A human antibody facilitates opsonophagocytic killing, inhibits attachment of Pseudomonas aeruginosa, and exerts protective effects in several animal models of P. aeruginosa infection.
Abstract
Pseudomonas aeruginosa is a leading cause of hospital-associated infections in the seriously ill, and the primary agent of chronic lung infections in cystic fibrosis patients. A major obstacle to effective control of P. aeruginosa infections is its intrinsic resistance to most antibiotic classes, which results from chromosomally encoded drug-efflux systems and multiple acquired resistance mechanisms selected by years of aggressive antibiotic therapy. These factors demand new strategies and drugs to prevent and treat P. aeruginosa infections. Herein, we describe a monoclonal antibody (mAb) selection strategy on whole P. aeruginosa cells using single-chain variable fragment phage libraries derived from healthy individuals and patients convalescing from P. aeruginosa infections. This approach enabled identification of mAbs that bind three distinct epitopes on the product of the Psl. This exopolysaccharide is important for P. aeruginosa attachment to mammalian cells, and for the formation and maintenance of biofilms produced by nonmucoid and mucoid P. aeruginosa isolates. Functional screens revealed that mAbs to one epitope exhibit superior activity in opsonophagocytic killing and cell attachment assays, and confer significant protection in multiple animal models. Our results indicate that Psl is an accessible serotype-independent surface feature and promising novel protective antigen for preventing P. aeruginosa infections. Furthermore, our mAb discovery strategy holds promise for application to other bacterial pathogens.
Antibody therapy for serious bacterial infections using polyclonal immune antitoxin or anticapsule horse serum actually predates antibiotic use. The development of broader-spectrum antibiotics rapidly supplanted the use of horse serum for reasons of safety, convenient empirical use, and cost (Casadevall and Scharff, 1994; Casadevall and Scharff, 1995; Buchwald and Pirofski, 2003). However, widespread drug resistance is quickly reducing the number of effective antibiotics available for treatment of severe bacterial infections. Although much effort has been spent on new antibacterial target and antibiotic lead discovery, none of the currently approved antibiotic classes were derived from target-focused efforts (Fernandes, 2006; Lange et al., 2007). Indeed, there is little in the antibiotic pipeline other than next generation compounds focusing on the same targets identified decades ago using whole-cell screening (Payne et al., 2007). This worsening antibiotic resistance predicament, coupled with advancements in human mAb technologies, has led to serious consideration of returning to specific antibody-based prophylaxis or therapy (Saylor et al., 2009). In particular, these drugs could be effective in preventing or treating high-risk hospital infections caused by bacterial pathogens such as Pseudomonas aeruginosa.
P. aeruginosa is a highly adaptable opportunistic bacterium that can cause life-threatening infections. Already intrinsically resistant to many antibiotics, reports of P. aeruginosa acquisition of multidrug resistance to late generation antibiotics are now common (Jovcic et al., 2011; Kunz and Brook, 2010). This reality demands new approaches and drugs to prevent and treat P. aeruginosa infections. Efforts to select protective antibodies to P. aeruginosa and other pathogens have been mostly target-centric, focusing on bacterial surface features or virulence factors correlated with disease. Antibodies targeting P. aeruginosa O-antigen, flagella, alginate, and components of the type 3 secretion system have all shown potential, and some are currently being tested in clinical studies (Döring et al., 1995, 2007; Sawa et al., 1999; Pier et al., 2004; Neely et al., 2005; DiGiandomenico et al., 2007). However, the development of antibody phage libraries and high-throughput capabilities to identify active leads has made it possible to take a more target-indifferent approach in which desirable mAb activities are first identified, followed by elucidation. In principal, this strategy is similar to the approach used to identify the targets for all antibiotics currently approved for human use, in which leads with desirable activities were selected before their targets were identified.
Here, we describe a phenotypic or target indifferent strategy based on selecting human single-chain variable fragment (scFv)–expressing phage on whole P. aeruginosa bacteria. After first enriching for P. aeruginosa whole-cell binding, phage derived from highly diverse antibody libraries constructed from multiple healthy subjects or convalescing P. aeruginosa–infected patients were converted to IgG1 mAbs and screened for desirable and potentially protective activities. Using this strategy, we identified mAbs that bind three distinct epitopes of P. aeruginosa Psl, an exopolysaccharide involved in host cell attachment and in the formation and maintenance of biofilms produced by both nonmucoid and mucoid strains (Friedman and Kolter, 2004; Jackson et al., 2004; Matsukawa and Greenberg, 2004; Byrd et al., 2009; Ma et al., 2009). The structure of Psl, which consists of a repeating pentasaccharide containing d-mannose, d-glucose, and l-rhamnose, was recently described (Byrd et al., 2009). Interestingly, visualization of Psl on the surface of P. aeruginosa indicates that it is anchored to the cell surface in a helical pattern; an organization that is thought to provide a scaffold for other biofilm-initiating components, as well as contributing to cell–cell interactions (Ma et al., 2009). Although synthesis and transport of Psl to the surface of P. aeruginosa has not been characterized, several proteins encoded by the Psl biosynthetic gene loci are homologous to proteins found in the Wzy-dependent biosynthesis pathway of Escherichia coli group 1 capsular polysaccharides (Franklin et al., 2011).
Our results indicate that Psl is a serotype-independent, antibody-accessible antigen that is prevalent among both nonmucoid and mucoid clinical isolates. Interestingly, mAbs that bound to one particular Psl epitope mediated potent opsonophagocytic killing of P. aeruginosa in vitro, inhibited bacterial attachment to cultured lung epithelial cells, and provided potent prophylactic protection in multiple animal models of P. aeruginosa infection. These data indicate that Psl is a novel protective antigen and that antibodies targeting Psl might be an effective measure for prophylaxis against P. aeruginosa infections in high-risk patients. Furthermore, the phenotypic whole-cell screening approach coupled with the early relevant mechanism of action screens shows significant promise for identifying new bacterial targets and antibodies that are active against other challenging bacterial pathogens.
RESULTS
Construction and screening of human antibody phage display libraries
Patients recently exposed and recovering from P. aeruginosa infection are a possible enriched source of protective P. aeruginosa antibodies. With this in mind, we used a target-indifferent P. aeruginosa whole-cell panning approach with human scFv phage libraries to identify novel protective antigens. An M13 phage-based human antibody library was constructed from peripheral blood B cells collected from subjects 7–10 d after documented P. aeruginosa infection. The final cloned scFv library contained 5.4 × 108 transformants, and sequencing revealed that 79% of scFv genes were full-length and in-frame. The VH CDR3 loops, often important for determining epitope specificity, were 84% diverse at the amino acid level before library selection (unpublished data).
After development and validation of the whole-cell affinity selection methodology, both the patient library and a previously constructed library derived from multiple healthy subjects (Vaughan et al., 1996) underwent affinity selection on suspensions of P. aeruginosa strain 3064 (possessing a complete O-antigen), as well as an isogenic wapR mutant strain that lacked surface expression of O-antigen. Output titers from successive patient library selections were found to increase at a greater rate for the patient library than for the naive library (107 vs. 3 × 105 at round 3, respectively; unpublished data). Duplication of VH CDR3 loop sequences in the libraries, which is a measure of clonal enrichment during selection, was also found to be higher in the patient library, reaching 88–92%, compared with 15–25% in the healthy subject library at round 3 (unpublished data). This suggests that donors contributing to the patient library were responding to infection by expanding B cell repertoires specific for P. aeruginosa. Individual scFv phage from affinity selections were next screened by ELISA for reactivity to P. aeruginosa heterologous serotype strains. As expected, the dominant phage type obtained from whole-cell selections with both libraries were clones that did not cross react with multiple O-antigen serotypes. However clones exhibiting serotype-independent binding to P. aeruginosa in the absence of nonspecific binding to E. coli or bovine serum albumin were identified and selected for further evaluation. Human IgG1 antibodies made with the variable regions encoded by the selected phage were further evaluated for specificity and prioritized for subsequent analysis by whole-cell binding to clinically relevant P. aeruginosa serotypes (O11, O6, O9, O1) by FACS analysis. FACS-positive mAbs confirmed to bind to P. aeruginosa independent of serotype (unpublished data) were prioritized for functional activity screening in opsonophagocytic killing (OPK) assays.
Identification of mAbs promoting OPK of P. aeruginosa
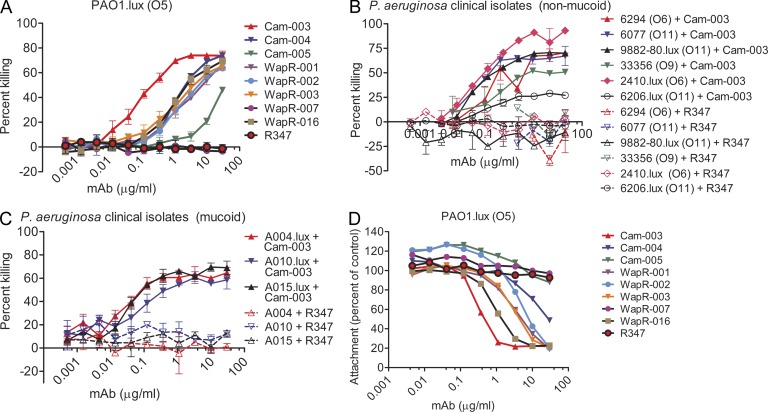
Antibodies that mediate phagocyte-dependent killing of P. aeruginosa have been correlated with in vivo efficacy (Pollack and Young, 1979; Pier and Markham, 1986). Human IgG1 antibodies selected as described in the previous paragraph, were evaluated for their potential to promote OPK of P. aeruginosa. With the exception of WapR-007 and the negative control antibody R347, all antibodies mediated concentration and complement-dependent killing of PAO1.lux (Fig. 1 A). Cam-003 exhibited superior OPK activity, having an EC50 ∼10-fold lower than the next best mAb, WapR-016 (Fig. 1 A; Table 1). We also evaluated the ability of Cam-003 to mediate OPK activity against representative nonmucoid strains from clinically relevant O-antigen serotypes and against mucoid P. aeruginosa strains that were derived from cystic fibrosis (CF) patients. Cam-003 mediated potent OPK of all nonmucoid clinical isolates tested, consistent with its ability to bind heterologous serotypes of P. aeruginosa (Fig. 1 B). In addition, Cam-003 mediated potent killing of all mucoid P. aeruginosa isolates that were tested (Fig. 1 C).
Figure 1.
