UNG activity repairs activation-induced deaminase-generated U:G mismatches via error-prone or error-free repair, depending on the sequence context of the deaminated cytosine.
Abstract
Secondary diversification of antibodies through somatic hypermutation (SHM) and class switch recombination (CSR) is a critical component of the immune response. Activation-induced deaminase (AID) initiates both processes by deaminating cytosine residues in immunoglobulin genes. The resulting U:G mismatch can be processed by alternative pathways to give rise to a mutation (SHM) or a DNA double-strand break (CSR). Central to this processing is the activity of uracil-N-glycosylase (UNG), an enzyme normally involved in error-free base excision repair. We used next generation sequencing to analyze the contribution of UNG to the resolution of AID-induced lesions. Loss- and gain-of-function experiments showed that UNG activity can promote both error-prone and high fidelity repair of U:G lesions. Unexpectedly, the balance between these alternative outcomes was influenced by the sequence context of the deaminated cytosine, with individual hotspots exhibiting higher susceptibility to UNG-triggered error-free or error-prone resolution. These results reveal UNG as a new molecular layer that shapes the specificity of AID-induced mutations and may provide new insights into the role of AID in cancer development.
Activation-induced deaminase (AID) is a critical component of the immune response, as it is essential for the reactions of antibody secondary diversification, namely somatic hypermutation (SHM) and class switch recombination (CSR; Muramatsu et al., 2000; Revy et al., 2000), which take place in B cells that have been activated by antigen. SHM introduces nucleotide substitutions in the variable, antigen binding region of immunoglobulins, allowing the generation of variants with higher affinity for antigen that are selected by affinity maturation (Di Noia and Neuberger, 2007; Peled et al., 2008). CSR is a region-specific recombination reaction that replaces the primary μ constant region by a downstream constant region, providing the antibody with specialized means of antigen removal (Stavnezer et al., 2008). AID deficiency prevents SHM and CSR and promotes a hyper-immunoglobulin M immunodeficiency in humans (Muramatsu et al., 2000; Revy et al., 2000).
AID initiates SHM and CSR by deaminating cytosine residues at the DNA of immunoglobulin genes (Petersen-Mahrt et al., 2002; Di Noia and Neuberger, 2007; Maul et al., 2011). The U:G mismatch thus generated can be processed by alternative pathways to give rise to the introduction of a mutation (SHM) or the generation of a DNA double-strand break and a recombination reaction (CSR). If the U:G mismatch is replicated over, it will give rise to transition mutations (C→T or G→A, depending on the DNA strand where the deamination took place). Alternatively, uracil can be excised from the DNA by uracil-N-glycosylase (UNG), an enzyme which is part of the base excision repair (BER) pathway. UNG inhibition in B cell lines and UNG deficiency in mice and humans give rise to an altered SHM pattern where the proportion of transition mutations is dramatically increased and transversion mutations (C→A or G and G→C or T) are decreased (Di Noia and Neuberger, 2002; Rada et al., 2002, 2004; Imai et al., 2003; Xue et al., 2006). In addition, UNG deficiency severely impairs CSR (Rada et al., 2002; Imai et al., 2003). These results showed that in the context of antibody diversification, UNG contributes to the mutagenic resolution of U:G mismatches rather than to their error-free repair by BER, but the basis for this error-prone activity of UNG is not understood. Alternatively, the U:G mispair can be recognized by the Msh2/Msh6 components of the mismatch repair machinery, which can trigger a patch DNA synthesis leading to nucleotide substitutions mostly at adenine or thymine nucleotides (also known as second phase of SHM; Rada et al., 1998, 2004; Schrader et al., 1999; Wiesendanger et al., 2000).
Somatic mutations are not randomly distributed along Ig variable genes; instead, they accumulate preferentially at mutational hotspots (Rogozin and Kolchanov, 1992; Dörner et al., 1998; Shapiro et al., 1999; Rogozin and Diaz, 2004). Analysis of Ig gene databases from hypermutating B cells revealed that SHM at cytosine residues is strongly favored when they are part of an RGYW (where R = A/G, Y = C/T, and W = A/T) or its reverse complement sequence WRCY (Rogozin and Kolchanov, 1992; Dörner et al., 1998). Among the RGYW/WRCY variants, AGCT has been proposed to be a primordial motif for CSR (Zarrin et al., 2004). A similar hotspot preference has been shown in non-Ig transgenes expressed in B cell lines (Martin and Scharff, 2002) and in fibroblasts with heterologous AID expression (Yoshikawa et al., 2002; McBride et al., 2004), suggesting that targeting of these motifs is a universal feature of the SHM machinery. In addition, biochemical studies showed that AID activity on DNA substrates in vitro displays a mutational preference for the WRC motif (Pham et al., 2003; Bransteitter et al., 2004; Yu et al., 2004).
AID activity is not exclusively restricted to immunoglobulin genes and can cause relatively widespread DNA lesions in the genome. Mutations bearing the hallmarks of SHM were detected on the Bcl6 gene in human memory B cells (Shen et al., 1998) and in several loci, including the proto-oncogenes PIM1, Pax5, and Myc in diffuse B cell lymphomas (Pasqualucci et al., 2001). More recently, an extensive sequencing study estimated that 25% of the genes expressed by germinal center B cells can accumulate AID-mediated mutations (Liu et al., 2008). A direct link between AID function and B cell neoplasias was established by the finding that lymphomagenic c-myc/IgH translocations (Adams et al., 1985) are initiated by AID, both in vivo and in vitro (Ramiro et al., 2004, 2006; Dorsett et al., 2007; Robbiani et al., 2008). Indeed, the absence of AID in several lymphoma mouse models delays the onset or shifts the nature of the neoplasia (Ramiro et al., 2004; Kovalchuk et al., 2007; Pasqualucci et al., 2008). In addition, there are evidences that AID activity is not confined to the B cell lineage and could contribute to non–B cell neoplasias. AID expression has been detected in several human cancers, which correlated with accumulation of mutations in various genes, including Trp53 or Myc (Endo et al., 2007; Kou et al., 2007; Matsumoto et al., 2007). Recently, it has been shown that AID deficiency exerts protection against the development of colitis-associated cancers (Takai et al., 2012). Therefore, AID specificity is a relevant issue for the understanding of both secondary diversification of antibodies and the role of this enzyme in cancer.
Here, we have addressed the contribution of UNG to the specificity of AID-induced mutations by combining gain- and loss-of-function approaches and mutation analysis using next generation sequencing (NGS) technology. We find that UNG can process U:G lesions generated by AID to give rise both to faithful and error-prone repair depending on the sequence context. Our results provide the first evidence that UNG activity shapes the sequence specificity of AID during SHM.
