Abstract
Pathogenesis of many bacterially-induced inflammatory diseases is driven by toll- like receptor (TLR) mediated immune responses following recognition of bacterial factors by different TLRs. Periodontitis is a chronic inflammation of the tooth supporting apparatus often leading to tooth loss, and is caused by a Gram-negative bacterial consortium that includes Tannerella forsythia. This bacterium expresses a virulence factor, the BspA, which drives periodontal inflammation by activating TLR2. The N- terminal portion of the BspA protein comprises a leucine-rich repeat (LRR) domain previously shown to be involved in the binding and activation of TLR2. The objective of the current study was to identify specific epitopes in the LRR domain of BspA that interact with TLR2. Our results demonstrate that a sequence motif GC(S/T)GLXSIT is involved in mediating the interaction of BspA with TLR2. Thus, our study has identified a peptide motif that mediates the binding of a bacterial protein to TLR2 and highlights the promiscuous nature of TLR2 with respect to ligand binding. This work could provide a structural basis for designing peptidomimetics to modulate the activity of TLR2 in order to block bacterially-induced inflammation.
Keywords: leucine-rich repeat protein, BspA, TLR-2, Tannerella forsythia
1. INTRODUCTION
Pattern-recognition receptors (PPRs) of the Toll-like receptor (TLRs) family recognize distinct microbial patterns to transduce intracellular signaling for release of inflammatory cytokines involved in orchestrating innate and adaptive immune responses [1]. For example, TLR2 primarily binds acylated peptides, and in cooperation with TLR1 or TLR6 is able to differentially discriminate tri- or di-acylated peptides, respectively. However, TLR2 is also quite unrestricted with respect to its ligand specificity, evidenced from recent studies demonstrating its ability to bind non-acylated peptides and proteins of diverse origin with no structural similarity [2–11]. In infectious diseases, such as periodontitis, a bacterially-induced chronic inflammation of the tooth supporting tissues often leading to tooth loss, TLRs modulate inflammatory responses of the host to oral bacteria during disease pathogenesis [12]. Tannerella forsythia, a pathogen strongly implicated in periodontitis, expresses a cell-surface associated and secreted protein BspA [13], which by activating TLR2 induces the secretion of inflammatory cytokines [14, 15]. Furthermore, TLR2 signaling leads to Th2 cell bias, which ultimately drives T. forsythia-induced periodontal inflammation and jaw associated bone loss in mice [16].
BspA comprises a horse-shoe shaped leucine-rich domain [17], which is formed by the 23 tandem repeats of a leucine-rich repeat (LRR) motif in the N-terminal portion, and four bacterial immunoglobulin-like domains in the C-terminal portion [13, 18]. The LRR motif in BspA belongs to a cysteine-containing subtype, which might allow the LRR domain to adapt a cysteine-ladder conformation reported in other cysteine containing LRR proteins [19]. We have previously shown that BspA activates TLR2/1 receptor heterodimer and that the LRR region comprising LRR repeats 1–16 is involved in direct binding to TLR2 [14].
The objective of the current study was to identify specific motif(s) within the LRR 1–16 region of BspA that bind and activate TLR2. Our approaches involved screening a series of overlapping synthetic peptides derived from the LRR 1–16 region for binding and activation of TLR2 by using reporter cell lines and macrophages from wild-type and TLR2 knockout mice.
2. MATERIALS AND METHODS
2.1. Reagents
Toll-like receptor agonists Pam3CSK4 lipopeptide, highly purified Escherichia coli K-12 LPS (Ec. LPS), polymyxin B sulfate, human monocytic THP1-Blue cells and HEK293 (hTLR2/hTLR1) cells stably expressing human TLR2 and TLR1 and a reporter plasmid, pNiFty2, expressing a NF-κB-inducible secreted alkaline phosphatase (SEAP) were purchased from InvivoGen (San Diego, CA). Monoclonal anti-TLR2 (MAb) and isotype control (IgG2a) antibodies were obtained from eBioscience. Synthetic peptides were obtained from GenScript (Piscataway, NJ). The recombinant His-tagged proteins were purified from E. coli lysate by metal-chelation chromatography followed by decontamination of LPS by polymixin B immobilized resin (Detoxi-Gel; Pierce) as described; BspA LRR1 domain (rBspLRR1) [14], TdLrrA [20], or YopM [21]. The purity of the recombinant proteins was routinely confirmed by SDS-PAGE and silver staining; this yielded single band of expected size with no detectable ladder-like pattern typical of LPS contamination. Furthermore, LPS levels of less than 0.010 ng/µg protein were detected using amebocyte based assay system (Lonza).
2.2. Trypsin digestion
The purified rBspLRR1 was digested for 4 h at 37°C with TPCK-treated trypsin (Promega) at a ratio of 1:100 (enzyme:protein) by weight, trypsin activity was then inactivated by adding phenylmethylsulfonyl fluoride to 1 mM final concentration and the mixture was desalted by gel filtration on a Sephadex G-25 (GE Biosciences) column equilibrated in PBS (10 mM phosphate buffer, 150 mM NaCl, pH 7.3). The protein fraction eluting in the void volume (proteins > 5 × 103 kDa) was collected and subjected to tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis on gradient gels (4–20%; Invitrogen, Carlsbad, CA), and the separated protein fragments were detected by silver staining. In-silico, BspLRR1 region is potentially cleaved by trypsin into 14 peptides (Fig. 1). The results indicated that the trypsin digestion was complete as no band migrating at the expected size of rBspLRR1 was visible but bands migrating at 6–8 kDa range expected to be cleaved from BspLRR1 were visible (data not shown).