Functional activity screening of antibodies derived from phage scFv patient libraries. In vitro functional screens included OPK assays and cell attachment assays using the lung epithelial cell line A549. R347, an isotype-matched human mAb that does not bind P. aeruginosa antigens, was used as a negative control. (A) Opsonophagocytosis assay with P. aeruginosa serogroup O5 strain PAO1, which was engineered to be luminescent (PAO1.lux), with dilutions of purified mAbs derived from phage panning. (B) Opsonophagocytosis assay with Cam-003 and heterologous serotype P. aeruginosa nonmucoid clinical isolates. Strains 9882–80, 2410, and 6206 were engineered to be luminescent (lux). (C) Opsonophagocytosis assay with mucoid P. aeruginosa clinical isolates that were engineered to be luminescent (lux). (A–C) The lines represent the mean percentage of killing and error bars represent the standard deviation. Percent killing was calculated relative to results obtained in assays run in the absence of antibody. (D) Log-phase PAO1.lux were added to a confluent monolayer of human cell line A549 cells after the addition of antibody at a MOI of 10 followed by analysis of RLUs after repeated washing to remove unbound P. aeruginosa. Wells lacking antibody were used as the comparative control. (A–D) Results are representative data from three independent experiments performed for each antibody and P. aeruginosa isolate.
Table 1.
Epitope mapping and identification of the relative affinity for each Psl mAb
| Epitope | mAb | Kd | Phage library | Cell attachment max. inhibition | OPK EC50 |
| nM | µg/ml | µg/ml | |||
| Class 1 | Cam-003 | 144.00 | Naive | 1.0 | 0.0220 |
| Cam-004 | 2,100.00 | Naive | >30.0 | 0.2771 | |
| Cam-005 | 8,400.00 | Naive | >30.0 | >30.0 | |
| Class 2 | WapR-001 | 0.84 | Patient | 30.0 | 0.3100 |
| WapR-003 | 12.20 | Patient | 30.0 | 0.2778 | |
| WapR-002 | 12.60 | Patient | 30.0 | 0.3960 | |
| Class 3 | WapR-016 | 75.00 | Patient | 10.0 | 0.2417 |
Epitope mapping was performed by competition ELISA and confirmed using an Octet flow system with Psl derived from the supernatant of an overnight culture of PAO1. The results indicate that Psl antibodies bind to at least three epitopes, which we refer to as class I, II, and III. Class I and II antibodies do not compete for binding; however, the class III antibody, WapR-016, partially inhibits binding of the class I and II antibodies. Antibody affinity was determined by Octet binding assays using Psl derived from the supernatant of overnight PAO1 cultures. Antibody KD was determined by averaging the binding kinetics of seven concentrations for each antibody. Also shown is a summary of cell attachment and OPK data experiments for each antibody. ELISA and antibody affinity was determined using three independent studies.
Serotype-independent anti–P. aeruginosa antibodies target the Psl exopolysaccharide
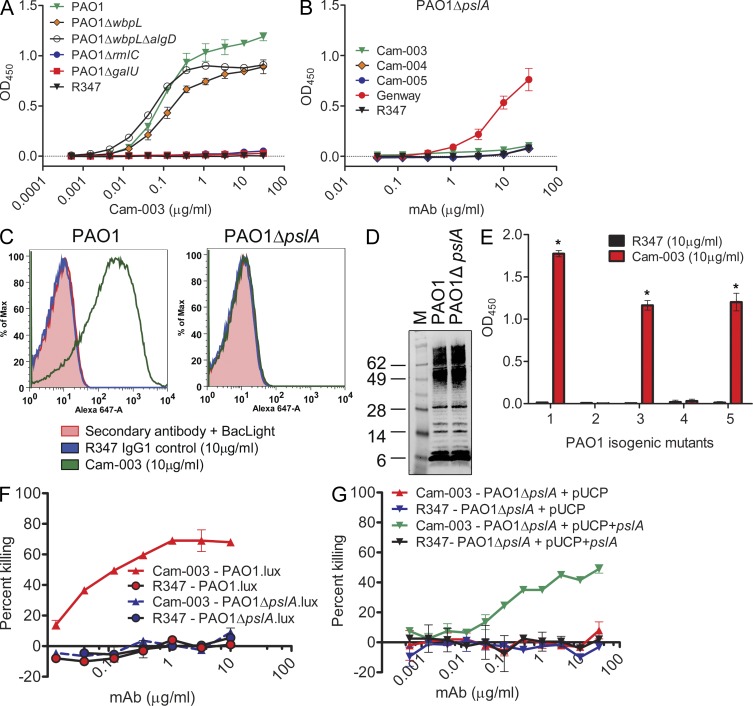
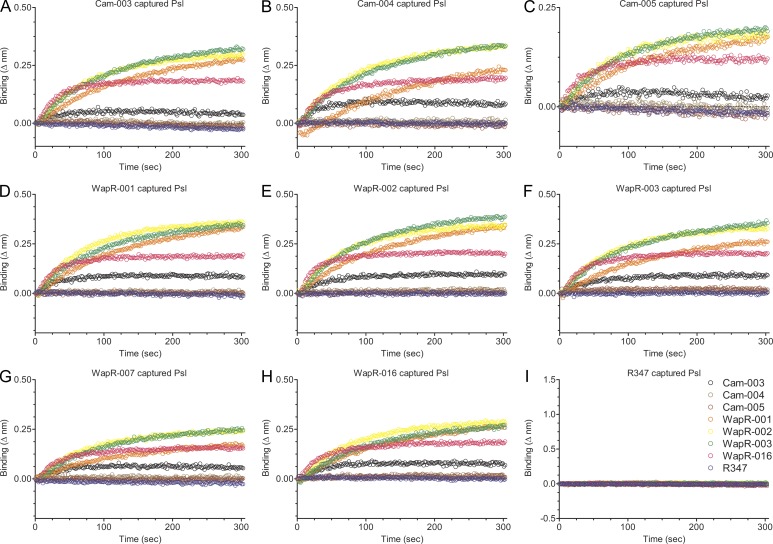
We first analyzed whether serotype-independent antibodies targeted protein or carbohydrate antigens on P. aeruginosa. No loss of binding was observed in ELISA of PAO1 whole-cell extracts that were exhaustively digested with proteinase K, suggesting that antibody reactivity targeted surface accessible carbohydrate residues (unpublished data). We initially hypothesized the antibodies targeted the LPS core region or alginate because these structures have conserved motifs among P. aeruginosa strains. Isogenic strains of PAO1 were constructed with mutations in genes responsible for O-antigen, alginate, and LPS core biosynthesis; PAO1ΔwbpL (O-antigen deficient); PAO1ΔwbpL/algD (O-antigen and alginate deficient); PAO1ΔrmlC (O-antigen deficient and truncated outer core); and PAO1ΔgalU (O-antigen deficient and truncated inner core). Cam-003 binding to PAO1ΔwbpL or the PAO1ΔwbpL/algD double mutant was comparable to wild-type PAO1; however, binding to the PAO1ΔrmlC and PAO1ΔgalU single mutants was negligible (Fig. 2 A). These results indicate that Cam-003 does not target the O-antigen or alginate. In addition, although these data suggest the antibodies could target the LPS core, which is a serotype-independent antigen, we were unable to observe binding of Cam-003 to LPS purified from wild-type PAO1 by Western blot (unpublished data). Interestingly, the rmlC and galU genes, in addition to their role in LPS core monosaccharide synthesis, were recently shown to be required for biosynthesis of components of the Psl exopolysaccharide, a repeating pentasaccharide polymer consisting of d-mannose, l-rhamnose, and d-glucose (Byrd et al., 2009). We tested Cam-003 binding to an isogenic PAO1ΔpslA knockout (O-antigen and alginate positive, Psl-negative) because pslA, which is predicted to encode for a nucleotide sugar dehydrogenase (Franklin et al., 2011), is required for Psl biosynthesis (Byrd et al., 2009). Cam-003 failed to bind PAO1ΔpslA when tested by ELISA and FACS (Fig. 2, B and C), and also failed to mediate OPK of this mutant (Fig. 2 F). In addition, Western blot analysis of LPS extracted from PAO1 and PAO1ΔpslA using antisera from PAO1-vaccinated mice revealed the LPS binding pattern of this mutant to be comparable to wild-type PAO1 (Fig. 2 D), confirming that the LPS of PAO1ΔpslA was intact and not the target of Cam-003. Binding of Cam-003 was restored in a PAO1ΔwbpL/algD/pslA (O-antigen–, alginate-, and Psl-deficient) triple mutant complemented with pslA expressed from a P. aeruginosa expression plasmid (Fig. 2 E), as was the ability of Cam-003 to mediate opsonic killing of complemented PAO1ΔpslA (Fig. 2 G). We also evaluated Cam-003 binding to PAO1ΔpelA, which is deficient in expression of the Pel exopolysaccharide, a glucose-rich polysaccharide polymer that is also known to play a role in P. aeruginosa biofilm formation (Friedman and Kolter, 2004; Colvin et al., 2011). Cam-003 binding to PAO1ΔpelA was similar to PAO1ΔwbpL/algD and the PAO1ΔwbpL/algD/pslA triple mutant complemented with pslA (Fig. 2 E). The remaining serotype-independent anti–P. aeruginosa mAbs selected during phenotypic screening were shown by ELISA to not bind PAO1ΔpslA, in contrast to wild-type PAO1, indicating that the remaining mAbs also targeted Psl (unpublished data). To confirm these results, we performed a Psl capture binding assay with proteinase K–treated enriched carbohydrate purified from PAO1ΔwbpL/algD/pelA (O-antigen, alginate, and Pel exopolysaccharide deficient) using a ForteBio Octet instrument. In this assay, individual antibodies were bound to aminopropylsilane biosensors, followed by blocking and the addition of enriched carbohydrate (capture of antigen). After washing to remove unbound antigen, binding of unlabeled mAbs was assessed. All antibodies (Cam-003, Cam-004, Cam-005, WapR-001, WapR-002, WapR-003, WapR-007, and WapR-016), with the exception of the control mAb R347, were capable of capturing antigen that strongly reacted with Cam-003, WapR-001, WapR-002, WapR-003, and WapR-016 (Fig. 3). Interestingly, minimal reactivity to captured Psl was observed with Cam-004, Cam-005, and WapR-007, even though all three of these antibodies captured sufficient Psl to potently react with Cam-003, WapR-001, WapR-002, WapR-003, and WapR-016 (Fig. 3). Collectively, these results suggest that all of the mAbs derived by phenotypic screening that bound P. aeruginosa independently of serotype target epitopes associated with Psl exopolysaccharide.