RESULTS
Assay to monitor AID mutational activity
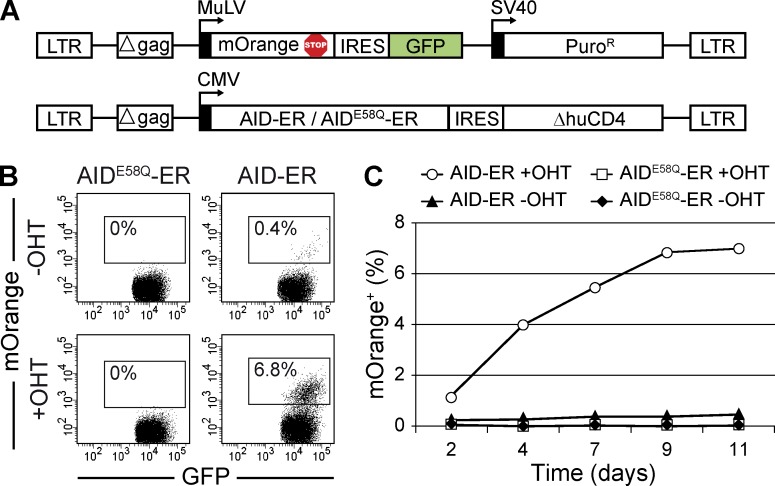
To monitor AID mutational activity we developed a sensitive fluorescence revertance assay. In brief, a stop codon overlapping with an AGCT AID mutational hotspot was introduced at positions 230–233 of the sequence encoding the mOrange fluorescent protein (mOrangeSTOP; Fig. 1 A and Fig. S1 A), a monomeric RFP1 variant which can be easily detected by flow cytometry (Shaner et al., 2004). This TAG stop codon generates a nonfluorescent truncated protein, but transversion mutations at its third nucleotide revert it to TAC or TAT tyrosine-encoding codons that reconstitute the full-length mOrange fluorescent protein. mOrangeSTOP was introduced into the GFP-containing retroviral vector pMX-PIE (Barreto et al., 2003) to allow the tracking of transduced cells (Fig. 1 A). Inducible AID activity was achieved by fusing AID to the estrogen-binding domain of estrogen receptor (ER; AID-ER), thus generating a protein that can be translocated into the nucleus—and therefore grant access to its DNA substrate—by tamoxifen (OHT) treatment (Doi et al., 2003). AID-ER, or the catalytically inactive mutant AIDE58Q-ER, was cloned into a second retroviral vector that contains a truncated, signaling-devoid form of the human CD4 molecule (ΔhuCD4) for tracking purposes (Fig. 1 A). To test the mOrangeSTOP revertance assay, we retrovirally transduced the mOrangeSTOP vector along with either AID-ER– or AIDE58Q-ER–containing vectors into NIH-3T3 mouse fibroblasts. After 3 d of puromycin selection, >95% of cells were GFP+ΔhuCD4+ (unpublished data). Cells were then cultured in the presence or absence of OHT for up to 11 d. We detected the appearance of mOrange+ cells in AID-ER transduced cultures as soon as 2 d after OHT treatment and their percentage increased with time (Fig. 1, B and C). In contrast, AIDE58Q-ER transduction failed to generate detectable mOrange+ cells and, in the absence of OHT AID-ER, only promoted marginal numbers of mOrange revertants (Fig. 1, B and C). These results show that AID mutational activity can be monitored by the generation of mOrange revertants in NIH-3T3 cells.
Figure 1.
Fluorescence revertance assay to monitor AID activity. (A) Representation of the pMX-PIE-mOrangeSTOP and AID-ER-ΔhuCD4 or AIDE58Q-ER-ΔhuCD4 retroviral vectors used to transduce NIH-3T3 cells. (B) Detection of AID activity. NIH-3T3 cells were co-transduced with the retroviral vectors depicted in A and cultured with (bottom) or without (top) 1 µM OHT. mOrange+ cells were monitored by flow cytometry. Representative FACS analyses at day 9 are shown. (C) Time-course analysis of mOrange+ cell appearance in NIH-3T3 cells co-transduced as described in B. One representative experiment is shown.
Detection of AID-induced mutations by NGS
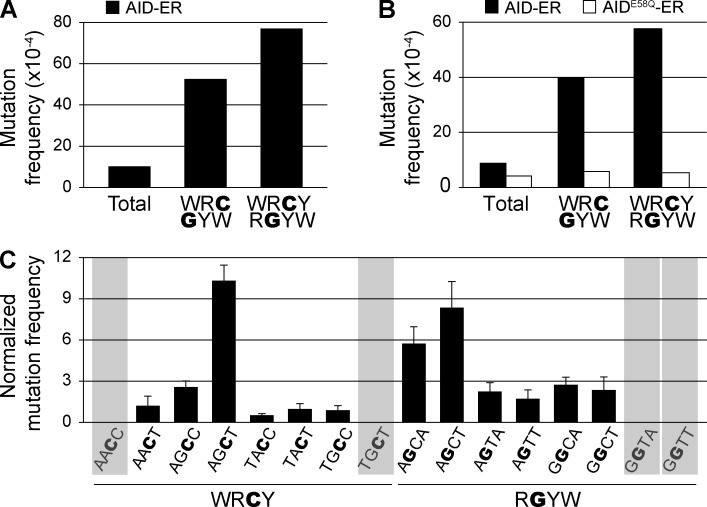
To perform a detailed analysis of AID-induced mutations, we set to develop an NGS approach that would allow mutational analysis at very high depth coverage. We PCR amplified the mOrangeSTOP transgene from total NIH-3T3 cells that had been co-transduced with mOrangeSTOP and AID-ER or AIDE58Q-ER and cultured for 11 d in the presence of OHT. PCR products were sheared, bar-coded, and sequenced in an Illumina platform. After high quality filtering and alignment, we obtained a depth of 8.7 × 104 reads per base position on average, i.e., ∼100–1,000-fold higher than in a typical experiment by Sanger sequencing, which provided the detection of thousands of mutations per experiment (Table 1). We compared the mutation pattern of the sequences obtained by Sanger and NGS in one individual experiment. As expected, the mean mutation frequency observed by conventional sequencing was increased at cytosines contained in WRC/GYW and WRCY/RGYW AID mutational hotspots (Fig. 2 A; analyzed hotspot cytosines and guanines are underlined throughout the paper). The same mutation preference was found in the sequences obtained by NGS, with roughly a fourfold and sixfold increase in mean mutation frequency at WRC/GYW and WRCY/RGYW hotspots, respectively (Fig. 2 B and Table 2). In contrast, sequences obtained from AIDE58Q-ER cells contained fewer total mutation frequencies that did not increase at these hotspots and instead harbored evenly distributed mutations, indicating background mutation levels (Fig. 2 B and Table 2). These results validate the use of NGS for the detection of AID mutations.
Table 1.
mOrangeSTOP sequencing by Sanger and NGS
| Sequencing method | Number of positionsa | Number of reads (mean)b | Mutation numberc | Frequency (×10−4)d |
| Sanger | 222 | 101 | 31 | 13.8 |
| NGS | 222 | 87,009 | 22,370 | 11.1 |
Number of total cytosine in mOrange sequence.
Mean number of reads per cytosine.
Total mutation number in cytosines.
Mutation frequency calculated as c/(b × a).
Figure 2.
Detection of AID-induced mutations by NGS. NIH-3T3 cells were co-transduced with mOrangeSTOP and AID-ER– or AIDE58Q-ER–expressing vectors. Transduced cells were cultured in the presence of OHT during 11 d. mOrangeSTOP transgene was PCR amplified from the total cell population and mutations were analyzed by Sanger sequencing (A) and NGS (B and C). (A) Mutation frequency in AID-ER–expressing cells in the complete mOrangeSTOP sequence (total) and in cytosines or guanines of AID hotspots (WRC/GYW and WRCY/RGYW) is represented. (B) Mutation frequency in AID-ER– or AIDE58Q-ER–expressing cells. Total mutation frequency was calculated as the mean of the mutation frequencies in all nucleotides of the amplified mOrangeSTOP sequence. WRC/GYW and WRCY/RGYW mutation frequencies were calculated as the mean of the mutation frequencies in G/C nucleotides contained in the respective hotspots. Values are from one representative of three independent experiments (Table 2). (C) Mutation frequency in individual WRCY and RGYW hotspots in mOrangeSTOP sequence, normalized to the mean mutation frequency at G/C nucleotides. Bars represent the mean of three independent experiments. Error bars show standard deviations. Shadowed hotspots (AACC, TGCT, GGTA, and GGTT) are not present in the mOrangeSTOP sequence. The reference mOrangeSTOP sequence is shown in Fig. S1 A.
Table 2.
Mutation analysis in AID and AIDE58Q expressing NIH-3T3 cells
| Experiment | Totala | WRC+GYWb | WRCY+RGYWc | |||
| AID | AIDE58Q | AID | AIDE58Q | AID | AIDE58Q | |
| 1 | 8.9 | 4.4 | 39.8 | 6.0 | 57.8 | 5.9 |
| 2 | 5.8 | 3.3 | 21.5 | 4.5 | 30.1 | 4.4 |
| 3 | 14.9 | 6.6 | 73.0 | 7.3 | 106.7 | 7.4 |
Total frequency of mutations (×10−4).
Frequency of mutations (×10−4) in C or G of WRC or GYW hotspots.