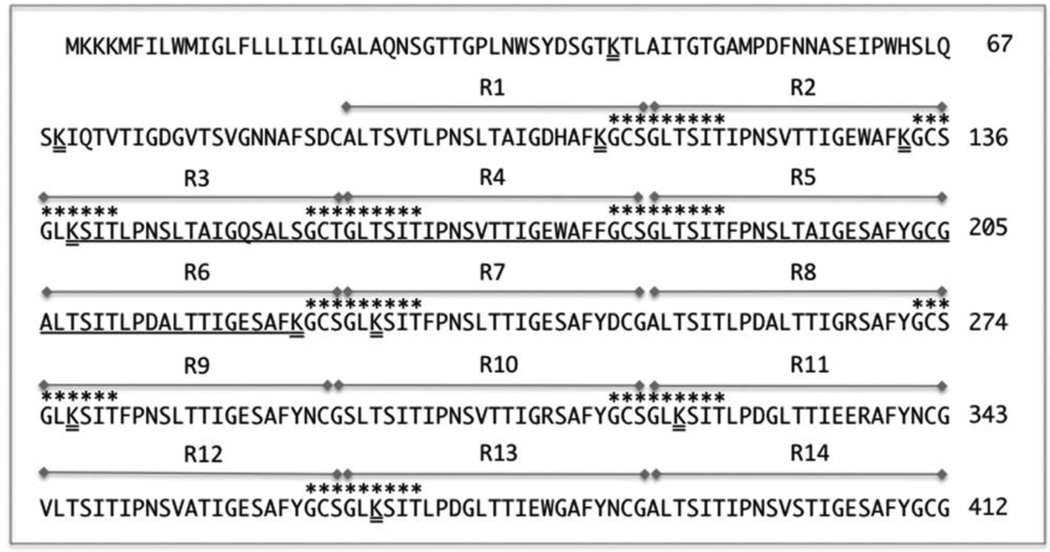
Fig. 1.
Amino acid sequence of the LRR1 domain of BspA (GenBank ID: AAC82625.1). LRR repeats 1–14 are indicated with double sides arrays over the repeats. The largest ~ 8-KD fragment to which overlapping peptides were synthesized (Table 1) is underlined. Trypsin prone lysine sites are double underlined. Sequences indicated with asterisks over the amino acid residues represent a motif GC(S/T)GLxSIT.
2.3. NF-κB activation in monocytic cell line THP-1
THP1-Blue cells stably expressing NF-κB-inducible secreted alkaline phosphatase were used to test the activity of peptides. Briefly, THP1-Blue cells were cultured in RPMI media containing 10% heat-inactivated FBS, 2 mM glutamine, 200µg/ml Zeocin at a density of 2 × 105 per ml in 48-well plates at 37°C in CO2 incubator. Cells were challenged with different agonists or synthetic peptides for 16–18 hrs and, culture supernatants were collected, clarified by centrifugation, diluted 1:50 or 1:100, and NF-κB activity was measured by quantification of SEAP activity using a colorimetric enzyme assay Quanti-Blue (Invivogen). Optical densities of colored end products were measured at 620 nm with a microplate reader.
2.4. Assessment of TLR2 activation with a reporter cell line
HEK293(hTLR2/hTLR1) cells were seeded at 6 × 104 cells per well in a 48-well plate in DMEM containing 10% heat inactivated fetal bovine serum,2 mM glutamine, 50 µg/ml gentamycin, and 10 µg/ml blasticidin. The cells were cultured at 37°C at 5% CO2 overnight to 60% confluency. The cells were transfected with the pNifty2 reporter plasmid using the FuGENE 6 transfection reagent according to the manufacturer's instructions (Roche Diagnostics, Indianapolis, IN). The total amount of transfected plasmid DNA was equalized by supplementation with an empty vector, pFLAG-CMV. Polymyxin B sulfate (10 µg/ml) was added to each recombinant protein prior to stimulation to block any contaminating LPS in the preparation. At 24 hr post transfection, the cells were preincubated with anti-TLR2 MAb (TLR2.1) or an Ig isotype-matched control (IgG2a) for 30 min and then incubated with Pam3CSK4 (1 µg/ml), rBspLRR1 and trypsin digest of BspLRR1 (D1 domain) for 16 hr at 37°C in humidified air containing 5% (vol/vol) CO2. rBspLRR1 and trypsin digests of rBspLRR1 were tested at 5 µg/ml final concentration based on our previous study [14]. The culture supernatants were collected, clarified by centrifugation, and SEAP activity was measured as described above.
2.5. Synthetic peptides
Synthetic peptides covering the entire length of BspA LRR repeats 3–6 (Fig. 1) with an overlap of 2 amino acids were synthesized and purified at a purity level of >95% (Table 1). Peptides with sequences in the reverse or scrambled order to the sequence of BspA peptide were used as negative controls. All peptides tested negative for endotoxin by the Limulus amebocyte lysate assay (Lonza). Selected peptides were obtained as fluorescently labeled with fluorescein isothiocyanate (FITC) (GenScript). All peptides were dissolved in culture grade dimethyl sulfoxide and tested at final concentrations of 5, 50 and 150 µg/ml.
