13.Nadler HL, Gerbie AB. Role of amniocentesis in the intrauterine detection of genetic disorders. N Engl J Med. 1970;282:596–9. doi: 10.1056/NEJM197003122821105. [DOI] [PubMed] [Google Scholar]
- 14.Modell B. Chorionic villus sampling. Evaluating safety and efficacy. Lancet. 1985;1:737–40. doi: 10.1016/S0140-6736(85)91272-3. [DOI] [PubMed] [Google Scholar]
- 15.Rafi I, Chitty L. Cell-free fetal DNA and non-invasive prenatal diagnosis. Br J Gen Pract. 2009;59:e146–8. doi: 10.3399/bjgp09X420572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, Foo CH, Xie B, Tsui NB, Lun FM, Zee BC, Lau TK, Cantor CR, Lo YM. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA. 2008;105:20458–63. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Basille C, Frydman R, El Aly A, Hesters L, Fanchin R, Tachdjian G, Steffann J, LeLorc’h M, Achour-Frydman N. Preimplantation genetic diagnosis: state of the art. Eur J Obstet Gynecol Reprod Biol. 2009;145:9–13. doi: 10.1016/j.ejogrb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Bach G, Tomczak J, Risch N, Ekstein J. Tay-Sachs screening in the Jewish Ashkenazi population: DNA testing is the preferred procedure. Am J Med Genet. 2001;99:70–5. doi: 10.1002/1096-8628(20010215)99:1<70::AID-AJMG1120>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 19.Evans JP, Skrzynia C, Burke W. The complexities of predictive genetic testing. BMJ. 2001;322:1052–6. doi: 10.1136/bmj.322.7293.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vizirianakis IS. Nanomedicine and personalized medicine toward the application of pharmacotyping in clinical practice to improve drug-delivery outcomes. Nanomedicine. 2011;7:11–17. doi: 10.1016/j.nano.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Vijverberg SJ, Pieters T, Cornel MC. Ethical and social issues in pharmacogenomics testing. Curr Pharm Des. 2010;16:245–52. doi: 10.2174/138161210790112700. [DOI] [PubMed] [Google Scholar]
- 22.Scriver CR, Waters PJ. Monogenic traits are not simple: lessons from phenylketonuria. Trends Genet. 1999;15:267–72. doi: 10.1016/S0168-9525(99)01761-8. [DOI] [PubMed] [Google Scholar]
- 23.Dror AA, Avraham KB. Hearing impairment: a panoply of genes and functions. Neuron. 2010;68:293–308. doi: 10.1016/j.neuron.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 24.Cutting GR. Modifier genetics: cystic fibrosis. Annu Rev Genomics Hum Genet. 2005;6:237–60. doi: 10.1146/annurev.genom.6.080604.162254. [DOI] [PubMed] [Google Scholar]
- 25.Dorfman R, Sandford A, Taylor C, Huang B, Frangolias D, Wang Y, Sang R, Pereira L, Sun L, Berthiaume Y, Tsui LC, Paré PD, Durie P, Corey M, Zielenski J. Complex two-gene modulation of lung disease severity in children with cystic fibrosis. J Clin Invest. 2008;118:1040–9. doi: 10.1172/JCI33754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hefferon TW, Groman JD, Yurk CE, Cutting GR. A variable dinucleotide repeat in the CFTR gene contributes to phenotype diversity by forming RNA secondary structures that alter splicing. Proc Natl Acad Sci USA. 2004;101:3504–9. doi: 10.1073/pnas.0400182101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2011;11:786–99. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–58. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kerem B, Rommens JM, Buchanan JA, Markiewicz D, Cox TK, Chakravarti A, Buchwald M, Tsui LC. Identification of the cystic fibrosis gene: genetic analysis. Science. 1989;245:1073–80. doi: 10.1126/science.2570460. [DOI] [PubMed] [Google Scholar]