Figure 2.
Identification of the P. aeruginosa Psl exopolysaccharide as the target of antibodies derived from phenotypic screening. (A and B) Reactivity of antibodies was determined by whole-cell ELISA on plates coated with indicated P. aeruginosa strains: (A) wild-type PAO1, PAO1ΔwbpL, PAO1ΔwbpLΔalgD, PAO1ΔrmlC, and PAO1ΔgalU, and (B) PAO1ΔpslA. In B, mAb (Genway Biosciences) specific to a P. aeruginosa outer membrane protein was used as a positive control. (C) FACS binding analysis of Cam-003 to PAO1 and PAO1ΔpslA. Cam-003 is indicated by a green line; an isotype matched non–P. aeruginosa–specific human IgG1 antibody was used as a negative control and is indicated by the blue line. Washed cells were stained with BacLight to differentiate live from dead cells. Staining with the secondary antibody alone plus BacLight was used as an additional control. (D) LPS purified from PAO1 and PAO1ΔpslA was resolved by SDS-PAGE and immunoblotted with antisera derived from mice vaccinated with PAO1ΔwapRΔalgD, a double mutant strain deficient in O-antigen transport to the outer membrane and alginate production. (E) Cam-003 ELISA binding data with isogenic mutants of PAO1. Mutant 1, PAO1ΔwbpLΔalgD; mutant 2, PAO1ΔwbpLΔalgDΔpslA; mutant 3, PAO1ΔwbpLΔalgDΔpelA; mutant 4, PAO1ΔwbpLΔalgDΔpslA + pUCP; mutant 5, PAO1ΔwbpLΔalgDΔpslA + pUCP+pslA. * indicates P < 0.005 using the Mann-Whitney U test when comparing Cam-003 versus R347 binding. (F and G) Opsonophagocytosis assay using Cam-003 and negative control R347 with indicated luminescent (lux) strains of P. aeruginosa (F) or strains complemented with pUCP+pslA (G). R347 was used as a negative control in all experiments. (A–C, F, and G). (A–G) Results are representative data from three independent experiments.
Figure 3.
Anti-Psl antibody capture of enriched Psl isolated from whole P. aeruginosa cells. To confirm that all of the antibodies bound the same antigen, we developed a capture binding assay using an Octet platform with total carbohydrate extracts prepared from P. aeruginosa, which was deficient in O-antigen, alginate, and Pel polysaccharide production. In addition, the enriched carbohydrate extract was exhaustively digested with proteinase K. Antibodies were bound to aminopropylsilane sensors, followed by the addition of enriched carbohydrate. After washing, binding of the other indicated mAbs to mAb-captured carbohydrate (Psl exopolysaccharide) was assessed. (A–I) Data are representative results from three independent experiments.
Anti-Psl mAbs block attachment of P. aeruginosa to cultured epithelial cells
Psl is important in facilitating P. aeruginosa colonization of host tissues and establishing/maintaining biofilms (Borlee et al., 2010). Given these characteristics, we tested whether anti-Psl antibodies prevented P. aeruginosa association with epithelial cells. With the exception of Cam-005 and WapR-007, all antibodies reduced association of PAO1.lux with A549 human pulmonary epithelial cells (Fig. 1 D). As in the OPK assays, Cam-003 was also the most active at inhibiting cell attachment, providing an ∼80% reduction compared with the negative control (Fig. 1 D and Table 1). WapR-016 was the second most active antibody, exhibiting similar inhibitory activity as Cam-003 but at ∼10-fold higher antibody concentration.
Psl expression among clinical isolates in vitro and in vivo
A survey of Cam-003 binding by ELISA to 173 P. aeruginosa clinical isolates from diverse infection types and geographical locations revealed Psl expression/accessibility in 85% of all strains tested (unpublished data). Subgroup analysis revealed that Psl/Cam-003 reactivity is more frequently observed with isolates from confirmed nonchronic infections (96%) relative to strains obtained from the lungs of CF patients (73%). Psl was detected on 91.7% of confirmed ventilator-associated pneumonia isolates and 96.1% of isolates derived from diverse tissues (i.e., urinary tract, eye, sinus, skin and soft tissue, and intestinal tract infections). Among confirmed CF isolates, 73% were Psl positive (40 of 55 strains), including 10 of 11 mucoid strains (91%).
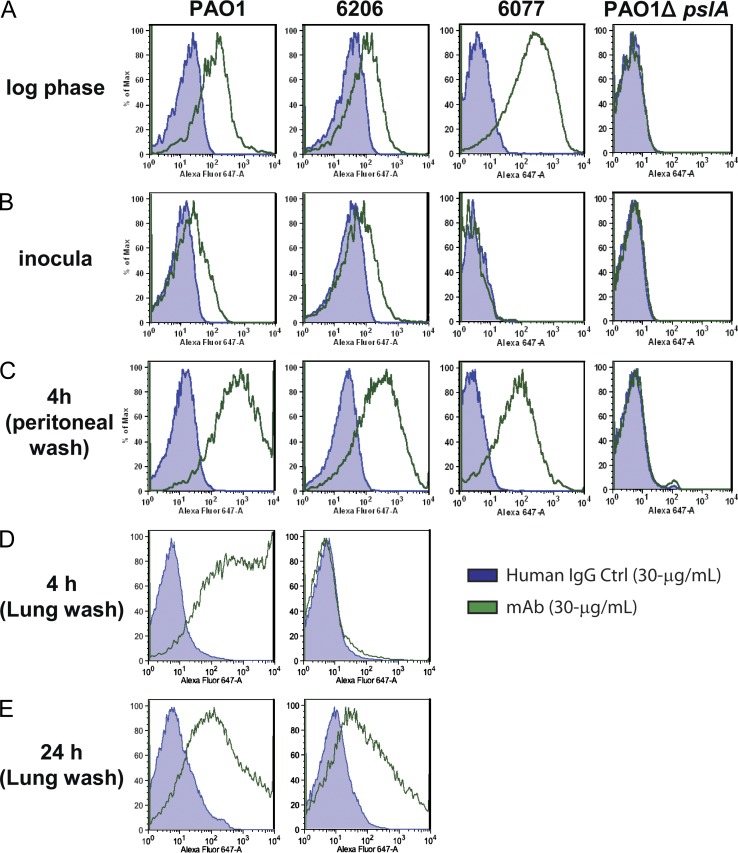
We next determined if the relative Psl expression levels were maintained on the surface of P. aeruginosa after in vivo passage in mice. For these experiments, mice were initially infected by intraperitoneal injection with wild-type P. aeruginosa strains PAO1, 6206, and 6077 or with a pslA deletion mutant of PAO1 (PAO1ΔpslA) prepared from confluent overnight cultures on plates. PAO1, 6206, and 6077 prepared in this manner all displayed reduced or no Cam-003 staining in comparison to bacteria grown to log phase in liquid culture, suggesting that Psl was either expressed at lower levels or was less accessible to the antibody on the bacterial preparations used to infect mice (Fig. 4, A and B). As expected, PAO1ΔpslA exhibited no Cam-003 staining under in vitro growth conditions (Fig. 4 A and B). At 4 or 24 h after infection, bacteria were recovered from the mice by peritoneal lavage and analyzed for Cam-003 staining by FACS. All of the wild-type P. aeruginosa strains collected 4 h after infection showed strong Cam-003 staining, which was comparable to log phase cultured bacteria (Fig. 4, A and C). In contrast, no Cam-003 binding was observed with PAO1ΔpslA collected from the peritoneal cavities of infected mice (Fig. 4 C). At 24 h after infection, an insufficient number of intact viable bacteria were harvested from the mice to allow FACS analysis of Psl expression/accessibility. However, Cam-003 reactivity was observed by ELISA for peritoneal lavage samples of PAO1, 6206, and 6077 at 24 h after infection, indicating that Psl was still present in these samples (unpublished data).
Figure 4.
Anti-Psl binding to P. aeruginosa passaged in vivo. To test if Psl expression is maintained in vivo, BALB/c mice were injected intraperitoneally with P. aeruginosa isolates, followed by harvesting of bacteria by peritoneal lavage 4 h after infection. The presence of Psl was analyzed with a control antibody and Cam-003 by FACS. (A) For the positive control, Cam-003 was assayed for binding to strains grown in vitro to log-phase from an overnight culture (∼5 × 108/ml). (B) The inocula for each strain were prepared to 5 × 108 CFU/ml, which were suspended from an overnight TSA plate grown to lawn and tested for reactivity to Cam-003. (C) 4 h after intraperitoneal challenge, bacteria was harvested from mice by peritoneal lavage and assayed for Cam-003 binding. (D) 4 h after intranasal challenge, bacteria was harvested from mice by BAL and assayed for Cam-003 binding. (E) 24 h after intranasal challenge, bacteria was harvested from mice by BAL and assayed for Cam-003 binding. Three animals were used in each group for the peritoneal lavage, and eight mice were used for BAL at each time point. (A–E) Results are representative data from five independent experiments.
We next assessed the level of Psl expression/accessibility on the surface of P. aeruginosa strains PAO1 and 6206 in the acute pneumonia model. Bacteria were prepared from overnight-incubated, confluent plates as described in the previous paragraph, intranasally administered to mice, and then recovered from the lungs by bronchoalveolar lavage (BAL) at 4 and 24 h after infection. Strong Cam-003 staining was observed for PAO1 at 4 h after infection, but was minimal for 6206 at this time point (Fig. 4 D). However, for both strain PAO1 and 6206, strong Cam-003 staining was observed at 24 h after infection (Fig. 4 E). Collectively, these data suggest that Psl expression or accessibility increases after exposure of the bacteria to the in vivo environment.