Frequency of mutations (×10−4) in C or G of WRCY or RGYW hotspots.
We next analyzed the mutation distribution at cytosines and guanines contained in each of the WRCY and RGYW hotspots for SHM in the mOrangeSTOP sequence by NGS (Fig. 2 C). All hotspot sequences are shown on the coding strand, and therefore, in the case of RGYW motifs, the context of the mutated cytosine is that of the reverse complement sequence. Mutation frequencies are shown after normalization to the mean mutation at G/C nucleotides in each individual experiment (Fig. 2 C). We found that the mutation susceptibility differed widely across the different WRCY and RGYW combinations, with the highest mutation frequency accumulating at cytosines at the AGCT motif and guanines at AGCA and AGCT motifs. We conclude that the mutability of different motifs conforming to the canonical WRCY/RGYW mutational hotspots is highly variable and that AGCT appears to be a major target for SHM in non–B cells.
UNG repairs faithfully a fraction of the mutations induced by AID in non–B cells
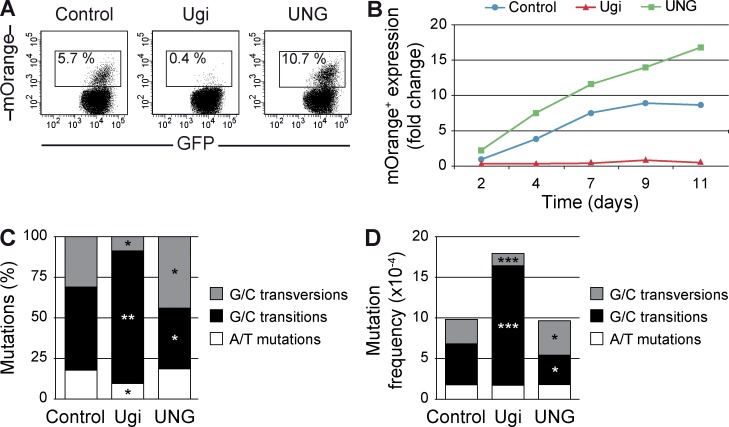
UNG plays a crucial role in the processing of AID-induced lesions in B cells. In particular, UNG ablation biases the SHM pattern at G/C base pairs toward transition mutations, indicating that transversions at G/C pairs are dependent on UNG (Di Noia and Neuberger, 2002; Rada et al., 2002). To address the role of UNG in the resolution of AID-induced lesions in non–B cells, we made use of the uracil-DNA glycosylase inhibitor (Ugi) protein from the bacteriophage PSB2. First, we examined the contribution of UNG to the mutational activity of AID in the mOrange-based revertance assay. We co-transduced Ugi together with mOrangeSTOP and AID-ER into NIH-3T3 cells, induced AID activity by OHT treatment, and analyzed the appearance of mOrange+ revertant cells for up to 11 d of culture. We found that Ugi expression prevented the generation of mOrange+ cells, suggesting that UNG activity is needed to promote the transversion mutations that revert the STOP codon at the mOrangeSTOP sequence (Fig. 3, A and B).
Figure 3.
UNG can promote both error-prone and error-free repair of AID-induced lesions. NIH-3T3 cells were co-transduced with mOrangeSTOP- and AID-ER–expressing vectors, and Ugi, UNG, or empty vector as control. Transduced cells were cultured in the presence of OHT during 11 d. (A) Percentage of mOrange+ cells was monitored by flow cytometry. Representative FACS analyses at day 9 are shown. (B) Time-course analysis of mOrange+ cells appearance in co-transduced NIH-3T3 cells, relative to the percentage of mOrange+ cells in control cultures after 2 d of OHT treatment. Means from three to seven experiments are represented (P < 0.05 at all days). (C and D) mOrangeSTOP transgene was PCR amplified from the total population of co-transduced NIH-3T3 cells after 11 d of OHT treatment and sequenced by NGS. Proportion (C) and absolute mutation frequency (D) of G/C transversions, G/C transitions, and A/T mutations are represented. Frequencies were calculated as the mean of the mutation frequencies in all nucleotides of the amplified mOrangeSTOP sequence. Error bars show the mean values of three different experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Complete mutation data are shown in Table S1.
To explore the contribution of UNG to the mutational pattern induced by AID in non–B cells, we analyzed mutations at the entire mOrangeSTOP sequence in the presence or absence of Ugi. NIH-3T3 cells were co-transduced with AID-ER or AIDE58Q-ER, mOrangeSTOP, and either Ugi or control retroviral vectors, cultured in the presence of OHT, and the mOrangeSTOP transgene was amplified from the total population of cells and sequenced by NGS. We found that Ugi expression reduced the proportion of transversions at G/C pairs (control 31.8 vs. Ugi 12.5%, P = 0.02, Student’s t test) and increased the proportion of G/C transitions (control 51.1 vs. Ugi 77.1%, P = 0.01, Student’s t test; Fig. 3 C and Table S1). Indeed, although we observed a considerable variability among different experiments, presumably as a result of the different expression levels achieved by the transduced vectors, we consistently found a dramatic bias toward transition generation and transversion impairment in Ugi-expressing cells. For instance, in experiment 3, transitions increased from 51.9% (control) to 95.1% (Ugi) and transversions decreased from 27.4% (control) to 3.9% (Ugi; Table S1). However, we did not observe differences in viability of Ugi (or UNG) transduced cells (unpublished data). These data show that, as in B cells (Di Noia and Neuberger, 2002; Rada et al., 2002), in non–B cells UNG is required for the generation of transversion mutations, in agreement with the data on immunoglobulin gene mutations in UNG-deficient or inhibited B cells. The proportion of mutations at A/T pairs was reduced in Ugi-expressing cells (Fig. 3 C), although the absolute frequency of these mutations was not altered (Fig. 3 D). Interestingly, we found that UNG inhibition resulted in a significant increase in the overall mutation load (control 9.9 × 10−4 vs. Ugi 18 × 10−4 mutations/bp, P < 0.0001, Student’s t test), which is accounted for by an increase in the frequency of transitions at G/C pairs (control 5.1 × 10−4 vs. Ugi 14.8 × 10−4 mutations/bp, P < 0.0001, Student’s t test; Fig. 3 D; similar results were obtained by Sanger sequencing, not depicted). We conclude that UNG repairs faithfully a significant fraction of the U:G lesions. Therefore, these results show that UNG can trigger two alternative resolutions of the deamination events initiated by AID: it promotes transversions at G/C pairs and it faithfully repairs a fraction of the lesions.
Sequence context influences UNG-triggered error-prone versus error-free repair of AID-induced deaminations
The finding that Ugi expression increases the mutation load in the mOrangeSTOP transgene opened the question of whether enforcing UNG expression can lead to a more efficient repair of U:G lesions initiated by AID and whether this repair activity displays any sequence preference. To approach the contribution of UNG activity in the processing of AID-mediated deaminations, we performed gain-of-function experiments. NIH-3T3 cells were co-transduced with AID-ER, mOrangeSTOP, and either UNG or control vectors and cultured in the presence of OHT. The effect of UNG overexpression was first assessed with the mOrangeSTOP reversion assay. In contrast to the dramatic blockade in the generation of mOrange+ cells after Ugi expression, we found that cells transduced with UNG generated a higher percentage of mOrange+ revertants (Fig. 3, A and B). Next, we analyzed mutations at the mOrangeSTOP transgene by NGS. We found that UNG overexpression promoted a higher frequency of transversions at G/C nucleotides (control 3 × 10−4 vs. UNG 4.2 × 10−4, P = 0.04, Student’s t test) at the expense of G/C transitions (control 5 × 10−4 vs. UNG 3.6 × 10−4, P = 0.02, Student’s t test; Fig. 3 D). However, the overall mutation frequency in UNG overexpressing cells was similar to that observed in control cells (Fig. 3 D). These results show that UNG overexpression does not significantly alter the mutation load, but it does contribute to the final outcome of the resolution of the lesions induced by AID and increases the generation of transversions at G/C pairs.