Table 1.
| Peptide* | Sequence |
|---|---|
| P140–155 | SITLPNSLTAIGQSAL |
| P154–168 | ALSGCTGLTSITIPN |
| P167–181 | PNSVTTIGEWAFFGC |
| P180–194 | GCSGLTSITFPNSLT |
| P193–208 | LTAIGESAFYGCGALT |
| P207–221 | LTSITLPDALTTIGE |
| P220–234 | GESAFKGCSGLKSIT |
| P215–226 | ALTTIGESAFKG |
| P149–169 | AIGQSALSGCTGLTSITIPNS |
| Scram | GIISTQPTLGINTSLACASGS |
numbers indicate residue positions based on the BspA protein sequence (GenBank ID: AAC82625.1)
2.6. Isolation of macrophages and stimulation
Specific-pathogen free BALB/cJ mice (WT) were obtained from The Jackson Laboratory (Bar Harbor, ME). TLR2−/− mice on BALB/cJ background are bred and maintained at our animal facility [16]. All animal procedures were conducted in compliance with the protocols approved by the University at Buffalo Institutional Animal Care and Use Committee. Mouse peritoneal macrophages were prepared as previously described [16, 22]. A total of 2 × 105 single suspensions cells/well were added to 48-well tissue culture plates and allowed to adhere to the substrate by culturing them for 3–4 hr at 37°C. Non- adherent cells were removed by gentle washings with warm PBS, and adhered cells were used for stimulation.
2.7. Flow cytomtery-based binding assay
HEK 293 (hTLR1/2) cells or mouse peritoneal macrophages were seeded at a density of 2 × 106 /ml in 96 well U-bottom microplates in PBS/2% FBS and incubated with increasing concentrations of FITC-labeled peptides for 1 h at 4°C or 37°C. Unbound peptides were removed by three washes of PBS/2% FBS at 4°C. Cells were resuspended in 300 µl of PBS/2% FBS and immediately analyzed by flow cytometry. For the competition assay, cells were co-incubated with 5 µg/ml FITC-labeled peptide in the presence of increasing concentrations of unlabeled peptide (5–25 µg/ml). Cell-bound fluorescence was analyzed after gating on intact cells with a FACScan flow cytometer using Cell Quest acquisition and analysis software (BD Biosciences).
2.8. Cytokine ELISA
The levels of IL-6, TNF-α and IL-1β cytokines in culture supernatants were measured by using enzyme-linked immunosorbent assay (ELISA) kits purchased from eBiosciences.
2.9. Statistical Analysis
Data were evaluated by analysis of variance and the Dunnett multiple comparison test using the Prism software (GraphPad Software, San Diego, CA). Where appropriate (comparisons involving two groups only), two-tailed t tests were performed. Statistical differences were considered significant at the level of p < 0.05.
3. RESULTS
3.1. Trypsin treated LRR1 domain retains the ability to activate TLR2
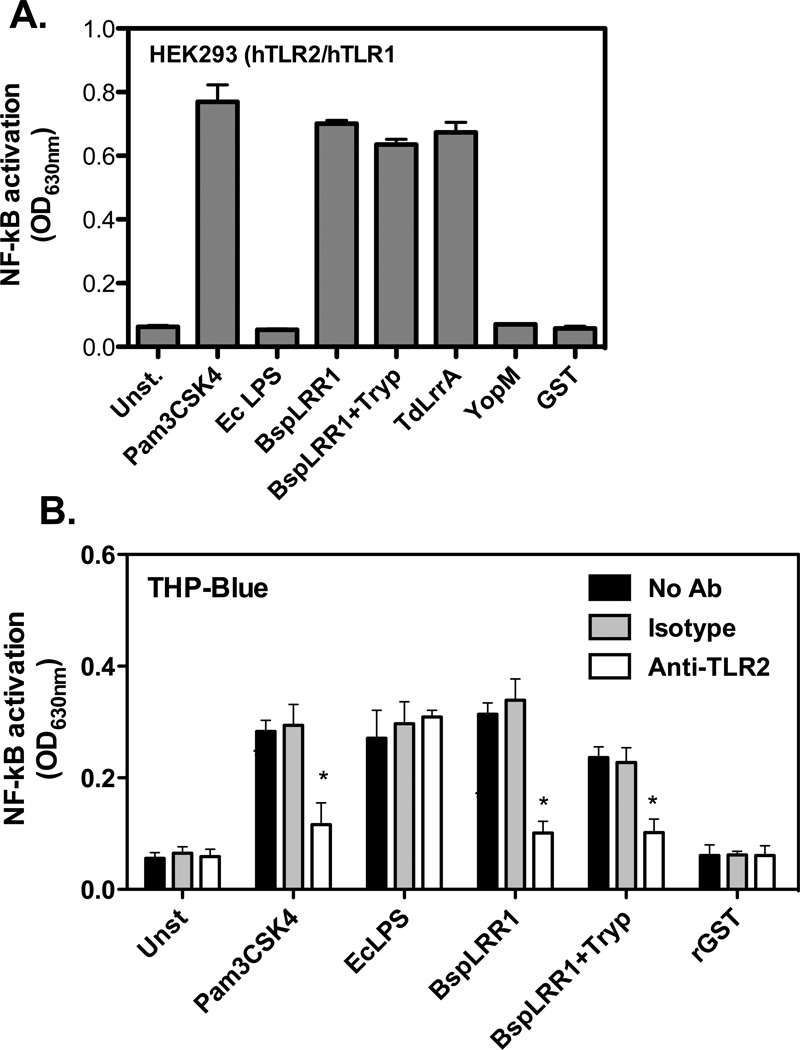