In vivo protection afforded by anti-Psl mAb Cam-003
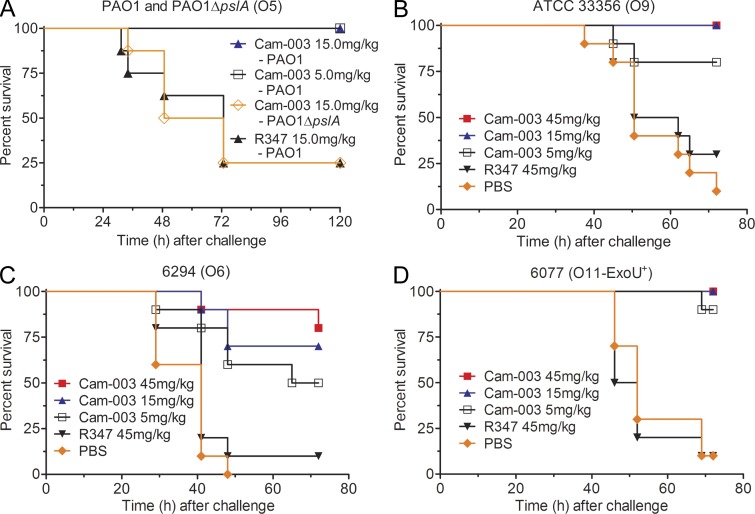
Given the superior activity of Cam-003 in the OPK and cell attachment assays (Fig. 1, A–D), we focused on evaluation of this mAb in vivo. Cam-003 was evaluated in a mouse acute lethal pneumonia model against P. aeruginosa strains representing the most dominant serotypes associated with clinical disease. In these experiments, mice received a single intraperitoneal injection of mAb 24 h before intranasal infection with different strains of P. aeruginosa including the pslA knockout strain, PAO1ΔpslA. Cam-003 provided potent protection to strain PAO1 at both 15 and 5mg/kg while providing significant concentration-dependant survival in mice infected with strain 6294 compared with control mAb-treated animals (Fig. 5, A and C). No protection was observed to the Psl mutant strain, PAO1ΔpslA, which was used as a negative control in the PAO1 acute pneumonia study (Fig. 5 A), confirming the lack of Cam-003 activity against strains deficient in Psl expression. It should be noted that in this high challenge dose acute lethal pneumonia model, endotoxic shock is a substantial component of the pathogenicity and an isogenic Psl null strain is comparable or even more lethal in comparison to the wild-type strain. Complete protection from challenge with strain ATCC 33356 or the cytotoxic strain 6077 was afforded by Cam-003 at 45 and 15 mg/kg while 80 and 90% survival was observed at 5mg/kg for ATCC 33356 and 6077, respectively (Fig. 5, B and D). The level of protection afforded by Cam-003 against strain 6077 was particularly impressive since this strain is ∼200-fold more pathogenic than strain PAO1, ATCC 33356, and 6294. The increased pathogenicity of 6077 is due to the presence of the type 3 secretion system protein, ExoU, which harbors potent phospholipase activity when delivered to host cells (Sato et al., 2003). The enhanced protection observed against this strain is likely explained by the fact that a lower required challenge dose, and thus, less LPS, is also administered to initiate the infection, allowing the anti-Psl mAb’s mechanisms more time to clear the pathogen while the bacteria is proliferating and spreading before endotoxic shock ensues.
Figure 5.
Psl mAb Cam-003 protects mice against lethal challenge in a P. aeruginosa acute pneumonia model. In each experiment, BALB/c mice were treated with PBS (B–D) or antibody (Cam-003 or negative control R347; A–D) 24 h before intranasal infection with PAO1 (4.4e7 CFU) and PAO1ΔpslA (3e7 CFU; A), 33356 (3e7 CFU; B), 6294 (7e6 CFU; C), or 6077 (1e6 CFU; D). Animals were monitored for survival between 72 and 120 h after infection. In all experiments, PBS and R347 served as negative controls. (A) A PBS control was not tested in this experiment because previous results indicated no difference in survival versus mice treated with R347. In its place, we used challenge with PAO1ΔpslA as an additional control. Results are represented as Kaplan-Meier survival curves; differences in survival were calculated by the log-rank test for Cam-003 versus R347. (A) Cam-003 (15 mg/kg, P = 0.0028; 5 mg/kg, P = 0.0028); (B) Cam-003 (45 mg/kg, P = 0.0012; 15 mg/kg, P = 0.0012; 5 mg/kg, P = 0.0373); (C) Cam-003 (45 mg/kg, P = 0.0007; 15 mg/kg, P = 0.0019; 5 mg/kg, P = 0.0212); (D) Cam-003 (45 mg/kg, P < 0.0001; 15 mg/kg, P < 0.0001; 5 mg/kg, P = 0.0001). 10 animals were used in each group. Results are representative data from three (A and D) and two independent experiments (B and C).
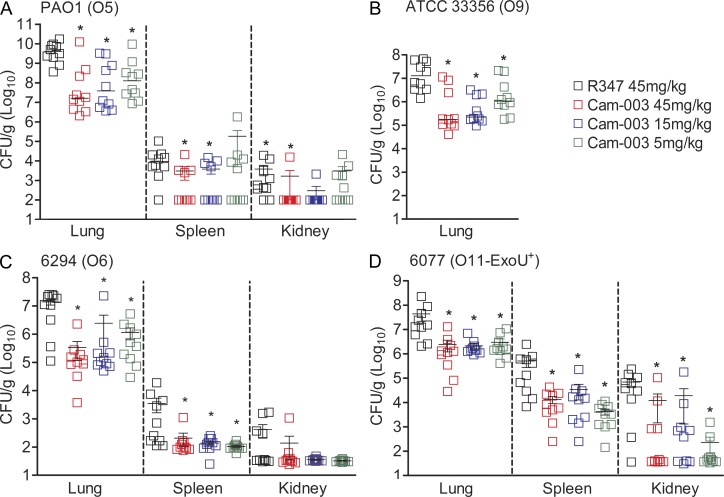
Cam-003 was next evaluated for its ability to reduce P. aeruginosa organ burden in the lung, and dissemination to distal organs with the same bacterial strains evaluated in the survival studies. Cam-003 significantly reduced P. aeruginosa lung burden for all strains tested (Fig. 6, A–D). Interestingly, Cam-003 was significantly effective against the highly pathogenic cytotoxic strain, 6077 (Fig. 6 D). Cam-003 also markedly reduced dissemination to the spleen and kidneys in mice infected with PAO1, 6294, and 6077, while dissemination to these organs for all treatment groups was not observed in ATCC 33356-infected mice (Fig. 6, A–D).
Figure 6.
Anti-Psl mAb, Cam-003, reduces organ burden after induction of acute pneumonia. BALB/c mice were treated with antibody 24 h before infection with PAO1 (1.1e7 CFU; A), 33356 (1e7 CFU; B), 6294 (6.25e6; C), and 6077 (1e6 CFU; D). 24 h after infection, animals were euthanized, followed by harvesting of organs for identification of viable CFUs. Differences in viable CFU were determined by the Mann-Whitney U test for Cam-003 versus R347. (A) Lung, Cam-003 (45 mg/kg, P = 0.0015; 15 mg/kg, P = 0.0021; 5 mg/kg, P = 0.0015); spleen, Cam-003 (45 mg/kg, P = 0.0120; 15 mg/kg, P = 0.0367); kidneys, Cam-003 (45 mg/kg, P = 0.0092; 15 mg/kg, P = 0.0056; (B) lung, Cam-003 (45 mg/kg, P = 0.0010; 15 mg/kg, P < 0.0001; 5 mg/kg, P = 0.0045); (C) lung, Cam-003 (45 mg/kg, P = 0.0003; 15 mg/kg, P = 0.0039; 5 mg/kg, P = 0.0068); spleen, Cam-003 (45 mg/kg, P = 0.0057; 15 mg/kg, P = 0.0230; 5 mg/kg, P = 0.0012); (D) lung, Cam-003 (45 mg/kg, P = 0.0005; 15 mg/kg, P = 0.0003; 5 mg/kg, P = 0.0007); spleen, Cam-003 (45 mg/kg, P = 0.0015; 15 mg/kg, P = 0.0089; 5 mg/kg, P = 0.0089); kidneys, Cam-003 (45 mg/kg, P = 0.0191; 15 mg/kg, P = 0.0355; 5 mg/kg, P = 0.0021). (A–D) The lungs, spleen, and kidneys from 10 animals (for each antibody-treated group) were used. Results are representative data from two independent experiments for each P. aeruginosa strain.
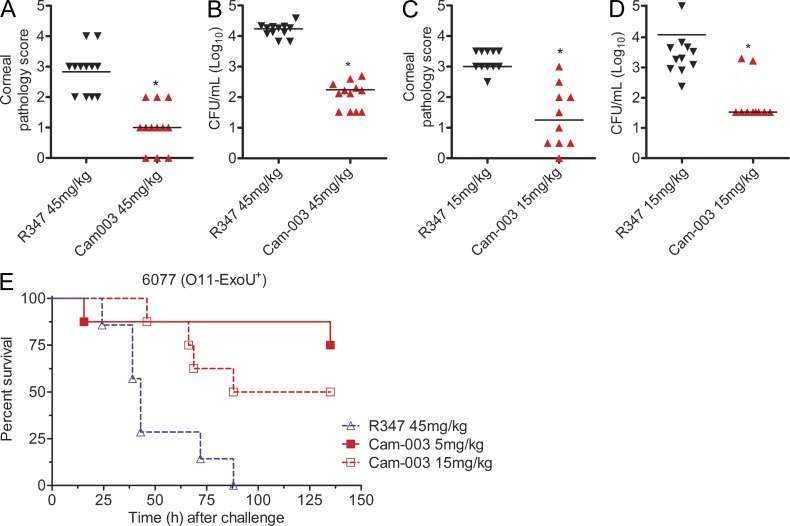
Cam-003 efficacy was next evaluated in a P. aeruginosa corneal infection model which emphasizes the pathogen’s ability to attach and colonize damaged epithelial cells. Mice receiving systemically dosed (IP) Cam-003 showed significantly less pathology and reduced bacterial colony forming units (CFU) in total eye homogenates than was observed in the R347 control mAb-treated animals (Fig. 7, A–D). When tested in a P. aeruginosa thermal injury model, Cam-003 provided significant protection at 15 and 5 mg/kg when compared with animals treated with the control R347 mAb (Fig. 7 E).
Figure 7.
Anti-Psl mAb Cam-003 is active in P. aeruginosa keratitis and thermal injury models. C3H/HeN mice were treated with control R347 antibody or Cam-003 at 45 mg/kg (A and B) or 15 mg/kg (C and D) 24 h before infection with 6077 (O11-cytotoxic, 2e6 CFU). The corneal infection model was performed as previously described (DiGiandomenico et al., 2007). Differences in pathology scores and viable CFU were determined by the Mann-Whitney U test. P = 0.0001 (A); P < 0.0001 (B); P = 0.0003 (C); P = 0.0015 (D). (E) The thermal injury model was performed using CD-1 mice as previously described (DiGiandomenico et al., 2007). Survival analysis from Cam-003– and R347-treated CF-1 mice in a P. aeruginosa thermal injury model after 6077 infection (2 × 105 CFU; log-rank: R347 vs. Cam-003 15 mg/kg, P = 0.0094; R347 vs. Cam-003 5 mg/kg, P = 0.0017). (A–D) 12 animals were used in each group. (E) Number of animals in each treatment group (R347, n = 7; Cam-003 5 mg/kg, n = 8; Cam-003 15 mg/kg, n = 8). Results are representative data from five (A–D) and from three independent experiments (E).