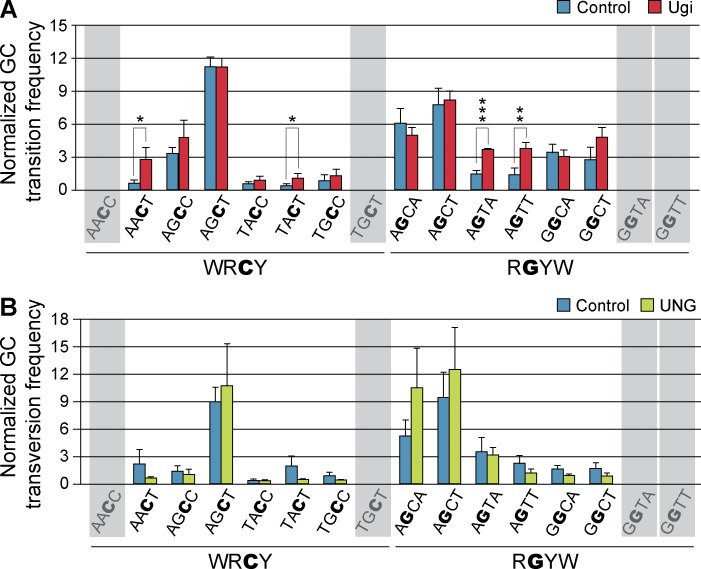
To determine the distribution of mutations in the mOrangeSTOP transgene, we analyzed the frequency of transition and transversion mutations at cytosine and guanine residues contained in WRCY/RGYW hotspots from control, Ugi, and UNG expressing cells. Transition and transversion mutations were analyzed separately, and mean mutation frequencies for individual hotspots were calculated. For the sake of comparison, mutation frequencies were normalized to the mean transition or transversion frequency at G/C nucleotides, respectively, for each individual experiment. As expected (Fig. 2 C), we found an uneven distribution of mutations across different hotspots (Fig. 4, A and B). We reasoned that given that UNG inhibition by Ugi increases the frequency of transition mutations, finding increased normalized frequencies in Ugi-expressing cells would reflect the preferred sites for UNG-mediated faithful repair. Conversely, preferred hotspots for error-prone repair by UNG would show a higher frequency of transversions in UNG overexpressing cells. We found that four individual hotspots (AACT, TACT, AGTA, and AGTT) accumulated a significantly higher frequency of mutations in Ugi-expressing cells, indicating that AID-generated uracils contained in these sequence contexts are particularly predisposed to be processed by error-free repair initiated by UNG (Fig. 4 A). In addition, we found that of all sixteen WRCY/RGYW hotspots only AGCT, AGCA, and AGCT showed a trend to accumulate transversion mutations when UNG is overexpressed (Fig. 4 B), suggesting that UNG preferentially promotes the error-prone processing of uracils when they are in these particular contexts.
Figure 4.
UNG displays sequence preference to promote error-prone or high fidelity repair at AID mutational hotspots. NIH-3T3 cells were co-transduced with mOrangeSTOP- and AID-ER–expressing vectors and Ugi, UNG, or empty vector as control. Transduced cells were cultured in the presence of OHT during 11 d and mOrangeSTOP transgene was PCR amplified and sequenced by NGS. (A) G/C transition frequencies in control (blue bars) and Ugi-expressing (red bars) cells normalized to the mean G/C transition frequency are shown for each of the WRCY and RGYW hotspots in mOrangeSTOP sequence. (B) G/C transversion frequencies in control (blue bars) and UNG-overexpressing (green bars) cells normalized to the mean G/C transversion frequency are represented as in A. Error bars represent the mean values of three independent experiments. Standard deviations are shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Shadowed hotspots (AACC, TGCT, GGTA, and GGTT) are not present in the mOrangeSTOP sequence. Complete mutation data are shown in Table S1.
UNG shapes the specificity of AID-induced mutations in B cells
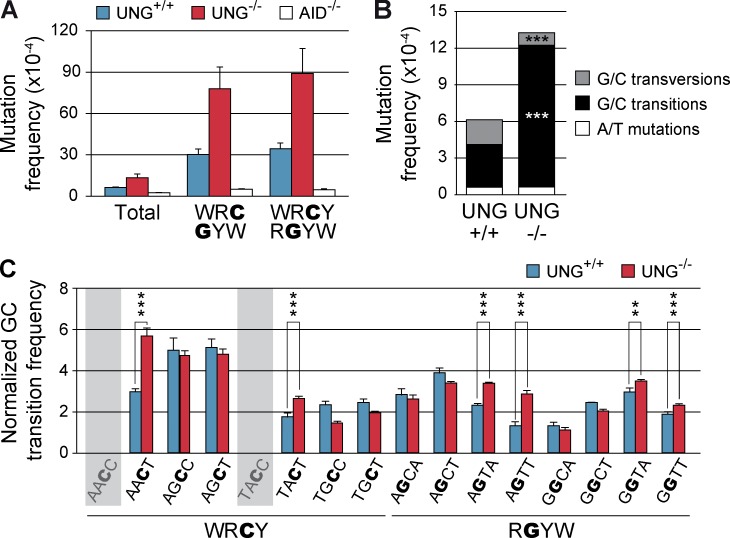
To address the contribution of UNG to error-free and error-prone resolution of AID-induced lesions in B cells, we performed NGS analysis of UNG-proficient and -deficient mice. Spleen B cells from UNG+/+, UNG−/−, and AID−/− mice were labeled with CFSE and stimulated in vitro in the presence of LPS, IL4, and BAFF. Under these conditions, B cells undergo CSR and accumulate mutations in the μ switch region (Sμ) of the immunoglobulin heavy chain locus (Reina-San-Martin et al., 2003). After 4 d, cells that had undergone more than five divisions were sorted and a region immediately upstream of Sμ was amplified and sequenced by NGS. We detected a higher total mutation frequency in wild-type (UNG+/+) B cells than in AID−/− B cells (UNG+/+ 6.2 × 10−4 vs. AID−/− 2.6 × 10−4 mutations/bp, P < 0.0001, Student’s t test), and this difference became increasingly larger in G/C nucleotides in WRC/GYW and WRCY/RGYW hotspots (Fig. 5 A, Table 3, Table S2), indicating that mutations induced by endogenous AID on immunoglobulin genes can be reliably detected by NGS. As expected, UNG deficiency resulted in a decrease in the frequency of transversions and an increase in the frequency of transitions (Di Noia and Neuberger, 2002; Rada et al., 2002; Fig. 5 B). Notably, UNG−/− B cells harbored a higher total mutation frequency than UNG+/+ B cells (UNG+/+ 6.2 × 10−4 vs. UNG−/− 13.4 × 10−4 mutations/bp, P < 0.0001, Student’s t test), which were the result of an absolute increase in the frequency of transitions (UNG+/+ 3.5 × 10−4 vs. UNG−/− 11.6 × 10−4 mutations/bp, P < 0.0001, Student’s t test; Fig. 5 B). These data reveal that, in agreement with our findings in non–B cells (Fig. 3 D), a fraction of the lesions induced by AID are faithfully repaired by UNG also in B cells.
Figure 5.
UNG sequence preference for error-free repair is conserved in B cells. Primary spleen B cells from UNG+/+ (n = 4), UNG−/− (n = 4), and AID−/− (n = 2) mice were isolated, stained with CFSE, and cultured in the presence of LPS, IL-4, and BAFF for 96 h. Cells that had undergone five or more divisions were sorted and Sµ region was PCR amplified and sequenced by NGS. (A) Mutation frequency in UNG+/+, UNG−/−, or AID−/− was calculated as in Fig. 2 B. Standard deviations are shown. Frequency values are shown in Table 3. (B) Absolute frequency of G/C transversions, G/C transitions, and A/T mutations calculated as in Fig. 3 D. (C) G/C transition frequencies in UNG+/+ and UNG−/− mice in individual WRCY and RGYW hotspots in Sµ sequence, normalized to the mean G/C transition frequency in each mouse. Standard deviations are shown. Shadowed hotspots (AACC and TACC) are not present in the Sμ sequence. **, P < 0.01; ***, P < 0.001. Reference Sμ sequence is shown in Fig. S1 B. Complete mutation data are shown in Table S2.