- 30.Zielenski J, Tsui LC. Cystic fibrosis: genotypic and phenotypic variations. Annu Rev Genet. 1995;29:777–807. doi: 10.1146/annurev.ge.29.120195.004021. [DOI] [PubMed] [Google Scholar]
- 31.Kreindler JL. Cystic fibrosis: exploiting its genetic basis in the hunt for new therapies. Pharmacol Ther. 2010;125:219–29. doi: 10.1016/j.pharmthera.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malik V, Rodino-Klapac LR, Viollet L, Wall C, King W, Al-Dahhak R, Lewis S, Shilling CJ, Kota J, Serrano-Munuera C, Hayes J, Mahan JD, Campbell KJ, Banwell B, Dasouki M, Watts V, Sivakumar K, Bien-Willner R, Flanigan KM, Sahenk Z, Barohn RJ, Walker CM, Mendell JR. Gentamicin-induced readthrough of stop codons in Duchenne muscular dystrophy. Ann Neurol. 2010;67:771–80. doi: 10.1002/ana.22024. [DOI] [PubMed] [Google Scholar]
- 33.Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, Wilde RG, Karp G, Takasugi J, Chen G, Jones S, Ren H, Moon YC, Corson D, Turpoff AA, Campbell JA, Conn MM, Khan A, Almstead NG, Hedrick J, Mollin A, Risher N, Weetall M, Yeh S, Branstrom AA, Colacino JM, Babiak J, Ju WD, Hirawat S, Northcutt VJ, Miller LL, Spatrick P, He F, Kawana M, Feng H, Jacobson A, Peltz SW. Sweeney HL.PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 34.Ezzeldin HH, Diasio RB. Genetic testing in cancer therapeutics. Clin Cancer Res. 2006;12:4137–41. doi: 10.1158/1078-0432.CCR-06-0707. [DOI] [PubMed] [Google Scholar]
- 35.Cheok MH, Evans WE. Acute lymphoblastic leukaemia: a model for the pharmacogenomics of cancer therapy. Nat Rev Cancer. 2006;6:117–29. doi: 10.1038/nrc1800. [DOI] [PubMed] [Google Scholar]
- 36.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell, 4th edition. New York: Garland Science. 2002.
- 37.Müller UR, Nicolau DV. (Eds.). Microarray Technology and its Application. Berlin Heidelberg New York: Springer; 2005.
- 38.Manolio TA, Collins FS. The HapMap and genome-wide association studies in diagnosis and therapy. Annu Rev Med. 2009;60:443–56. doi: 10.1146/annurev.med.60.061907.093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.H’mida-Ben Brahim D, M’zahem A, Assoum M, Bouhlal Y, Fattori F, Anheim M, Ali-Pacha L, Ferrat F, Chaouch M, Lagier-Tourenne C, Drouot N, Thibaut C, Benhassine T, Sifi Y, Stoppa-Lyonnet D, N’Guyen K, Poujet J, Hamri A, Hentati F, Amouri R, Santorelli FM, Tazir M, Koenig M. Molecular diagnosis of known recessive ataxias by homozygosity mapping with SNP arrays. J Neurol. 2011;258:56–67. doi: 10.1007/s00415-010-5682-5. [DOI] [PubMed] [Google Scholar]
- 40.Wordsworth S, Papanicolas I, Buchanan J, Frayling I, Taylor J, Tomlinson I. Molecular testing for somatic cancer mutations:a survey of current and future testing in UK laboratories. J Clin Pathol. 2008;61:373–6. doi: 10.1136/jcp.2007.048819. [DOI] [PubMed] [Google Scholar]
- 41.Yagi T, Morimoto A, Eguchi M, Hibi S, Sako M, Ishii E, Mizutani S, Imashuku S, Ohki M, Ichikawa H. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102:1849–56. doi: 10.1182/blood-2003-02-0578. [DOI] [PubMed] [Google Scholar]