Epitope mapping and relative affinity of anti-Psl antibodies
Competition binding experiments between anti-Psl mAbs possessing functional activity (OPK and anti–cell attachment activity) demonstrated that they targeted at least three unique epitopes on Psl. Based on this analysis, the mAbs were categorized as class I, II, and III antibodies (Table 1). Class I and II antibodies were noncompetitive with each other, whereas the lone class III antibody, WapR-016, partially competed with both class I and II antibodies. We next determined the relative affinity of each anti-Psl antibody (Table 1). Although class II mAbs had the highest affinities of all the anti-Psl antibodies, they exhibited substantially less functional activity in vitro than the class I mAb Cam-003. Among the class II mAbs, affinity did not correlate with in vitro potency. Although WapR-003 exhibited lower affinity for Psl than WapR-001, these mAbs were equally active in the OPK and cell attachment inhibition assays (Table 1 and Fig. 1, A and D). In contrast, higher affinity did correlate with enhanced functional activity among the class I mAbs, with Cam-003 exhibiting the highest affinity for Psl and also demonstrating the greatest potency in the OPK and cell attachment assays when compared with Cam-004 and Cam-005 (Table 1 and Fig. 1, A and D).
Cam-003 Fc mutant has diminished activity
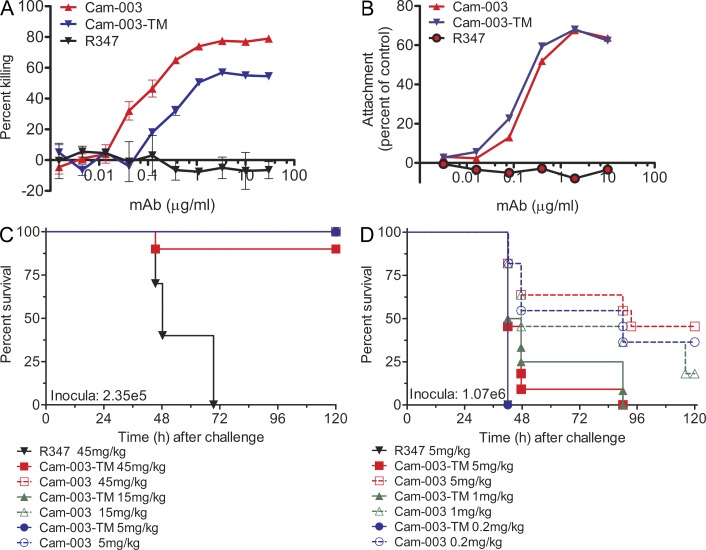
Given the potential for dual mechanisms of action (inhibition of cell attachment and OPK activity), we constructed a Cam-003 Fc mutant, Cam-003-TM, which contains sequence changes that reduce its ability to interact with Fcγ receptors (Oganesyan et al., 2008), to test whether in vivo protection against P. aeruginosa was correlated more with blocking cell attachment (likely to be Fcγ receptor independent) or with OPK activity (Fcγ receptor dependent). Although Cam-003-TM exhibited a fivefold reduction in OPK activity compared with Cam-003 (EC50 of 0.354 and 0.064, respectively), inhibition of cell attachment was comparable for Cam-003 and Cam-003-TM (Fig. 8, A and B). When tested in the mouse acute pneumonia model, Cam-003-TM was similar in potency to Cam-003 at a low infectious inoculum of 6077 (2.4 × 105 CFU; Fig. 8 C). However, further titration of the antibody dose followed by challenge with a larger infectious inoculum (1.07 × 106) revealed that Cam-003 activity was superior to Cam-003-TM, suggesting that OPK activity significantly contributes to optimal protection in vivo (Fig. 8 D).
Figure 8.
A Cam-003 Fc mutant antibody, Cam-003-TM, has diminished OPK and in vivo efficacy but maintains anti-cell attachment activity. (A) PAO1.lux OPK assay with Cam-003 and Cam-003-TM, which harbors mutations in the Fc domain that reduces Fc interactions with Fcγ receptors (Oganesyan et al., 2008). R347 was used as a negative control. (B) PAO1 cell attachment assay with Cam-003 and Cam-003-TM. Wells lacking antibody was used as the comparative control. (C, D) P. aeruginosa strain 6077 acute pneumonia model using BALB/c mice inoculated with (C) 2.35e5 or (D) 1.07e6 comparing efficacy of Cam-003 versus Cam-003-TM. Mice were treated with antibody 24 h before challenge. (C-D) Ten animals were used in each group. Results are representative data from (A) three independent experiments. (B-D) two independent experiments.
DISCUSSION
In this study, we used a phenotypic whole-cell selection approach to identify serotype-independent mAbs that mediated OPK activity against P. aeruginosa and inhibited attachment to epithelial cells. The most active antibody in both the OPK and cell attachments assays described in this study, Cam-003, was also shown to be prophylactically protective in three distinct clinically relevant P. aeruginosa murine infection models: pneumonia, thermal injury, and ocular keratitis, which emphasizes the different aspects of opportunistic P. aeruginosa infections. Although whole-cell phage panning approaches have previously been attempted in the identification of mAbs that bind other microorganisms and cancer cells (Reiche et al., 2002; Zou et al., 2007; Beerli et al., 2008; Fukuchi et al., 2010), this is the first example using this approach with both healthy subject and convalescent patient donors in combination with mechanistic activity screening and target identification for a bacterial pathogen. All antibodies shown to bind multiple P. aeruginosa serotypes were determined to target the Psl exopolysaccharide, suggesting that Psl is ubiquitous and readily accessible on the surface of P. aeruginosa. Although the greatest clonal enrichment, broadest coverage of multiple Psl epitopes, and highest affinity anti-Psl mAbs were isolated from the phage library derived from P. aeruginosa–infected convalescing patients, it is noteworthy that the convalescent patient library did not yield significantly more OPK active antibodies. It is tempting to speculate that expansion of pathogen-specific antibody subsets in infected patients are not necessarily directed to the most protective epitopes. Therefore, to identify mAbs with the greatest protective activity, it is important to implement functional activity assays, such as those described herein, as early as possible in the screening procedure while monitoring the progressive enrichment of specific phage by binding. Furthermore, our data suggests that it is most advantageous to use multiple independent phage libraries possessing high diversity. Cam-003 was selected from the phage library derived from healthy human subjects; however, it is possible that the individuals from which these libraries were constructed could have been exposed to environmental P. aeruginosa at some point in their lives.
One of the greatest technical challenges for the development of mAbs targeting bacterial pathogens is identification of serotype-independent antigens consistently expressed by the majority of clinical isolates and in the different tissues or disease stages caused by the pathogen. For P. aeruginosa, serotype-specific protection against acute infections can be achieved with O-antigen specific mAbs, although mAbs targeting serotype-independent protective targets, such as PcrV and alginate, have also been investigated (Sawa et al., 1999; Pier et al., 2004; Neely et al., 2005). In the case of PcrV, the surface-associated expression levels are not as robust as Psl (Ma et al., 2009; Sato et al., 2011), and antibodies targeting PcrV, in contrast to anti-Psl mAbs, do not promote opsonophagocytic killing of P. aeruginosa, which is an important mechanism of clearance, particularly in a mucosal setting. In contrast to alginate expression, which arises in vivo in chronically infected patients because of mutations in the negative regulator mucA (Martin et al., 1993), Psl expression is thought to be constitutively present on the surface of planktonic cells (Overhage et al., 2005; Campisano et al., 2006; Ghafoor et al., 2011). Importantly, the planktonic phenotype of P. aeruginosa, which is characterized by expression of smooth-LPS, as well as other cell surface–associated virulence factors, is believed to be important for initial colonization (Hancock et al., 1983; Burns et al., 2001). Furthermore, we show expression of Psl was maintained or up-regulated on the surface of all wild-type strains recovered 4 h after intraperitoneal infection. Likewise, Psl was accessible on the surface of P. aeruginosa recovered from the lungs of mice 24 h after intranasal administration. These results indicate that Psl expression is maintained or accessible during acute pneumonia at least 24 h after infection (Fig. 4).
P. aeruginosa is a ubiquitous, extremely versatile free-living environmental bacterium. Among its diverse lifestyles, enabled by its comparatively large bacterial genome, is its ability to colonize, persist, proliferate, and cause serious disease in individuals with damaged tissue or compromised immune defense mechanisms (Stover et al., 2000). Given the very high frequency of Psl expression observed in primary P. aeruginosa clinical isolates associated with acute clinical disease, it is likely that this unique exopolysaccharide offers general survival advantages to this environmental bacterium in the variety of conditions it encounters in its lifestyle including the human host. In addition to prevalent expression in planktonic, free-living P. aeruginosa, Psl has been shown in to be present in the biofilm matrix secreted by nonmucoid sessile bacteria in vitro (Ma et al., 2009; Overhage et al., 2005; Ryder et al., 2007) and has more recently been shown to serve as an essential scaffold in biofilms produced by alginate-secreting mucoid bacteria (Ma et al., 2012; Yang et al., 2012). Psl is also reported to be involved in facilitating attachment of P. aeruginosa to epithelial cells and in forming/maintaining biofilms in vivo (Jackson et al., 2004; Ma et al., 2009; Borlee et al., 2010). In addition, very recent studies show that Psl inhibits efficient opsonization, resulting in reduced neutrophil production of reactive oxygen species and decreased killing by phagocytic cells, providing a distinct survival advantage in vivo (Mishra et al., 2012). The preponderance of evidence cited above and elsewhere certainly suggests potential rationale and data for survival advantages conferred to P. aeruginosa by Psl during infection. Furthermore, the current literature and our own studies support Psl as a colonization or persistence-by-enhancing determinant for the bacteria, and not a virulence- or acute lethality–enhancing feature. We showed that strain PAO1ΔpslA is at least comparable or even marginally more lethal than wild-type PAO1 when used to induce acute pneumonia in mice. In this model, which requires a high challenge dose of bacteria, onset of endotoxic shock is considered a substantial component contributing to lethality. Our observation with the Psl mutant strain in the acute pneumonia model is supported by recent findings using a chinchilla model of otitis media, which emphasizes a longer term infection to study in vivo biofilm formation. Using this chronic infection model, Byrd et al. (2011) reported that P. aeruginosa proliferates from a low-challenge dose in which high levels of Psl expression were observed within in vivo biofilms, supporting the persistence observed with Psl-positive strains. In comparison, Psl knockouts are comparably or marginally more virulent in this chronic model while producing less bulky biofilms (Byrd et al., 2011). Moreover, studies investigating regulation of Psl exopolysaccharide expression indicate that type 3 secretion expression for injection of known cytotoxic virulence factors are conversely regulated at least in part by RsmA, further suggesting a coordinated balance between persistence and more virulent cytotoxic mechanisms (Irie et al., 2010; Moscoso et al., 2011).