Table 3.
Mutation analysis in UNG+/+ and UNG−/− B cells
| Mouse ID | Totala | WRC+GYWb | WRCY+RGYWc |
| UNG+/+ 1 | 6.5 | 35.1 | 40.0 |
| UNG+/+ 2 | 6.3 | 31.0 | 34.8 |
| UNG+/+ 3 | 6.2 | 26.9 | 30.6 |
| UNG+/+ 4 | 5.9 | 27.8 | 32.0 |
| UNG−/− 1 | 14.6 | 87.5 | 100.6 |
| UNG−/− 2 | 15.5 | 92.7 | 105.8 |
| UNG−/− 3 | 13.1 | 74.4 | 84.1 |
| UNG−/− 4 | 10.3 | 57.6 | 65.7 |
| AID−/− 1 | 2.5 | 4.8 | 4.3 |
| AID−/− 2 | 2.6 | 5.5 | 5.1 |
Total frequency of mutations (×10−4).
Frequency of mutations (×10−4) in C or G of WRC or GYW hotspots.
Frequency of mutations (×10−4) in C or G of WRCY or RGYW hotspots.
Next, we analyzed the distribution of mutations at WRCY/RGYW hotspots in UNG+/+ and UNG−/− as described (Fig. 4). We found that different hotspots showed a differential accumulation of transitions in the absence of UNG. Specifically, UNG−/− B cells showed a significant increase in transition mutations in cytosines contained in AACT, TACT, AGTA, AGTT, GGTA, and GGTT hotspots (Fig. 5 C). Remarkably, AACT, TACT, AGTA, and AGTT were also found to accumulate transition mutations in Ugi-expressing NIH-3T3 cells (Fig. 4 A). GGTA and GGTT hotspots are not present in the mOrangeSTOP transgene and could not be evaluated. Together, these results indicate that UNG can trigger error-free resolution of AID-induced lesions in a sequence-dependent fashion. We conclude that UNG activity displays sequence preference that biases the error-free versus error-prone resolution of U:G mismatches and that this bias is conserved in different cell types and target sequences.
DISCUSSION
We have addressed in this study the role of UNG activity in the resolution of the U:G mismatches generated by AID-induced cytosine deamination. To that aim, we implemented a fluorescence reversion assay to monitor AID activity. In contrast to other monitoring systems published previously (Bachl and Olsson, 1999; Yoshikawa et al., 2002), the mOrangeSTOP reversion assay includes a second fluorescent protein (GFP) that allows tracking of transduced cells, providing a more accurate and sensitive detection of AID-induced revertants. This assay will be particularly useful in primary cells where transduction efficiency is typically low. In addition, we have made use of NGS technology to detect AID mutations. NGS is being widely used for the study of clonal nucleotide changes, such as the detection of cancer-associated mutations (Meyerson et al., 2010; Puente et al., 2011). However, AID-initiated mutations occur at relatively low frequencies (10−5–10−3 mutations per base pair) and widespread in a given sequence, which makes depth coverage and fidelity critical for their detection. We have shown that the PCR-based NGS approach yields a mean depth close to 100 thousand readings per base pair and that the fidelity of the technique allows the detection of mutations at frequencies at the range of 10−4 per base pair. We believe this powerful approach will be extremely valuable for future SHM studies.
Here, we have performed a thorough analysis of AID mutation spectrum by NGS. In contrast to previous analogous analyses, we are scoring only the mutations at the cytosine (or guanine, when in the opposite strand) residues contained in WRCY/RGYW hotspots. In addition, it is worth mentioning that in NIH-3T3, as in other cell lines, AID promotes low mutation frequency at A/T residues (second phase of SHM; Martin and Scharff, 2002; Yoshikawa et al., 2002). Although the reasons for this mutation pattern are not well understood, it most likely reflects the combined activity of AID and UNG in the absence of error-prone mismatch repair (Martin et al., 2002; Yoshikawa et al., 2002; Pham et al., 2008). We show here that UNG inhibition in NIH-3T3 cells results in a severe block of transversions at G/C pairs, revealing that as in B cells (Rada et al., 2004; Di Noia et al., 2006), UNG is in non–B cells the major glycosylase activity responsible for the channeling of AID generated U:G mismatches toward G/C transversions. Hence, this seems a universal pathway for the generation of this type of AID-initiated mutations.
UNG normally removes uracil from DNA and initiates error-free BER, usually by short-patch repair that involves the activity of polymerase β (Hagen et al., 2006; Visnes et al., 2009). However, in the case of AID-generated U:G mismatches, UNG triggers an error-prone resolution of these lesions that leads to the generation of mutations (SHM) or DNA double-strand breaks (CSR). This noncanonical outcome of UNG activity remains one of the most puzzling issues in the field. Here, we find that besides impairing transversions at G/C pairs, UNG inhibition both in B and non–B cells gives rise to an increased load of mutations, which is in agreement with previous observations made on UNG-deficient primary B cells and in Ugi-expressing DT40 cells (Di Noia and Neuberger, 2002; Rada et al., 2002; Robbiani et al., 2008). Although the mutation increase observed in the Sμ region could be partially explained by ongoing AID targeting as a result of the block in CSR, our finding that mutations are likewise increased in a heterologous target in non–B cells shows that not all of the U:G lesions generated by AID are subject to error-prone repair. This observation does not rule out that other glycosylases, in particular SMUG1, can repair a proportion of the deamination events (Nilsen et al., 2001; Di Noia et al., 2006; Doseth et al., 2011), but it reveals that UNG can promote error-free repair of AID-induced U:G mismatches.
In addition, we find that enforcing UNG activity in overexpression experiments does not significantly reduce the overall mutation frequency; however, it does shift the mutation pattern, leading to an increase in the transversion and a decrease in the transition mutation frequencies. It is worth mentioning that although processing of the abasic site left by UNG may presumably lead to both transitions and transversions, only the latter are unmistakably a result of UNG activity, as transitions can also arise from direct replication of the U:G mismatch. Together these results suggest that UNG overexpression enhances two alternative pathways that repair the initial U:G lesion: faithful repair, which leads to the decrease of the fraction of lesions that would generate transitions through replication; and error-prone repair, which increases the frequency of transversion mutations, presumably coupled to translesion synthesis activities.
These alternative outcomes of UNG activity prompted us to approach the sequence specificity of error-prone versus error-free repair of AID deaminations. Importantly, we find that not all of the WRCY/RGYW AID mutational hotspots are equally susceptible to both pathways. Indeed, AACT, TACT, AGTA, and AGTT hotspots were particularly prone to accumulate transition mutations when UNG activity was impaired, both in B and non–B cells, strongly suggesting that UNG has a preference to promote error-free repair of uracils at these sequence contexts. Notably, the fact that AACT/AGTT and TACT/AGTA are reverse complement sequences suggests that this specificity is consistent in both strands of DNA. In addition, GGTA and GGTT hotspots also showed an increase in transitions in UNG-deficient cells. Although these hotspots could not be evaluated in the mOrangeSTOP transgene in NIH-3T3 cells, this finding may hint that an adenine located at the −1 position of the deaminated cytosine would promote error-free resolution triggered by UNG. Conversely, all three hotspots that tend to accumulate transversions in UNG-overexpressing cells (AGCT, AGCA, and AGCT) contain a guanine at the −1 position of the targeted cytosine, suggesting that error-prone resolution of uracils could be favored in this sequence context. Analysis of a wider array of target sequences may help to define additional sequence contexts that favor error-prone versus error-free resolution of U:G lesions.