To ascertain whether smaller discrete peptide epitopes for TLR2 activation are present in the LRR1 domain of BspA (Fig. 1), we tested the activity of the trypsin digest of rBspLRR1. For this purpose, the fraction eluted in the exclusion volume of the G-25 column from the trypsin digest of BspLRR1 was tested for TLR2 activation using reporter cell lines HEK293 (hTLR2/hTLR1) and THP-Blue cells. The results showed that the tryptic digest of BspLRR1 induced NF-κB in HEK293 (hTLR2/hTLR1) (Fig. 2A) and THP-Blue reporter cell lines (Fig 2B). In addition, the TdLrrA protein of oral spirochete Treponema denticola that shares a strong sequence identity with the BspA LRR1 domain [20] as a recombinant protein also induced NF-κB activity (Fig. 2 A & B). As expected, Pam3CSK4 and rBspLRR1 induced NF-κB activity in HEK293 (hTLR2/hTLR1) cells. On the other hand, the recombinant YopM protein of Yersinia entercolitica that belongs to a LRR family different from that of BspA [21] as well as recombinant glutathione S-transferase protein did not induce NF- κB in TLR2 expressing reporter cell lines (Fig. 2 A). Since THP-Blue cells express multiple receptors in addition to TLR2, the specificity of the response was confirmed with the function-blocking monoclonal antibody to TLR2. The results showed that anti-TLR2 MAb significantly blocked NF-κB activity in THP-Blue cells induced by Pam3, rBspLRR1, or trypsin digested rBspLRR1 compared to the non-antibody treated or the isotype matched controls (Fig. 2 B). We inferred from the above results that smaller discrete epitopes were present in the peptide digest able to activate TLR2. Since trypsin cleaves the LRR1 domain into 14 peptides, with the largest peptide of approximately 8-kDa retaining nearly complete LRR repeats 3 to 6 (Fig. 1), we predicted that this fragment would have the high probability of retaining the minimal functional motifs for TLR2 binding and activation.
Fig. 2.
Trypsin digested BspLRR1 retains the ability to activate TLR2. HEK293 (hTLR2/hTLR1) (A) or THP-Blue (B) reporter cells with NF-κB inducible alkaline phosphatase were incubated with Pam3Cys (0.1 µg/ml), Ec LPS (1 µg/ml) or rBspLRR1 (5 µg/ml), rBspLRR1 trypsin digest (5 µg/ml), TdLrrA (5 µg/ml) rYopM (5 µg/ml) and rGST (5 µg/ml). For THP-Blue cells, pretreatment with either anti-TLR2 or isotype control antibody prior to stimulation was also carried out. The secreted alkaline phosphatase activity (NF-κB induction) in the medium was measured after 16–18 h. The data represent averages ± standard deviations for triplicate determinations from one three independent experiments yeilding similar results; *, p < 0.05.
3.2. Peptides derived from the LRRs 3–6 activate and bind TLR2
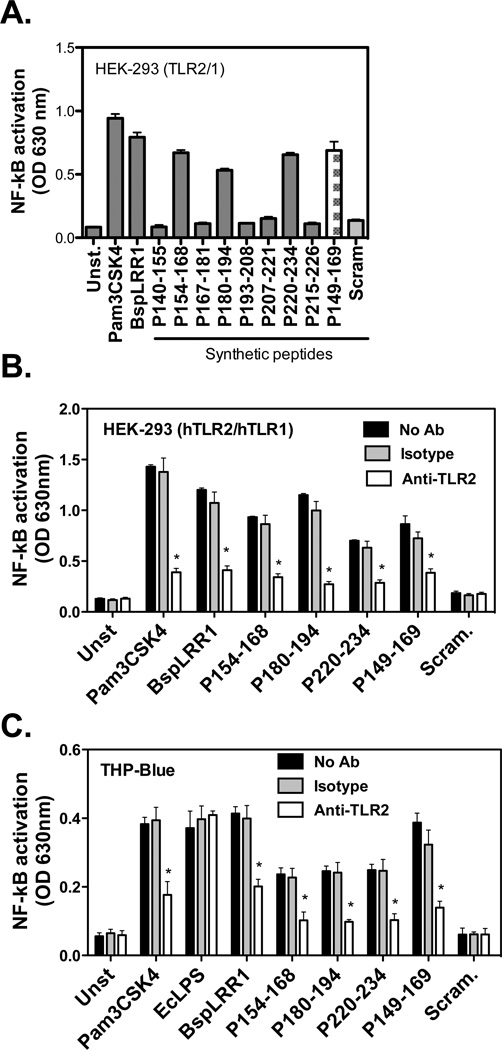
We tested the activity of overlapping synthetic peptides covering the LRRs 3–6 (region underlined; Fig. 1) based on assumption mentioned above. 7 synthetic peptides were generated with an overlap of 2 amino acids (Table 1). These peptides were tested for NF-κB inducing activity via TLR2 activation in HEK293 (hTLR2/hTLR1) and THP-Blue cells as described above. The results showed that peptides P154–168, P180–194 and P220–234 induced NF-κB activity in both reporter cell lines (Fig. 3 A & B). Moreover, the peptide-induced NF-κB activity was significantly inhibited with an anti-TLR2 monoclonal antibody but not with an isotype matched control.