Before this study, protective immune responses to Psl or mAbs targeting Psl have not been reported. The structure of Psl was only recently elucidated as a mannose-rich repeating pentasaccharide (Byrd et al., 2009; Kocharova et al., 1988). Despite its repetitive nature, our data demonstrated the presence of at least three distinct Psl epitopes. Antibody Cam-003, which bound the class I epitope, had the greatest anti–P. aeruginosa activity in the OPK and cell attachment assays of all the anti-Psl mAbs tested. Interestingly, mAbs targeting the class I epitope had lower affinities for Psl than mAbs to other epitopes (Table 1). Given the repetitive nature of the Psl structure, which likely would lead to an increase in mAb avidity upon binding, it was surprising that mAbs harboring the greatest affinities (class II and III) did not correlate with increased OPK activity. Opsonophagocytosis mechanisms are exquisitely complex and involve a variety of opsonins and receptors that work in concert to promote internalization of microorganisms (Underhill and Ozinsky, 2002). Opsonophagocytosis of P. aeruginosa requires complement fixation, particularly C3b, and engagement of both complement and Fcγ receptors. Whether other cellular receptors are involved or if the different classes of Psl antibodies facilitate interactions with complement, Fcγ, or other receptors that mediate activity are currently unknown. Work is under way to identify the Psl binding requirements for each anti-Psl mAb class, which could shed light on their differing ability to promote killing of P. aeruginosa.
Given the potential for dual mechanisms of action (inhibition of cell attachment and/or OPK activity), we tested which activity might be most correlative for protection by constructing an Fc mutant of Cam-003, Cam-003-TM, which reduces interaction with Fcγ1 receptors (Oganesyan et al., 2008). Cam-003-TM had reduced OPK activity but inhibited cell attachment to the same level as the parental mAb. When compared in the mouse pneumonia model to Cam-003, Cam-003-TM efficacy was inferior, suggesting that full competency in the ability to mediate OPK is necessary for optimal anti-Psl–mediated protection against P. aeruginosa in the lung. Although inhibition of cell attachment activity may contribute to protection, these data underscore OPK activity to the Psl epitope as an important in vivo mechanism of action.
With the promising protection observed with Cam-003 in multiple and disparate P. aeruginosa mouse models of acute infection using multiple P. aeruginosa strains, it was also important to further confirm that Psl expression was observed in representative clinical isolates from geographically and disease diverse sources. In this study, we observed Psl expression in a high percentage (96%) of clinical isolates derived from confirmed acute infections. Although only a small fraction of the total cases worldwide are chronic infections in CF patients, P. aeruginosa is the direst threat to this patient population, as most CF patients eventually succumb to the progressive damage caused by chronic P. aeruginosa infections. Although Psl expression was also observed in a majority of both mucoid and nonmucoid CF clinical isolates, overall Psl elaboration in diverse CF isolates was less prevalent (73%) than observed for acute isolates. The long-term establishment of a chronic P. aeruginosa infection in the CF lung occurs by a process termed adaptive radiation in which the originally infecting strain undergoes a wide variety of genotypic and phenotypic changes in the face of antibiotic and host selective pressures (Hogardt and Heesemann, 2010). This process results in both metabolic and antigenic changes, conferring an overall advantage for the P. aeruginosa population in the diverse microenvironmental niches of the inflamed CF lung (Hogardt and Heesemann, 2010). Although this could suggest less potential for anti-Psl monotherapeutic treatment of an active CF or other established chronic P. aeruginosa infections, mounting evidence strongly suggests that in addition to the prevalent Psl expression in the primary infecting strain, Psl has been implicated in establishing a persistent infection and is expressed in a variety of the diverse phenotypes, including P. aeruginosa in vivo biofilms and small colony variants (Byrd et al., 2011; Ma et al., 2012; Yang et al., 2012). Thus, targeting Psl to prevent infections in patients known to be at high risk of contracting acute or chronic P. aeruginosa infections likely holds more promise than actual treatment of an established, phenotypically diverse complex chronic infection. Ultimately, effective treatment of active persistent P. aeruginosa infections, particularly in the CF lung, will likely require multiple drug entities (small molecule antibiotic and biological) with distinct mechanisms of action to cover the strain diversity and phenotypes associated with the different microenvironments, stages of infection and spectrum of diseases caused by the pathogen. The potential and rationale for an adjunctive use of an anti-Psl mAb in combination with antibiotics seems attractive, particularly for acute infections. First, adding OPK clearance to an antibiotic’s direct antibacterial activity could reduce excessive harmful pathogen–host interactions that elicit hyperinflammatory host innate immune responses, leading to sepsis. Second, the inhibition of adherence or formation of denser biofilms could aid in preventing the establishment of a more persistent biofilm-based, antibiotic-refractory infections from a primary acute infection. However, more study is required in multiple animal models with multiple antibiotics and diverse strain types to investigate this potential anti-Psl application in adjunctive therapy. It is conceivable that the best approach to controlling P. aeruginosa infections in high risk patients may indeed be preventing them. Although the current study’s findings characterizing protective anti-Psl antibodies also support Psl as a vaccine antigen, further definition of the class I Psl-associated epitope could advance efforts to focus the most protective immune response on the right epitope.
In summary, we have also demonstrated that through a combination of advanced antibody library technologies and a target-agnostic whole-cell panning approach, coupled with early engagement of informative functional screens, promising protective mAbs to conserved bacterial surface antigen epitopes can be identified. Moreover, this process holds promise for the development of antibodies targeting other challenging bacterial pathogens.
MATERIALS AND METHODS
Construction of patient antibody phage display library.
After informed consent, blood samples for the isolation of B cells were obtained from six patients with clinical and microbiological evidence of acute infection (pneumonia, pyelonephritis, or bacteremia) caused by Pseudomonas aeruginosa. Blood samples were obtained 7–10 d after the onset of infection. The protocol and informed consent were approved by the Christiana Care Health System Investigational Review Board (Newark, DE). RNA extraction and phage library construction from whole blood was performed as previously described (Vaughan et al., 1996; Wrammert et al., 2008).
Antibody isolation using phage display technology.
In addition to the patient library, a naive human scFv phage display library containing up to 1011 binding members (Lloyd et al., 2009) was used for antibody isolation (Vaughan et al., 1996). Heat-killed P. aeruginosa (109) was immobilized in Immuno Tubes (Nunc; MaxiSorp) followed for phage display selections as previously described (Vaughan et al., 1996), with the exception of 100 nM triethanolamine being used as the elution buffer. For selection on P. aeruginosa in suspension, heat-killed cells were blocked, followed by addition of blocked phage to cells. After washing, eluted phage was used to infect E. coli cells as previously described (Vaughan et al., 1996). Rescue of phage from E. coli and binding to heat-killed P. aeruginosa by ELISA was performed as previously described (Vaughan et al., 1996).
Expression of scFv and IgG1 antibodies.
scFvs were purified from E. coli as previously described (Vaughan et al., 1996). For IgG expression, the VH and VL chains of selected antibodies were cloned into human IgG1 expression vectors, coexpressed in HEK293 cells, and purified by protein A affinity chromatography as previously described (Persic et al., 1997).
Mice.
6-wk-old wild-type BALB/c and C3H/HeN mice and 10-wk-old non–Swiss albino (CD-1) mice were purchased from The Jackson Laboratory. All mice were housed under pathogen-free conditions and fed autoclaved rodent feed and water.
P. aeruginosa clinical isolates.
173 unique P. aeruginosa clinical isolates were collected from AstraZeneca Pharmaceuticals LP, University of Pittsburgh Medical Center–Mercy Hospital via P. Castric (Duquesne University, Pittsburgh, PA), and Seattle Children’s Hospital (Seattle, WA) via J. Burns (Seattle Children’s Research Institute, Seattle, WA). Strains 6077 and 6294 were provided by J.B. Goldberg (University of Virginia, Charlottesville, VA).
Indirect and competition ELISAs.
ELISA plates (Nunc MaxiSorp) were coated with P. aeruginosa strains from overnight cultures, as previously described (DiGiandomenico et al., 2004). Diluted antibodies were added to PBS + 1% BSA (PBS-B)–blocked plates for 1 h, washed with PBS supplemented with 0.1% Tween 20 (PBS-T), and treated with HRP-conjugated anti–human secondary antibodies for 1 h, followed by development and analysis as previously described (Ulbrandt et al., 2006). For competition ELISA, antibodies were biotinylated using the EZ-Link Sulfo-NHS-Biotin and Biotinylation kit (Thermo Fisher Scientific). Unbound biotin was removed using a Zeba Spin 7 MWCO desalting column (Thermo Fisher Scientific). Antigen-coated plates were treated with the EC50 of biotinylated antibodies co-incubated with unlabeled antibodies. After incubation with HRP-conjugated streptavidin (Thermo Fisher Scientific), plates were developed as previously described (Ulbrandt et al., 2006).
Isolation of P. aeruginosa mutants by allelic exchange and mutant complementation.
P. aeruginosa mutants were constructed based on the allele replacement strategy previously described by Schweizer (1993, 1992). Vectors were mobilized from E. coli strain S17.1 into P. aeruginosa strain PAO1; recombinants were isolated as described (Hoang et al., 1998). Gene deletion was confirmed by PCR. P. aeruginosa mutants were complemented with pUCP30T-based constructs harboring wild-type genes.
Enrichment of Psl exopolysaccharide from P. aeruginosa overnight cultures.