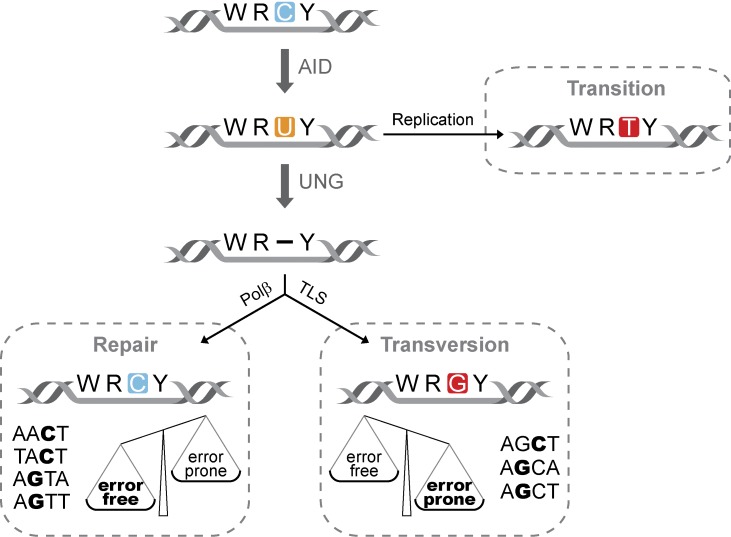
Our data raise the question of how specific sequence motifs can influence the molecular pathways acting downstream of the glycosylation reaction. It is tempting to speculate that the sequence surrounding the uracil could influence the dynamics of UNG-mediated uracil removal, allowing the specific recruitment of translesion synthesis polymerases and leading to transversion mutations in some sequence contexts, while favoring the recruitment of polβ and error-free repair in others. Although our study has only addressed this issue in the context of AID-mediated deaminations, this sequence dependency could be inherent to UNG activity itself (Nilsen et al., 1995). Alternatively, it remains possible that specific sequence contexts could increase the residence time of AID itself, thus preventing repair at those sites, as proposed previously (Delbos et al., 2007). Regardless of the molecular mechanism responsible for this effect, this is to our knowledge the first evidence that sequence environment influences the outcome of UNG activity and contributes to the mutagenic resolution of AID-induced U:G mismatches (Fig. 6).
Figure 6.
UNG shapes the specificity of AID-induced mutations. The diagram summarizes the main findings of this work (see text for details). Alternative processing pathways leading to the generation of double-strand breaks or A/T mutations are purposely excluded from the model for the sake of clarity (polβ, DNA polymerase β; TLS, translesion synthesis).
In summary, our work unveils a new layer of regulation of the specificity of AID-induced mutations that is driven by the sequence-dependent resolution of the U:G mismatch by UNG. These results open new perspectives in the understanding of AID function in antibody diversification and cancer development.
MATERIALS AND METHODS
Cell cultures and mice.
293T and NIH-3T3 cell lines were cultured in 10% FCS DME medium supplemented with 1 µM OHT when indicated. Mouse primary B cells were purified from the spleens of the indicated strains of mice by immunomagnetic depletion with anti-CD43 beads (Miltenyi Biotec) and were cultured in 50 µM 2-β-Mercaptoethanol (Invitrogen), 10 mM Hepes (Invitrogen), 10 ng/ml IL4 (PeproTech), 25 µg/ml LPS (Sigma-Aldrich), 20 ng/ml BAFF (R&D Systems), and 10% FCS RPMI medium. All mice were housed under pathogen-free conditions in accordance with the recommendations of the Federation of European Laboratory Animal Science Associations. All experiments were performed according to the Animal Bioethics and Comfort Committee protocols approved by the Instituto de Salud Carlos III.
Retroviral constructs.
The AID-ER and AIDE58Q-ER retroviral vectors were described in Ramiro et al. (2006). Ugi cDNA was provided by S.E. Bennett (Oregon State University, Corvallis, OR) and was PCR amplified using the oligonucleotides (forward) 5′-GGATCCCGCCACCATGACAAATTTATCTGACATCATTGAAAAAG-3′ and (reverse) 5′-CTCGAGTTATAACATTTTAATTTTATTTTCTCCAT-3′. PCR product was cloned in EcoRI site of pBABE-Hygro retroviral vector (Cell Biolabs). The full-length cDNA for the nuclear isoform of the mouse UNG (UNG2) was obtained from the Integrated Molecular Analysis of Genomes and their Expression (I.M.A.G.E.) consortium (I.M.A.G.E. clone 4009947). The coding region was PCR amplified using the oligonucleotides (forward) 5′-AAGGATCCCACCATGATCGGCCAGAAGACCCT-3′ and (reverse) 5′-AAGGATCCGTCACAGCTCCTTCCAGTTGA-3′ and cloned using pGEM-T easy vector kit (Promega). UNG2 was excised from this vector using flanking EcoRI sites and subcloned into pBABE-Hygro retroviral vector. mOrange point mutations were introduced by standard site-directed mutagenesis to create the STOP codon, using the oligonucleotides (forward) 5′-AAGGATCCCCACCATGGTGAGCAAGGGCGAGGAGAATAACA-3′, (reverse) 5′-TGTCGGCGGGGTGCTTCAGCTAGGCCTTGGAGCC-3′, (forward) 5′-GGCTCCAAGGCCTAGCTGAAGCACCCCGCCGACA-3′, and (reverse) 5′-AAGGATCCTTACTTGTACAGCTCGTCCATGC-3′ and cloned using the pGEM-T easy vector kit. mOrangeSTOP was excised from this vector using flanking BamHI sites and subcloned into a pMX-PIE retroviral vector (Barreto et al., 2003).
Retroviral infection and flow cytometry analysis.
Retroviral supernatants were produced by transient calcium phosphate cotransfection with pCL-ECO (Imgenex) and a combination of AID-ER, AIDE58Q-ER, pMX-PIE-OrangeSTOP, pBABE-Ugi, and pBABE-UNG retroviral vectors. NIH-3T3 cells were transduced with retroviral supernatants for 16 h in the presence of 8 µg/ml polybrene (Sigma-Aldrich) and selected with 0.4 µg/ml puromycin or 100 µg/ml hygromycin. Transduced NIH-3T3 cells were cultured in the presence of 1 µM OHT, and GFP+ mOrange+ cells were monitored by flow cytometry (FACSCanto; BD) at the indicated time points.
mOrangeSTOP amplification and mutation analysis
For analysis of mutations at mOrangeSTOP gene, co-transduced NIH-3T3 cells were cultured for 11 d in the presence of OHT. DNA was extracted and 104 cells were amplified using the oligonucleotides (forward) 5′-AATAACATGGCCATCATCAAGGA-3′ and (reverse) 5′-ACGTAGCGGCCCTCGAGTCTCTC-3′ with 2.5 U of Pfu Ultra (Stratagene) in a 50-µl reaction under the following conditions: 94°C for 5 min, followed by 25 cycles at 94°C for 10 s, 60°C for 30 s, and 72°C for 1 min. For Sanger sequencing, PCR products were cloned using the pGEM-T easy vector kit and plasmids from individual colonies were sequenced using SP6 universal primer. Sequence analysis was performed using SeqMan software (Lasergene). For NGS sequencing, five independent PCR reactions were pooled and equimolar amounts of each sample were mixed and fragmented to a range of 100-300 pb (Covaris S2). DNA was processed through successive enzymatic treatments of end repair, dA tailing, and ligation to adapters according to the manufacturer’s instructions (Illumina). Adapter-ligated libraries were further amplified by PCR with Phusion High-Fidelity DNA Polymerase (Finnzymes) using Illumina PE primers for 10 cycles. The resulting purified DNA libraries were multiplexed and applied to an Illumina flow cell for cluster generation, and sequenced on the Genome Analyzer IIx with SBS TruSeq v5 reagents according to the manufacturer’s protocols. High-quality reads (Phred Quality Score >20) were aligned using Novoalign (Novocraft Technologies) and were processed with SAMtools (Li et al., 2009) and custom scripting.
Sμ amplification and mutation analysis.