Fig. 3.
Peptides derived from Lrr repeats 3–6 activate TLR2. (A) HEK293 (hTLR2/hTLR1) reporter cells were stimulated with synthetic peptides (50 µg/ml each) for 16 h. (B & C) The active peptides inducing TLR2 were further tested in reporter cell lines HEK293 (HTLR2/hTLR1) and THP-Blue in the absence or presence of TLR2 blocking (or isotype control) antibody and NF-κB induction was assessed by measuring secreted alkaline phosphatase as in Fig. 2. The data represent averages ± standard deviations for triplicate determinations from one two independent experiments yielding similar results; asterisks (p < 0.05) in panel A indicate significant activation of NF-κB activation by agonist treatment compared to non treatment, and in panel B they indicate significant blocking of activation by anti-TLR2 antibody compared to isotype control (B).
3.3. Active peptide sequence binds TLR2
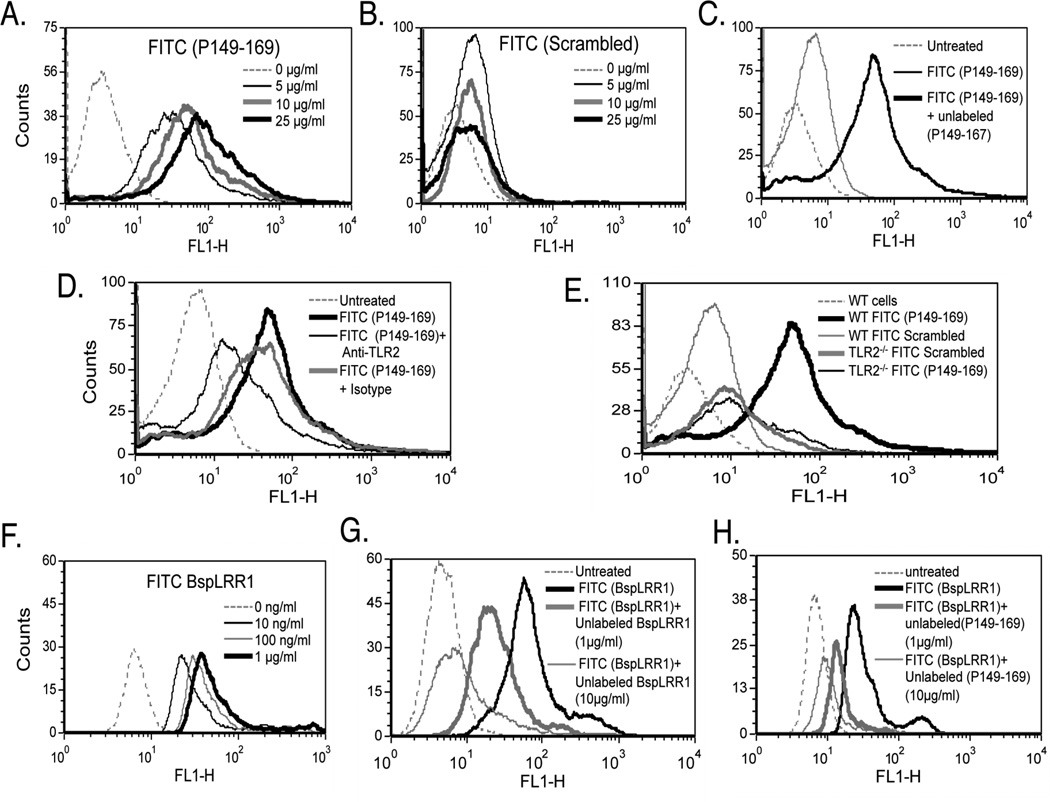
Interestingly, examination of amino acid sequences of the TLR2 activating peptides P154–168, P180–194 and P220–234 revealed that a sequence motif GC(S/T)GLXSIT is shared among these peptides. Interestingly, this motif is represented a total of eight times within the BspLRR1 region (Fig. 1). To investigate the direct binding of this motif to TLR2, peptide P149–169 was designed whose sequence extends five amino acids to the N-terminus of P154–168. As expected, the peptide P149–169 induced NF-κB activity (Fig. 3 A & B). This peptide was designed to avoid any possible hindrance to binding motif due to FITC label. To investigate binding, HEK293(hTLR2/hTLR1) cells were incubated with increasing concentrations (5, 10, 15, 25, 50 µg/ml) of FITC labeled P149- 167 or scrambled peptides at 4°C for 1 h, followed by flow cytometry. The results showed that binding of P149–169 was dose-dependent, indicated by an increase in the mean fluorescence intensities (MFI) of the cells incubated with increasing concentrations of the peptide P149–169 (Fig. 4 A). On the other hand, no significant increase in the MFI of the cells incubated with the FITC labeled scrambled peptide was observed (Fig. 4 B). To further confirm that the peptide P149–169 was binding specifically to TLR2, competitive inhibition and antibody blocking assays were performed. The results showed that the excess unlabeled peptide P149–169 (25 µg/ml) but not scrambled peptide significantly blocked the binding of labeled P149–169 peptide to HEK293 (hTLR2/hTLR1) cells (5 µg/ml) (Fig. 4 C). Furthermore, anti-TLR2 MAb blocked the binding of labeled P149–169 peptide significantly whereas the isotype matched control antibody had no affect on the binding (Fig. 4 D). In addition, mouse peritoneal macrophages from WT and TLR2−/− mice were assessed for binding to the FITC labeled P149–169 or scrambled peptide by flow cytometry. The WT, but not the TLR2−/−, macrophages significantly bound the peptide P149–169, and the scrambled peptide did not bind macrophages of either type (Fig. 4 E).