- 42.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, Gallia GL, Jallo GI, Binder ZA, Nikolsky Y, Hartigan J, Smith DR, Gerhard DS, Fults DW, VandenBerg S, Berger MS, Marie SK, Shinjo SM, Clara C, Phillips PC, Minturn JE, Biegel JA, Judkins AR, Resnick AC, Storm PB, Curran T, He Y, Rasheed BA, Friedman HS, Keir ST, McLendon R, Northcott PA, Taylor MD, Burger PC, Riggins GJ, Karchin R, Parmigiani G, Bigner DD, Yan H, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–9. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hardin J, Finnell RH, Wong D, Hogan ME, Horovitz J, Shu J, Shaw GM. Whole genome microarray analysis, from neonatal blood cards. BMC Genet. 2009;10:38. doi: 10.1186/1471-2156-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ropers HH. New perspectives for the elucidation of genetic disorders. Am J Hum Genet. 2007;81:199–207. doi: 10.1086/520679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human.disease pathogenesis. Genomics. 2009;93:22–6. doi: 10.1016/j.ygeno.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pani AM, Hobart HH, Morris CA, Mervis CB, Bray-Ward P, Kimberley KW, Rios CM, Clark RC, Gulbronson MD, Gowans GC, Gregg RG. Genome rearrangements detected by SNP microarrays in individuals with intellectual disability referred with possible Williams syndrome. PLoS ONE. 2010;5:e12349. doi: 10.1371/journal.pone.0012349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, Church DM, Crolla JA, Eichler EE, Epstein CJ, Faucett WA, Feuk L, Friedman JM, Hamosh A, Jackson L, Kaminsky EB, Kok K, Krantz ID, Kuhn RM, Lee C, Ostell JM, Rosenberg C, Scherer SW, Spinner NB, Stavropoulos DJ, Tepperberg JH, Thorland EC, Vermeesch JR, Waggoner DJ, Watson MS, Martin CL, Ledbetter DH. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 49.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. 1000 Genomes Project Consortium. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mardis ER. A decade’s perspective on DNA sequencing technology. Nature. 2011;470:198–203. doi: 10.1038/nature09796. [DOI] [PubMed] [Google Scholar]
- 51.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Committee on Bioethics (American Academy of Pediatrics): Ethical Issues With Genetic Testing in Pediatrics. Pediatrics. 2001;101:1451–5. [DOI] [PubMed]
- 54.The universal Declaration on the Human Genome and Human Rights :from theory to practice. UNESCO. 2000.
- 55.International Declaration on Human Genetic Data. UNESCO. 2004. [DOI] [PubMed]
- 56.Botkin JR, Teutsch SM, Kaye CI, Hayes M, Haddow JE, Bradley LA, Szegda K. EGAPP Working Group. Outcomes of interest in evidence-based evaluations of genetic tests. Genet Med. 2010;12:228–35. doi: 10.1097/GIM.0b013e3181fe5d50. [DOI] [PubMed] [Google Scholar]
- 57.Hall J, Viney R, Haas M. Taking a count: the evaluation of genetic testing. Aust N Z J Public Health. 1998;22(7):754–8. doi: 10.1111/j.1467-842X.1998.tb01488.x. [DOI] [PubMed] [Google Scholar]
- 58.25 Recommendations on the ethical, legal and social implications of genetic testing. The ECExpertGroup. 2004 (http://ec.europa.eu/research/conferences/2004/genetic/pdf/recommendations_en.pdf)
- 59.Malpas PJ. Predictive genetic testing of children for adult-onset diseases and psychological harm. J Med Ethics. 2008;34:275–8. doi: 10.1136/jme.2006.019802. [DOI] [PubMed] [Google Scholar]
- 60.On behalf of the European Society of Human Genetics’ PPPC, S Ayme. Provision of genetic services in Europe: current practices and issues. Eur J Hum Genet. 2003;11:900–2. [DOI] [PubMed]
- 61.Quality assurance and Proficiency Testing for Molecular Genetic Testing. OECD. 2005.
- 62.Gregory-Jones S, Wright C. Size of the direct-to-consumer genomic testing market. Genet Med. 2010;12:594. doi: 10.1097/GIM.0b013e3181ead743. [DOI] [PubMed] [Google Scholar]
- 63.European Society of Human Genetics. Statement of the ESHG on direct-to-consumer genetic testing for health-related purposes. Eur J Hum Genet. 2010;18:1271–3. [DOI] [PMC free article] [PubMed]