For analysis of mutations at the Sμ region, mouse primary B cells from the spleens of UNG+/+ (n = 4), UNG−/− (Nilsen et al., 2000; n = 4), and AID−/− (n = 2) mice were purified, labeled with 5 µM CFSE (Invitrogen), and cultured for 4 d in the presence of LPS, IL4, and BAFF as described earlier in this section. B cells that had undergone five or more cell divisions were isolated by sorting (FACSAria; BD). DNA was extracted and an Sμ fragment was PCR amplified using the oligonucleotides (forward) 5′-AATGGATACCTCAGTGGTTTTTAATGGTGGGTTTA-3′ and (reverse) 5′-GCGGCCCGGCTCATTCCAGTTCATTACAG-3′. For each sample, 1.3 × 104 cells were amplified using Pfu Ultra for 26 cycles (94°C for 30 s, 60°C for 30 s, and 72°C for 50 s). Eight independent PCR amplification reactions were pooled, processed, sequenced by NGS, and analyzed as described for mOrangeSTOP gene.
Statistical analysis.
Revertant assays in NIH-3T3 cells were analyzed with unpaired Student’s t tests. In Sanger sequencing experiments, mean mutation frequency of sequenced clones was analyzed with unpaired Student’s t tests. Mutation percentages were analyzed with a Fisher’s exact test.
Mutation frequencies in NGS experiments were analyzed with unpaired Student’s t tests as follows: total, G/C transition, G/C transversion, and A/T mean mutation frequencies were calculated for all nucleotides and Student’s t tests were performed (NIH-3T3, n = 3; B cells, n = 4); total, G/C transition, G/C transversion, and A/T mean mutation percentages were analyzed with an unpaired Student’s t test (NIH-3T3, n = 3); and normalized G/C transition and transversion frequencies for individual hotspots were analyzed with unpaired Student’s t tests (NIH-3T3, n = 3; B cells, n = 4).
Online supplemental material.
mOrangeSTOP and Sμ sequences analyzed by NGS are shown in Fig. S1. Tables S1 and S2 include the complete mutation data obtained from the mOrangeSTOP and Sμ sequences, respectively. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20112253/DC1.
Supplementary Material
Acknowledgments
We would like to thank V.M. Barreto, L. Blanco, O. Fernández-Capetillo, and J. Méndez for critical reading of the manuscript, S.E. Bennett and C. Rada for kindly providing the PSB2 cDNA and UNG−/− mice, respectively, and O. Domínguez, J.M. Ligos, M.D. Martinez, and A. Dopazo for technical advice.
Part of this work was conducted at the Spanish National Cancer Research Center (CNIO). P. Pérez-Durán was a fellow of the research-tra ining program (FPI) funded by the Ministerio de Ciencia e Innovación, and L. Belver and A.R. Ramiro were supported by CNIO. At present, P. Pérez-Durán, L. Belver, and A.R. Ramiro are funded by the Centro Nacional de Investigaciones Cardiovasculares (CNIC). V.G. de Yébenes is a Ramón y Cajal Investigator (Ministerio de Ciencia e Innovación), and P. Delgado is funded by the European Research Council Starting Grant program (BCLYM-207844). This work was funded by grants from Ministerio de Ciencia e Innovación (SAF2010-21394) and European Research Council Starting Grant program (BCLYM-207844).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AID
- activation-induced deaminase
- BER
- base excision repair
- CSR
- class switch recombination
- ER
- estrogen receptor
- NGS
- next generation sequencing
- OHT
- tamoxifen
- SHM
- somatic hypermutation
- Sμ
- μ switch region
- Ugi
- uracil-DNA glycosylase inhibitor
- UNG
- uracil-N-glycosylase
References
- Adams J.M., Harris A.W., Pinkert C.A., Corcoran L.M., Alexander W.S., Cory S., Palmiter R.D., Brinster R.L. 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 318:533–538 10.1038/318533a0 [DOI] [PubMed] [Google Scholar]
- Bachl J., Olsson C. 1999. Hypermutation targets a green fluorescent protein-encoding transgene in the presence of immunoglobulin enhancers. Eur. J. Immunol. 29:1383–1389 [DOI] [PubMed] [Google Scholar]
- Barreto V., Reina-San-Martin B., Ramiro A.R., McBride K.M., Nussenzweig M.C. 2003. C-terminal deletion of AID uncouples class switch recombination from somatic hypermutation and gene conversion. Mol. Cell. 12:501–508 10.1016/S1097-2765(03)00309-5 [DOI] [PubMed] [Google Scholar]
- Bransteitter R., Pham P., Calabrese P., Goodman M.F. 2004. Biochemical analysis of hypermutational targeting by wild type and mutant activation-induced cytidine deaminase. J. Biol. Chem. 279:51612–51621 10.1074/jbc.M408135200 [DOI] [PubMed] [Google Scholar]
- Delbos F., Aoufouchi S., Faili A., Weill J.C., Reynaud C.A. 2007. DNA polymerase eta is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 204:17–23 10.1084/jem.20062131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J., Neuberger M.S. 2002. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 419:43–48 10.1038/nature00981 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Rada C., Neuberger M.S. 2006. SMUG1 is able to excise uracil from immunoglobulin genes: insight into mutation versus repair. EMBO J. 25:585–595 10.1038/sj.emboj.7600939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi T., Kinoshita K., Ikegawa M., Muramatsu M., Honjo T. 2003. De novo protein synthesis is required for the activation-induced cytidine deaminase function in class-switch recombination. Proc. Natl. Acad. Sci. USA. 100:2634–2638 10.1073/pnas.0437710100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörner T., Foster S.J., Farner N.L., Lipsky P.E. 1998. Somatic hypermutation of human immunoglobulin heavy chain genes: targeting of RGYW motifs on both DNA strands. Eur. J. Immunol. 28:3384–3396 [DOI] [PubMed] [Google Scholar]
- Dorsett Y., Robbiani D.F., Jankovic M., Reina-San-Martin B., Eisenreich T.R., Nussenzweig M.C. 2007. A role for AID in chromosome translocations between c-myc and the IgH variable region. J. Exp. Med. 204:2225–2232 10.1084/jem.20070884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doseth B., Visnes T., Wallenius A., Ericsson I., Sarno A., Pettersen H.S., Flatberg A., Catterall T., Slupphaug G., Krokan H.E., Kavli B. 2011. Uracil-DNA glycosylase in base excision repair and adaptive immunity: species differences between man and mouse. J. Biol. Chem. 286:16669–16680 10.1074/jbc.M111.230052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Marusawa H., Kinoshita K., Morisawa T., Sakurai T., Okazaki I.M., Watashi K., Shimotohno K., Honjo T., Chiba T. 2007. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 26:5587–5595 10.1038/sj.onc.1210344 [DOI] [PubMed] [Google Scholar]
- Hagen L., Peña-Diaz J., Kavli B., Otterlei M., Slupphaug G., Krokan H.E. 2006. Genomic uracil and human disease. Exp. Cell Res. 312:2666–2672 10.1016/j.yexcr.2006.06.015 [DOI] [PubMed] [Google Scholar]
- Imai K., Slupphaug G., Lee W.I., Revy P., Nonoyama S., Catalan N., Yel L., Forveille M., Kavli B., Krokan H.E., et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 4:1023–1028 10.1038/ni974 [DOI] [PubMed] [Google Scholar]
- Kou T., Marusawa H., Kinoshita K., Endo Y., Okazaki I.M., Ueda Y., Kodama Y., Haga H., Ikai I., Chiba T. 2007. Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. Int. J. Cancer. 120:469–476 10.1002/ijc.22292 [DOI] [PubMed] [Google Scholar]
- Kovalchuk A.L., duBois W., Mushinski E., McNeil N.E., Hirt C., Qi C.F., Li Z., Janz S., Honjo T., Muramatsu M., et al. 2007. AID-deficient Bcl-xL transgenic mice develop delayed atypical plasma cell tumors with unusual Ig/Myc chromosomal rearrangements. J. Exp. Med. 204:2989–3001 10.1084/jem.20070882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., Schatz D.G. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845 10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Martin A., Scharff M.D. 2002. Somatic hypermutation of the AID transgene in B and non-B cells. Proc. Natl. Acad. Sci. USA. 99:12304–12308 10.1073/pnas.192442899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., Bardwell P.D., Woo C.J., Fan M., Shulman M.J., Scharff M.D. 2002. Activation-induced cytidine deaminase turns on somatic hypermutation in hybridomas. Nature. 415:802–806 10.1038/nature714 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Marusawa H., Kinoshita K., Endo Y., Kou T., Morisawa T., Azuma T., Okazaki I.M., Honjo T., Chiba T. 2007. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 13:470–476 10.1038/nm1566 [DOI] [PubMed] [Google Scholar]