Fig. 4.
TLR2-activating epitope of BspA specifically binds TLR2. (A & B) HEK293 (hTLR2/hTLR1) cells were incubated with increasing concentrations of the fluorescently labeled peptide P149–169 (A) or the scrambled peptide (B) and analyzed by flow cytometry. (C & D) Flow cytometry of HEK293 (hTLR2/hTLR1) cells preincubated with the unlabeled peptide P149–169 (25 µg/ml; 5 min) (C) or preincubated with anti-TLR2 or isotype control (IgG2a) antibody (each at 5 µg/ml; 15 min) (D) followed by incubation with labeled P149–169 peptide (5 µg/ml; 15 min). (E) WT and TLR2−/− mouse macrophages were incubated with fluorescently labeled peptides (P149–169 or scrambled). (F) HEK293 (hTLR2/hTLR1) cells were incubated with increasing concentrations of the fluorescently labeled peptide rBspLRR1. (G & H) Cells were preincubated with either the unlabeled rBspLRR1 (1 & 10 µg/ml; 5 min) (G) or unlabeled peptide P149–169 (1 & 10 µg/ml; 5 min) (H) prior to incubation with labeled rBspLRR1-(100 ng/ml; 15 min). Data in each panel are representative of three to four independent experiments.
Next, to confirm that the sequence motif GC(S/T)GLXSIT is involved in the interactions of BspA with TLR2, we tested the peptide P149–167 in inhibiting the binding of FITC labeled BspLRR1 to TLR2 (hTLR2/hTLR1). Firstly, as expected, FITC labeled rBspLRR1 bound cells in a dose dependent manner (Fig. 4 F). Importantly, the results showed significant inhibition of BspLRR1 binding to TLR2 by unlabeled P149–169 peptide (Fig. 4 H). As compared to unlabeled BspLRR1 (Fig. 4 G), higher concentration of unlabeled peptide P149–169 (Fig. 4 H) was required to significantly block the binding of BspLRR1 to TLR2. The requirement for higher peptide concentration suggest that either the peptides are present in unfavorable conformational states and/or the presence of multiple binding motifs in BspLRR1 result in multivalent binding.
Taken together, these data demonstrate that the peptide motif GC(S/T)GLXSIT of BspA mediates TLR2 binding.
3.4. TLR2-binding peptides stimulate cytokine secretion in macrophages
To assess the TLR2-dependent activity of the synthetic peptides in relation to cytokine secretion, peritoneal macrophages isolated from the WT and TLR2−/− mice were stimulated with synthetic peptides and supernatants were assayed for cytokines by ELISAs. In agreement with the data above, peptides P149–169, P154–168, P180–194 and P220–234 induced IL-1β and TNF-α secretion in wild-type macrophage, whereas TLR2−/− were unresponsive to these peptide (supplementary Fig. S1).
4. DISCUSSION
The current study was undertaken to identify the peptide motif(s) in the T. forsythia BspA virulence protein that mediate TLR2 binding and activation. Our study described here showed that a peptide motif GC(S/T)GLXSIT present in the LRR1 domain of the BspA protein is responsible for TLR2 binding and activation. Synthetic peptides derived from the LRR1 region of BspA containing this motif bound and activated reporter cell lines and mouse macrophages in a TLR2-dependnet manner. Moreover, a synthetic peptide with this motif competitively blocked the binding of the BspA protein to TLR2. Interestingly, the BspA homologue TdLrrA protein of T. denticola, which also activated TLR2 contains GC(S/T)GLXSIT. On the other hand, the LRR YopM protein with no sequence similarity with BspA did not activate TLR2. Thus, our study has identified a motif mediating the binding of a peptide/protein ligands to TLR2, and as a corollary highlights the promiscuous nature of TLR2 with respect to its ligand binding,
TLR2 has been considered the primary receptor for acylated peptides/proteins ligands and can discriminate tri- and di-acylated peptide ligands by using either TLR1 or TLR6 as a coreceptor, respectively [1]. TLR2 agonists induce intracellular signaling through activation of nuclear factor NF-κB and mitogen-activated protein (MAP) kinase dependent pathways, inducing secretion of immune mediators involved in the development of innate and adaptive immunity [1]. It has been shown that different subdomains on the extra-cellular domain (ECD) of TLR2 bind distinct TLR2 ligands such as peptidoglycans, lipoteichoic acid and palmitoylated peptides/proteins [23]. Conceivably, these multiple subdomains may allow activation of distinct intracellular pathways following engagement with different ligands. Although, TLR2 was initially thought to bind only acylated ligands, several recent studies have revealed its ability to recognize non-acylated proteins as well; for example Yersinia spp virulence determinant LcrV and its peptides [2, 11], Mycobacterium bovis protein MPB83 [3], fibrillar β-amyloid peptide [5], Neisseria lactamica PorB [7], Neisseria meningitides PorB [8, 24], Mycobacterium tuberculosis proline-proline-glutamic acid family protein PPE18 [9], Escherichia coli enterotoxin [6], and Porphyromonas gingivalis fimbrial protein FimA and its synthetic peptides [4, 10]. A peptide epitope represented by the sequence ALTTE present in P. gingivalis FimA protein has been shown to activate TLR2 [4, 10]. Thus, TLR2 appears to be quite promiscuous in terms of its ligand biding activity. These factors have been shown to play critical roles in immune modulation and pathogenesis through their interactions with TLR2.