Psl was enriched from P. aeruginosa cell culture supernatant based on the method described by Byrd et al. (2009) with modifications. In brief, PAO1ΔwbpL/algD/pelA (O-antigen–, alginate-, and Pel exopolysaccharide–deficient) triple mutant was grown by shaking at 37°C for 18 h in M63 minimal medium ([NH4]2SO4, 2 g/l; KH2PO4, 13.6 g/l; FeCl3, 0.5 mg/l, pH 7) supplemented with 0.5% Casaminoacids (BD), 1 mM MgCl2, and 0.2% glucose. After removing P. aeruginosa cells by centrifugation, the culture supernatant was concentrated through a 3K Tangential flow filtration membrane (Pall), washed extensively with PBS, and lyophilized. Lyophilized material was suspended in distilled water and treated with proteinase K for 60 min at 60°C. The enriched Psl extract was subjected to anti-Psl antibody capture by ELISA and the Octet platform.
OPK assays.
Assays were performed as previously described, with modifications (DiGiandomenico et al., 2004). In brief, assays were performed in 96-well plates using 0.025 ml of each OPK component: P. aeruginosa strains from log-phase cultures diluted to ∼2 × 106 cells/ml; diluted baby rabbit serum (1:10); differentiated HL-60 cells (2e7 cells/ml) as the polymorphonuclear leukocyte source; and mAb (final concentrations as indicated in Fig. 1). In some OPK assays, luminescent P. aeruginosa, which were constructed as previously described (Choi et al., 2005). Luminescent OPK assays were performed as previously described (DiGiandomenico et al., 2004), but with determination of relative luciferase units (RLUs) using an Envision Multilabel plate reader (PerkinElmer). The percentage of killing was determined by comparing the number of colonies or RLUs derived from assays lacking mAb to the number of colonies or RLUs obtained from assays with anti-Psl mAbs or the control R347 mAb.
P. aeruginosa cell attachment assay.
Antibodies were added to confluent A549 cells grown in opaque 96-well plates (Nunc Nunclon Delta). Log-phase luminescent PAO1 was added at an MOI of 10. After incubation at 37°C for 1 h, cells were washed, followed by the addition of 0.05 ml of LB + 0.5% glucose. Bacterial RLUs were quantified using an Envision Multilabel plate reader (PerkinElmer) after a 15-min incubation at 37°C. Measurements from wells without A549 cells were used to correct for nonspecific binding.
Flow cytometry based binding assays.
Mid-log phase P. aeruginosa strains were concentrated in PBS to an OD650 of 2.0. After incubation of antibody (10 µg/ml) and bacteria (∼1e7 cells) for 1 h at 4°C with shaking, washed cells were incubated with an Alexa Fluor 647 goat anti–human IgG antibody (Invitrogen) for 0.5 h at 4°C. Washed cells were stained with BacLight Green Bacterial Stain as recommended (Invitrogen). For ex vivo binding, 0.1 ml of bacterial inocula was prepared from an overnight TSA plate and delivered intraperitoneally to BALB/c mice. 4 h after challenge, bacteria were harvested, and RBCs were lysed, sonicated, and resuspended in PBS supplemented with 0.1% Tween-20 and 1% BSA. Samples were stained and run on an LSR II flow cytometer (BD) and analyzed using FACSDiva (v. 6.1.3; BD) and FlowJo (v. 9.2; Tree Star).
Octet measurements.
Affinity of antibodies was analyzed using the ForteBio Octet 384 instrument with 384 slanted well plates (ForteBio). PAO1 was grown overnight at 37°C at 200 RPM, followed by removal of cells by centrifugation. The culture supernatant, which served as the Psl source, was then loaded onto Octet aminopropylsilane (hydrated in PBS) sensors (300 s). The sensors were blocked with PBS-B (300 s), followed by the addition of anti-Psl mAbs that ranged from 7.8 to 10,000 nM (400 s). Disassociation was performed with PBS-B (400 s). For Octet carbohydrate capture experiments, antibodies were loaded onto aminopropylsilane (hydrated in PBS) sensors (300 s) at 10 µg/ml, followed by blocking with PBS-B (100 s). Enriched carbohydrate solution was then flowed over the sensors (300 s), followed by washing to remove unbound material (100 s). Unlabeled antibodies (10 µg/ml) were then flowed over the sensors. All steps were performed using a 3-mm sensor offset with 0.6 Hz sensitivity. Data were exported to Prism (GraphPad) for analysis of binding and for Global Association/Disassociation affinity curve fitting.
In vivo P. aeruginosa models.
All procedures were performed in accordance with federal, state, and institutional guidelines and were approved by the MedImmune Institutional Animal Care and Use Committee in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited facility. Antibodies or PBS were administered 24 h before infection in each model. P. aeruginosa acute pneumonia, keratitis, and thermal injury infection models were performed as described (DiGiandomenico et al., 2007) with modifications. In the acute pneumonia model, BALB/c mice (The Jackson Laboratory) were infected with P. aeruginosa strains suspended in a 0.05-ml inoculum. In the thermal injury model, CF-1 mice (Charles River) received a 10% total body surface area thermal injury with a metal brand heated to 92°C for 10 s. Animals were infected subcutaneously with P. aeruginosa strain 6077 at the indicated dose. For organ burden experiments, acute pneumonia was induced in mice followed by harvesting of lungs, spleens, and kidneys 24 h after infection for determination of CFU.
Statistical analyses.
All statistical analyses were performed using GraphPad Prism version 5.01 software. For survival studies, data are presented as Kaplan-Meier survival curves and were analyzed by the log-rank test. For comparison of the numbers of viable bacteria obtained from lung, spleen, kidney, and eye homogenates, Cam-003–treated animals were compared with R347-treated animals using the Mann-Whitney U test.
Acknowledgments
The authors thank Joanna B. Goldberg (University of Virginia) for kindly providing P. aeruginosa clinical isolates 6294 and 6077. In addition, the authors thank John Reinhardt (Christiana Hospital) for his help in coordinating the collection of patient blood samples.
This work was funded by MedImmune, a wholly owned subsidiary of AstraZeneca Pharmaceuticals. In addition, all authors with the exception of D.A. Melnick (AstraZeneca Pharmaceuticals LP) are employed by MedImmune.
Footnotes
Abbreviations used:
- BAL
- bronchoalveolar lavage
- CF
- cystic fibrosis
- OPK
- opsonophagocytic killing
- RLU
- relative luciferase unit
- scFv
- single-chain variable fragment
References
- Beerli R.R., Bauer M., Buser R.B., Gwerder M., Muntwiler S., Maurer P., Saudan P., Bachmann M.F. 2008. Isolation of human monoclonal antibodies by mammalian cell display. Proc. Natl. Acad. Sci. USA. 105:14336–14341 10.1073/pnas.0805942105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlee B.R., Goldman A.D., Murakami K., Samudrala R., Wozniak D.J., Parsek M.R. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75:827–842 10.1111/j.1365-2958.2009.06991.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald U.K., Pirofski L. 2003. Immune therapy for infectious diseases at the dawn of the 21st century: the past, present and future role of antibody therapy, therapeutic vaccination and biological response modifiers. Curr. Pharm. Des. 9:945–968 10.2174/1381612033455189 [DOI] [PubMed] [Google Scholar]
- Burns J.L., Gibson R.L., McNamara S., Yim D., Emerson J., Rosenfeld M., Hiatt P., McCoy K., Castile R., Smith A.L., Ramsey B.W. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183:444–452 10.1086/318075 [DOI] [PubMed] [Google Scholar]
- Byrd M.S., Sadovskaya I., Vinogradov E., Lu H., Sprinkle A.B., Richardson S.H., Ma L., Ralston B., Parsek M.R., Anderson E.M., et al. 2009. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 73:622–638 10.1111/j.1365-2958.2009.06795.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd M.S., Pang B., Hong W., Waligora E.A., Juneau R.A., Armbruster C.E., Weimer K.E., Murrah K., Mann E.E., Lu H., et al. 2011. Direct evaluation of Pseudomonas aeruginosa biofilm mediators in a chronic infection model. Infect. Immun. 79:3087–3095 10.1128/IAI.00057-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisano A., Schroeder C., Schemionek M., Overhage J., Rehm B.H. 2006. PslD is a secreted protein required for biofilm formation by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72:3066–3068 10.1128/AEM.72.4.3066-3068.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Scharff M.D. 1994. Serum therapy revisited: animal models of infection and development of passive antibody therapy. Antimicrob. Agents Chemother. 38:1695–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Scharff M.D. 1995. Return to the past: the case for antibody-based therapies in infectious diseases. Clin. Infect. Dis. 21:150–161 10.1093/clinids/21.1.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.H., Gaynor J.B., White K.G., Lopez C., Bosio C.M., Karkhoff-Schweizer R.R., Schweizer H.P. 2005. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods. 2:443–448 10.1038/nmeth765 [DOI] [PubMed] [Google Scholar]
- Colvin K.M., Gordon V.D., Murakami K., Borlee B.R., Wozniak D.J., Wong G.C., Parsek M.R. 2011. The pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264 10.1371/journal.ppat.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiandomenico A., Rao J., Goldberg J.B. 2004. Oral vaccination of BALB/c mice with Salmonella enterica serovar Typhimurium expressing Pseudomonas aeruginosa O antigen promotes increased survival in an acute fatal pneumonia model. Infect. Immun. 72:7012–7021 10.1128/IAI.72.12.7012-7021.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGiandomenico A., Rao J., Harcher K., Zaidi T.S., Gardner J., Neely A.N., Pier G.B., Goldberg J.B. 2007. Intranasal immunization with heterologously expressed polysaccharide protects against multiple Pseudomonas aeruginosa infections. Proc. Natl. Acad. Sci. USA. 104:4624–4629 10.1073/pnas.0608657104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döring G., Pfeiffer C., Weber U., Mohr-Pennert A., Dorner F. 1995. Parenteral application of a Pseudomonas aeruginosa flagella vaccine elicits specific anti-flagella antibodies in the airways of healthy individuals. Am. J. Respir. Crit. Care Med. 151:983–985 [DOI] [PubMed] [Google Scholar]
- Döring G., Meisner C., Stern M., for the Flagella Vaccine Trial Study Group 2007. A double-blind randomized placebo-controlled phase III study of a Pseudomonas aeruginosa flagella vaccine in cystic fibrosis patients. Proc. Natl. Acad. Sci. USA. 104:11020–11025 10.1073/pnas.0702403104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes P. 2006. Antibacterial discovery and development—the failure of success? Nat. Biotechnol. 24:1497–1503 10.1038/nbt1206-1497 [DOI] [PubMed] [Google Scholar]