- Maul R.W., Saribasak H., Martomo S.A., McClure R.L., Yang W., Vaisman A., Gramlich H.S., Schatz D.G., Woodgate R., Wilson D.M., III, Gearhart P.J. 2011. Uracil residues dependent on the deaminase AID in immunoglobulin gene variable and switch regions. Nat. Immunol. 12:70–76 10.1038/ni.1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride K.M., Barreto V., Ramiro A.R., Stavropoulos P., Nussenzweig M.C. 2004. Somatic hypermutation is limited by CRM1-dependent nuclear export of activation-induced deaminase. J. Exp. Med. 199:1235–1244 10.1084/jem.20040373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M., Gabriel S., Getz G. 2010. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11:685–696 10.1038/nrg2841 [DOI] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Nilsen H., Yazdankhah S.P., Eftedal I., Krokan H.E. 1995. Sequence specificity for removal of uracil from U.A pairs and U.G mismatches by uracil-DNA glycosylase from Escherichia coli, and correlation with mutational hotspots. FEBS Lett. 362:205–209 10.1016/0014-5793(95)00244-4 [DOI] [PubMed] [Google Scholar]
- Nilsen H., Rosewell I., Robins P., Skjelbred C.F., Andersen S., Slupphaug G., Daly G., Krokan H.E., Lindahl T., Barnes D.E. 2000. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 5:1059–1065 10.1016/S1097-2765(00)80271-3 [DOI] [PubMed] [Google Scholar]
- Nilsen H., Haushalter K.A., Robins P., Barnes D.E., Verdine G.L., Lindahl T. 2001. Excision of deaminated cytosine from the vertebrate genome: role of the SMUG1 uracil-DNA glycosylase. EMBO J. 20:4278–4286 10.1093/emboj/20.15.4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Neumeister P., Goossens T., Nanjangud G., Chaganti R.S., Küppers R., Dalla-Favera R. 2001. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 412:341–346 10.1038/35085588 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Bhagat G., Jankovic M., Compagno M., Smith P., Muramatsu M., Honjo T., Morse H.C., III, Nussenzweig M.C., Dalla-Favera R. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40:108–112 10.1038/ng.2007.35 [DOI] [PubMed] [Google Scholar]
- Peled J.U., Kuang F.L., Iglesias-Ussel M.D., Roa S., Kalis S.L., Goodman M.F., Scharff M.D. 2008. The biochemistry of somatic hypermutation. Annu. Rev. Immunol. 26:481–511 10.1146/annurev.immunol.26.021607.090236 [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt S.K., Harris R.S., Neuberger M.S. 2002. AID mutates E. coli suggesting a DNA deamination mechanism for antibody diversification. Nature. 418:99–103 10.1038/nature00862 [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107 10.1038/nature01760 [DOI] [PubMed] [Google Scholar]
- Pham P., Zhang K., Goodman M.F. 2008. Hypermutation at A/T sites during G.U mismatch repair in vitro by human B-cell lysates. J. Biol. Chem. 283:31754–31762 10.1074/jbc.M805524200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente X.S., Pinyol M., Quesada V., Conde L., Ordóñez G.R., Villamor N., Escaramis G., Jares P., Beà S., González-Díaz M., et al. 2011. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 475:101–105 10.1038/nature10113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C., Ehrenstein M.R., Neuberger M.S., Milstein C. 1998. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 9:135–141 10.1016/S1074-7613(00)80595-6 [DOI] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755 10.1016/S0960-9822(02)01215-0 [DOI] [PubMed] [Google Scholar]
- Rada C., Di Noia J.M., Neuberger M.S. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171 10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Eisenreich T., Difilippantonio S., Chen-Kiang S., Muramatsu M., Honjo T., Nussenzweig A., Nussenzweig M.C. 2004. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 118:431–438 10.1016/j.cell.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Ramiro A.R., Jankovic M., Callen E., Difilippantonio S., Chen H.T., McBride K.M., Eisenreich T.R., Chen J., Dickins R.A., Lowe S.W., et al. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 440:105–109 10.1038/nature04495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B., Difilippantonio S., Hanitsch L., Masilamani R.F., Nussenzweig A., Nussenzweig M.C. 2003. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J. Exp. Med. 197:1767–1778 10.1084/jem.20030569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revy P., Muto T., Levy Y., Geissmann F., Plebani A., Sanal O., Catalan N., Forveille M., Dufourcq-Labelouse R., Gennery A., et al. 2000. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 102:565–575 10.1016/S0092-8674(00)00079-9 [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Bothmer A., Callen E., Reina-San-Martin B., Dorsett Y., Difilippantonio S., Bolland D.J., Chen H.T., Corcoran A.E., Nussenzweig A., Nussenzweig M.C. 2008. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 135:1028–1038 10.1016/j.cell.2008.09.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin I.B., Kolchanov N.A. 1992. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta. 1171:11–18 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Diaz M. 2004. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J. Immunol. 172:3382–3384 [DOI] [PubMed] [Google Scholar]
- Schrader C.E., Edelmann W., Kucherlapati R., Stavnezer J. 1999. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 190:323–330 10.1084/jem.190.3.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 10.1038/nbt1037 [DOI] [PubMed] [Google Scholar]
- Shapiro G.S., Aviszus K., Ikle D., Wysocki L.J. 1999. Predicting regional mutability in antibody V genes based solely on di- and trinucleotide sequence composition. J. Immunol. 163:259–268 [PubMed] [Google Scholar]
- Shen H.M., Peters A., Baron B., Zhu X., Storb U. 1998. Mutation of BCL-6 gene in normal B cells by the process of somatic hypermutation of Ig genes. Science. 280:1750–1752 10.1126/science.280.5370.1750 [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Guikema J.E., Schrader C.E. 2008. Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26:261–292 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai A., Marusawa H., Minaki Y., Watanabe T., Nakase H., Kinoshita K., Tsujimoto G., Chiba T. 2012. Targeting activation-induced cytidine deaminase prevents colon cancer development despite persistent colonic inflammation. Oncogene. 31:1733–1742 10.1038/onc.2011.352 [DOI] [PubMed] [Google Scholar]
- Visnes T., Doseth B., Pettersen H.S., Hagen L., Sousa M.M., Akbari M., Otterlei M., Kavli B., Slupphaug G., Krokan H.E. 2009. Uracil in DNA and its processing by different DNA glycosylases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364:563–568 10.1098/rstb.2008.0186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesendanger M., Kneitz B., Edelmann W., Scharff M.D. 2000. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2-MSH6 heterodimer in modulating the base substitution pattern. J. Exp. Med. 191:579–584 10.1084/jem.191.3.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue K., Rada C., Neuberger M.S. 2006. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med. 203:2085–2094 10.1084/jem.20061067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa K., Okazaki I.M., Eto T., Kinoshita K., Muramatsu M., Nagaoka H., Honjo T. 2002. AID enzyme-induced hypermutation in an actively transcribed gene in fibroblasts. Science. 296:2033–2036 10.1126/science.1071556 [DOI] [PubMed] [Google Scholar]
- Yu K., Huang F.T., Lieber M.R. 2004. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 279:6496–6500 10.1074/jbc.M311616200 [DOI] [PubMed] [Google Scholar]
- Zarrin A.A., Alt F.W., Chaudhuri J., Stokes N., Kaushal D., Du Pasquier L., Tian M. 2004. An evolutionarily conserved target motif for immunoglobulin class-switch recombination. Nat. Immunol. 5:1275–1281 10.1038/ni1137 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.