Protein docking studies have demonstrated that non-acylated ligands bind to TLR2 ECD subdomains outside of the binding pocket for synthetic lipopeptide Pam3CSk4. In this regard, the binding of a proline-proline-glutamic acid (PPE) family protein PPE18 from Mycoplasma tuberculosis to TLR2 involves TLR2 ECD encompassing LRRs11–15 [9]. Likewise, E. coli type II enterotoxin B subunit pentamer (LT-IIb-B5) binds TLR2/1 heterodimer at a site that only partially overlaps with that of Pam3CSK4 [6]. These studies show that binding sites for different non-acylated peptides reside in different regions of TLR2 ECD. However, the binding sites for other non-acylated ligands is currently unknown. For example, the structure-function mapping studies of P. gingivalis FimA protein using synthetic peptides have identified a peptide motif, ALTTE, as the functional epitope involved in TLR2 activation [4, 10], the binding site of FimA protein or its active peptides on TLR2 has not been determined.
Currently, the crystal structure for the BspA protein or its LRR region is not available, and therefore docking studies for the prediction of binding sites for BspA on TLR2 ECD are not possible. However, based on the above studies indicating distinct TLR2 ECD sites for structurally unrelated non-acylated ligands suggests that BspA may also bind to a exclusive site on the TLR2 ECD. Strikingly, the presence of multiple (eight) GC(S/T)GLXSIT motifs in the LRR region of BspA suggest that each of those motifs either binds TLR2 independently or these motifs together form a single binding domain in the context of the tertiary conformation of the protein. While these motifs are expected to conform to the cysteine ladder characteristic of LRRs in BspA, their exact organization and role in TLR2 binding await crystal structure of BspA and results of functional studies using site-specific mutants of BspA. These studies are currently in progress in our laboratory. Nevertheless, the data presented here have set the stage for future studies to define TLR2-BspA interactions in detail, and possibly for the development of therapy against periodontal and related mucosal disease through targeting these interactions with small molecule peptidomimetics in the future.
Supplementary Material
Leucine-rich repeat (LRR) domain of a bacterial protein activates TLR2
Peptide repeats in the LRR domain responsible for TLR2 activation were identified
A common peptide motif in these repeats interacts with TLR2
This peptide motif in a TLR2-dependnet manner activates macrophages.
This study identifies a novel non-acylated peptide ligand of TLR2
ACKNOWLEDGEMENTS
This study was supported by U.S. Public Health Research Service grant DE014749
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Abramov VM, Khlebnikov VS, Vasiliev AM, Kosarev IV, Vasilenko RN, Kulikova NL, Khodyakova AV, Evstigneev VI, Uversky VN, Motin VL, Smirnov GB, Brubaker RR. Attachment of LcrV from Yersinia pestis at dual binding sites to human TLR-2 and human IFN-gamma receptor. J. Proteome Res. 2007;6:2222–2231. doi: 10.1021/pr070036r. [DOI] [PubMed] [Google Scholar]
- 3.Chambers MA, Whelan AO, Spallek R, Singh M, Coddeville B, Guerardel Y, Elass E. Non-acylated Mycobacterium bovis glycoprotein MPB83 binds to TLR1/2 and stimulates production of matrix metalloproteinase 9. Biochem. Biophys. Res. Commun. 2010;400:403–408. doi: 10.1016/j.bbrc.2010.08.085. [DOI] [PubMed] [Google Scholar]
- 4.Hajishengallis G, Ratti P, Harokopakis E. Peptide mapping of bacterial fimbrial epitopes interacting with pattern-recognition receptors. J. Biol. Chem. 2005 doi: 10.1074/jbc.M507326200. [DOI] [PubMed] [Google Scholar]
- 5.Jana M, Palencia CA, Pahan K. Fibrillar Amyloid-{beta} Peptides Activate Microglia via TLR2: Implications for Alzheimer's Disease. J. Immunol. 2008;181:7254–7262. doi: 10.4049/jimmunol.181.10.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang S, Hosur KB, Lu S, Nawar HF, Weber BR, Tapping RI, Connell TD, Hajishengallis G. Mapping of a microbial protein domain involved in binding and activation of the TLR2/TLR1 heterodimer. J. Immunol. 2009;182:2978–2985. doi: 10.4049/jimmunol.0803737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Wetzler LM, Oliveira Nascimento L, Massari P. Human Airway Epithelial Cell Responses to Neisseria lactamica and Purified Porin via Toll-Like Receptor 2-Dependent Signaling. Infect. Immun. 2010;78:5314–5323. doi: 10.1128/IAI.00681-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massari P, Visintin A, Gunawardana J, Halmen KA, King CA, Golenbock DT, Wetzler LM. Meningococcal porin PorB binds to TLR2 and requires TLR1 for signaling. J. Immunol. 2006;176:2373–2380. doi: 10.4049/jimmunol.176.4.2373. [DOI] [PubMed] [Google Scholar]