- Franklin M.J., Nivens D.E., Weadge J.T., Howell P.L. 2011. Biosynthesis of the Pseudomonas aeruginosa Extracellular Polysaccharides, Alginate, Pel, and Psl. Front Microbiol. 2:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L., Kolter R. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675–690 10.1046/j.1365-2958.2003.03877.x [DOI] [PubMed] [Google Scholar]
- Fukuchi K., Steiniger S.C., Deryugina E., Liu Y., Lowery C.A., Gloeckner C., Zhou B., Kaufmann G.F., Quigley J.P., Janda K.D. 2010. Inhibition of tumor metastasis: functional immune modulation of the CUB domain containing protein 1. Mol. Pharm. 7:245–253 10.1021/mp900236t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghafoor A., Hay I.D., Rehm B.H. 2011. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77:5238–5246 10.1128/AEM.00637-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R.E., Mutharia L.M., Chan L., Darveau R.P., Speert D.P., Pier G.B. 1983. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypable strains deficient in lipopolysaccharide O side chains. Infect. Immun. 42:170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang T.T., Karkhoff-Schweizer R.R., Kutchma A.J., Schweizer H.P. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 212:77–86 10.1016/S0378-1119(98)00130-9 [DOI] [PubMed] [Google Scholar]
- Hogardt M., Heesemann J. 2010. Adaptation of Pseudomonas aeruginosa during persistence in the cystic fibrosis lung. Int. J. Med. Microbiol. 300:557–562 10.1016/j.ijmm.2010.08.008 [DOI] [PubMed] [Google Scholar]
- Irie Y., Starkey M., Edwards A.N., Wozniak D.J., Romeo T., Parsek M.R. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78:158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson K.D., Starkey M., Kremer S., Parsek M.R., Wozniak D.J. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466–4475 10.1128/JB.186.14.4466-4475.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovcic B., Lepsanovic Z., Suljagic V., Rackov G., Begovic J., Topisirovic L., Kojic M. 2011. Emergence of NDM-1 metallo-β-lactamase in Pseudomonas aeruginosa clinical isolates from Serbia. Antimicrob. Agents Chemother. 55:3929–3931 10.1128/AAC.00226-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocharova N.A., Knirel Y.A., Shashkov A.S., Kochetkov N.K., Pier G.B. 1988. Structure of an extracellular cross-reactive polysaccharide from Pseudomonas aeruginosa immunotype 4. J. Biol. Chem. 263:11291–11295 [PubMed] [Google Scholar]
- Kunz A.N., Brook I. 2010. Emerging resistant Gram-negative aerobic bacilli in hospital-acquired infections. Chemotherapy. 56:492–500 10.1159/000321018 [DOI] [PubMed] [Google Scholar]
- Lange R.P., Locher H.H., Wyss P.C., Then R.L. 2007. The targets of currently used antibacterial agents: lessons for drug discovery. Curr. Pharm. Des. 13:3140–3154 10.2174/138161207782110408 [DOI] [PubMed] [Google Scholar]
- Lloyd C., Lowe D., Edwards B., Welsh F., Dilks T., Hardman C., Vaughan T. 2009. Modelling the human immune response: performance of a 1011 human antibody repertoire against a broad panel of therapeutically relevant antigens. Protein Eng. Des. Sel. 22:159–168 10.1093/protein/gzn058 [DOI] [PubMed] [Google Scholar]
- Ma L., Conover M., Lu H., Parsek M.R., Bayles K., Wozniak D.J. 2009. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 5:e1000354 10.1371/journal.ppat.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Wang S., Wang D., Parsek M.R., Wozniak D.J. 2012. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol. [DOI] [PubMed] [Google Scholar]
- Martin D.W., Schurr M.J., Mudd M.H., Govan J.R., Holloway B.W., Deretic V. 1993. Mechanism of conversion to mucoidy in Pseudomonas aeruginosa infecting cystic fibrosis patients. Proc. Natl. Acad. Sci. USA. 90:8377–8381 10.1073/pnas.90.18.8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukawa M., Greenberg E.P. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449–4456 10.1128/JB.186.14.4449-4456.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Byrd M.S., Sergeant S., Azad A.K., Parsek M.R., McPhail L., Schlesinger L.S., Wozniak D.J. 2012. Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell. Microbiol. 14:95–106 10.1111/j.1462-5822.2011.01704.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscoso J.A., Mikkelsen H., Heeb S., Williams P., Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13:3128–3138 10.1111/j.1462-2920.2011.02595.x [DOI] [PubMed] [Google Scholar]
- Neely A.N., Holder I.A., Wiener-Kronish J.P., Sawa T. 2005. Passive anti-PcrV treatment protects burned mice against Pseudomonas aeruginosa challenge. Burns. 31:153–158 10.1016/j.burns.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Oganesyan V., Gao C., Shirinian L., Wu H., Dall’Acqua W.F. 2008. Structural characterization of a human Fc fragment engineered for lack of effector functions. Acta Crystallogr. D Biol. Crystallogr. 64:700–704 10.1107/S0907444908007877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overhage J., Schemionek M., Webb J.S., Rehm B.H. 2005. Expression of the psl operon in Pseudomonas aeruginosa PAO1 biofilms: PslA performs an essential function in biofilm formation. Appl. Environ. Microbiol. 71:4407–4413 10.1128/AEM.71.8.4407-4413.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne D.J., Gwynn M.N., Holmes D.J., Pompliano D.L. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6:29–40 10.1038/nrd2201 [DOI] [PubMed] [Google Scholar]
- Persic L., Roberts A., Wilton J., Cattaneo A., Bradbury A., Hoogenboom H.R. 1997. An integrated vector system for the eukaryotic expression of antibodies or their fragments after selection from phage display libraries. Gene. 187:9–18 10.1016/S0378-1119(96)00628-2 [DOI] [PubMed] [Google Scholar]
- Pier G.B., Markham R.B. 1986. Serotypes and immune responses to Pseudomonas aeruginosa. Manual of Clinical Laboratory Immunology. Rose N.R., Friedman H., Fahey J.L., American Society of Microbiology, Washington, DC: Page 92 [Google Scholar]
- Pier G.B., Boyer D., Preston M., Coleman F.T., Llosa N., Mueschenborn-Koglin S., Theilacker C., Goldenberg H., Uchin J., Priebe G.P., et al. 2004. Human monoclonal antibodies to Pseudomonas aeruginosa alginate that protect against infection by both mucoid and nonmucoid strains. J. Immunol. 173:5671–5678 [DOI] [PubMed] [Google Scholar]
- Pollack M., Young L.S. 1979. Protective activity of antibodies to exotoxin A and lipopolysaccharide at the onset of Pseudomonas aeruginosa septicemia in man. J. Clin. Invest. 63:276–286 10.1172/JCI109300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiche N., Jung A., Brabletz T., Vater T., Kirchner T., Faller G. 2002. Generation and characterization of human monoclonal scFv antibodies against Helicobacter pylori antigens. Infect. Immun. 70:4158–4164 10.1128/IAI.70.8.4158-4164.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder C., Byrd M., Wozniak D.J. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr. Opin. Microbiol. 10:644–648 10.1016/j.mib.2007.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Frank D.W., Hillard C.J., Feix J.B., Pankhaniya R.R., Moriyama K., Finck-Barbançon V., Buchaklian A., Lei M., Long R.M., et al. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959–2969 10.1093/emboj/cdg290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H., Hunt M.L., Weiner J.J., Hansen A.T., Frank D.W. 2011. Modified needle-tip PcrV proteins reveal distinct phenotypes relevant to the control of type III secretion and intoxication by Pseudomonas aeruginosa. PLoS ONE. 6:e18356 10.1371/journal.pone.0018356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa T., Yahr T.L., Ohara M., Kurahashi K., Gropper M.A., Wiener-Kronish J.P., Frank D.W. 1999. Active and passive immunization with the Pseudomonas V antigen protects against type III intoxication and lung injury. Nat. Med. 5:392–398 10.1038/7391 [DOI] [PubMed] [Google Scholar]
- Saylor C., Dadachova E., Casadevall A. 2009. Monoclonal antibody-based therapies for microbial diseases. Vaccine. 27(Suppl 6):G38–G46 10.1016/j.vaccine.2009.09.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer H.P. 1992. Allelic exchange in Pseudomonas aeruginosa using novel ColE1-type vectors and a family of cassettes containing a portable oriT and the counter-selectable Bacillus subtilis sacB marker. Mol. Microbiol. 6:1195–1204 10.1111/j.1365-2958.1992.tb01558.x [DOI] [PubMed] [Google Scholar]
- Schweizer H.D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques. 15:831–834 [PubMed] [Google Scholar]
- Stover C.K., Pham X.Q., Erwin A.L., Mizoguchi S.D., Warrener P., Hickey M.J., Brinkman F.S., Hufnagle W.O., Kowalik D.J., Lagrou M., et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 406:959–964 10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- Ulbrandt N.D., Ji H., Patel N.K., Riggs J.M., Brewah Y.A., Ready S., Donacki N.E., Folliot K., Barnes A.S., Senthil K., et al. 2006. Isolation and characterization of monoclonal antibodies which neutralize human metapneumovirus in vitro and in vivo. J. Virol. 80:7799–7806 10.1128/JVI.00318-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill D.M., Ozinsky A. 2002. Phagocytosis of microbes: complexity in action. Annu. Rev. Immunol. 20:825–852 10.1146/annurev.immunol.20.103001.114744 [DOI] [PubMed] [Google Scholar]
- Vaughan T.J., Williams A.J., Pritchard K., Osbourn J.K., Pope A.R., Earnshaw J.C., McCafferty J., Hodits R.A., Wilton J., Johnson K.S. 1996. Human antibodies with sub-nanomolar affinities isolated from a large non-immunized phage display library. Nat. Biotechnol. 14:309–314 10.1038/nbt0396-309 [DOI] [PubMed] [Google Scholar]
- Wrammert J., Smith K., Miller J., Langley W.A., Kokko K., Larsen C., Zheng N.Y., Mays I., Garman L., Helms C., et al. 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 453:667–671 10.1038/nature06890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Hengzhuang W., Wu H., Damkiaer S., Jochumsen N., Song Z., Givskov M., Høiby N., Molin S. 2012. Polysaccharides serve as scaffold of biofilms formed by mucoid Pseudomonas aeruginosa. FEMS Immunol. Med. Microbiol. [DOI] [PubMed] [Google Scholar]
- Zou N., Newsome T., Li B., Tsai S., Lo S.C. 2007. Human single-chain Fv antibodies against Burkholderia mallei and Burkholderia pseudomallei. Exp. Biol. Med. (Maywood). 232:550–556 [PubMed] [Google Scholar]