- 9.Nair S, Ramaswamy PA, Ghosh S, Joshi DC, Pathak N, Siddiqui I, Sharma P, Hasnain SE, Mande SC, Mukhopadhyay S. The PPE18 ofMycobacterium tuberculosis Interacts with TLR2 and Activates IL-10 Induction in Macrophage. J. Immunol. 2009;183:6269–6281. doi: 10.4049/jimmunol.0901367. [DOI] [PubMed] [Google Scholar]
- 10.Ogawa T, Asai Y, Hashimoto M, Uchida H. Bacterial fimbriae activate human peripheral blood monocytes utilizing TLR2, CD14 and CD11a/CD18 as cellular receptors. Eur. J. Immunol. 2002;32:2543–2550. doi: 10.1002/1521-4141(200209)32:9<2543::AID-IMMU2543>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Sing A, Reithmeier-Rost D, Granfors K, Hill J, Roggenkamp A, Heesemann J. A hypervariable N-terminal region of Yersinia LcrV determines Toll-like receptor 2-mediated IL-10 induction and mouse virulence. Proc. Nat. Acad. Sci. USA. 2005;102:16049–16054. doi: 10.1073/pnas.0504728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat Rev Immunol. 2011;11:187–200. doi: 10.1038/nri2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma A. Virulence mechanisms of Tannerella forsythia. Periodontol. 2000. 2010;54:106–116. doi: 10.1111/j.1600-0757.2009.00332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onishi S, Honma K, Liang S, Stathopoulou P, Kinane D, Hajishengallis G, Sharma A. Toll-like receptor 2-mediated interleukin-8 expression in gingival epithelial cells by the Tannerella forsythia leucine-rich repeat protein BspA. Infect. Immun. 2008;76:198–205. doi: 10.1128/IAI.01139-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hajishengallis G, Martin M, Sojar HT, Sharma A, Schifferle RE, DeNardin E, Russell MW, Genco RJ. Dependence of bacterial protein adhesins on toll-like receptors for proinflammatory cytokine induction. Clin. Diagn. Lab. Immunol. 2002;9:403–401. doi: 10.1128/CDLI.9.2.403-411.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Myneni SR, Settem RP, Connell TD, Keegan AD, Gaffen SL, Sharma A. TLR2 signaling and Th2 responses drive Tannerella forsythia-induced periodontal bone loss. J. Immunol. 2011;187:501–509. doi: 10.4049/jimmunol.1100683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem. Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 18.Sharma A, Sojar HT, Glurich I, Honma K, Kuramitsu HK, Genco RJ. Cloning, expression, sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect. Immun. 1998;66:5703–5710. doi: 10.1128/iai.66.12.5703-5710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bublitz M, Holland C, Sabet C, Reichelt J, Cossart P, Heinz DW, Bierne H, Schubert WD. Crystal structure and standardized geometric analysis of InlJ, a listerial virulence factor and leucine-rich repeat protein with a novel cysteine ladder. J. Mol. Biol. 2008;378:87–96. doi: 10.1016/j.jmb.2008.01.100. [DOI] [PubMed] [Google Scholar]
- 20.Ikegami A, Honma K, Sharma A, Kuramitsu HK. Multiple functions of the leucine-rich repeat protein LrrA of Treponema denticola. Infect. Immun. 2004;72:4619–4627. doi: 10.1128/IAI.72.8.4619-4627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loimaranta V, Hytonen J, Pulliainen AT, Sharma A, Tenovuo J, Stromberg N, Finne J. Leucine-rich repeats of bacterial surface proteins serve as common pattern recognition motifs of human scavenger receptor gp340. J. Biol. Chem. 2009;284:18614–18623. doi: 10.1074/jbc.M900581200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng X, Goncalves R, Mosser DM. The Isolation and Characterization of Murine Macrophages. Curr. Protoc. Immunol. 2008;83:4.1.1–14.1.14. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meng G, Grabiec A, Vallon M, Ebe B, Hampel S, Bessler W, Wagner H, Kirschning CJ. Cellular recognition of tri-/di-palmitoylated peptides is independent from a domain encompassing the N-terminal seven leucine-rich repeat (LRR)/LRR-like motifs of TLR2. J. Biol. Chem. 2003;278:39822–39829. doi: 10.1074/jbc.M304766200. [DOI] [PubMed] [Google Scholar]
- 24.Singleton TE, Massari P, Wetzler LM. Neisserial porin-induced dendritic cell activation is MyD88 and TLR2 dependent. J. Immunol. 2005;174:3545–3550. doi: 10.4049/jimmunol.174.6.3545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.