A genetic locus acting in hematopoietic cells regulates cytokine expression and granulocyte recruitment to confer susceptibility in a Helicobacter hepaticus–induced model of innate colitis and associated cancer.
Abstract
Chronic inflammation of the intestine has been associated with an elevated risk of developing colorectal cancer. Recent association studies have highlighted the role of genetic predisposition in the etiology of colitis and started to unravel its complexity. However, the genetic factors influencing the progression from colon inflammation to tumorigenesis are not known. We report the identification of a genetic interval Hiccs that regulates Helicobacter hepaticus–induced colitis and associated cancer susceptibility in a 129.RAG−/− mouse model. The 1.7-Mb congenic interval on chromosome 3, containing eight genes and five microRNAs, renders susceptible mice resistant to colitis and reduces tumor incidence and multiplicity. Bone marrow chimera experiments showed that resistance is conferred by the hematopoietic compartment. Moreover, the Hiccs locus controls the induction of the innate inflammatory response by regulating cytokine expression and granulocyte recruitment by Thy1+ innate lymphoid cells. Using a tumor-promoting model combining chronic Helicobacter hepaticus infection and the carcinogen azoxymethane, we found that Hiccs also regulates the frequency of colitis-associated neoplasia. Our study highlights the importance of innate immune cells and their genetic configuration in driving progression from inflammation toward cancer and opens the door for analysis of these pathways in human inflammatory disorders and associated cancers.
There is now abundant evidence that chronic inflammation can promote tumorigenesis and cancer (Balkwill and Coussens, 2004; Karin, 2006; Mantovani et al., 2008). An example of this is Helicobacter pylori–induced gastritis (Hatakeyama, 2004; Crowe, 2005) which in some individuals can progress to gastric carcinoma. Similarly, inflammatory bowel disease (IBD) patients have an increased risk of developing colon cancer (Ahmadi et al., 2009; Feagins et al., 2009; Ullman and Itzkowitz, 2011). In ulcerative colitis there is an increase in cumulative incidence of colorectal cancer from 7.6 to 18.4%, 30 yr after diagnosis (Eaden et al., 2001). Since the initial discovery of the association between NOD2 mutations and Crohn’s disease susceptibility (Hugot et al., 2001; Ogura et al., 2001), genome-wide association studies (GWASs) have identified novel pathways that impact on IBD susceptibility, such as the IL-23 axis involving IL-23R (Duerr et al., 2006) and STAT3 (Barrett et al., 2008), and the autophagy pathway with ATG16L1 (Hampe et al., 2007). However, little is known about the genetic factors that influence the progression to colorectal cancer in patients with IBD. A recent study has reported an association with specific HLA-DR and HLA-DQ alleles (Garrity-Park et al., 2009). Single nucleotide polymorphisms (SNPs) in CDH1, the gene coding for E-cadherin, have also been associated with both ulcerative colitis and colorectal cancer predisposition (Houlston et al., 2008; Barrett et al., 2009).
Animal models of IBD and colitis-associated cancer (CAC) of the colon are required to better define genetic susceptibility factors and to perform functional studies in a controlled setting (Kanneganti et al., 2011). One such model is the typhlocolitis induced in T and B cell–deficient (RAG−/−) 129S6(SvEv) mice upon infection with the intestinal bacterial pathogen Helicobacter hepaticus (Cahill et al., 1997; Kullberg et al., 1998; Maloy et al., 2003). Proinflammatory and regulatory pathways have been well characterized in this model and a key pathogenic role for IL-23 was recently described (Hue et al., 2006; Kullberg et al., 2006), as well as the involvement of a novel innate lymphoid cell (ILC) population that secretes IL-17 and IFN-γ in response to IL-23 (Buonocore et al., 2010). Beyond the inflammatory phenotype, long-term infection (6–12 mo) of 129S6.RAG−/− mice with H. hepaticus has been found to induce the development of a wide spectrum of dysplastic and neoplastic lesions, including noninvasive carcinoma and adenocarcinoma (Erdman et al., 2003a,b).
In contrast to the 129S6.RAG−/− strain, infection of C57BL/6.RAG−/− mice with H. hepaticus results in minimal disease in the cecum and no inflammation of the colon or CAC (Erdman et al., 2003a,b). So far, no genetic analysis has been performed to identify the susceptibility loci in this setting. However, in an IL-10−/− mouse model of spontaneous colitis, a major susceptibility locus on the distal part of chromosome 3 (designated Cdsc1) was found by comparing the susceptible C3H/HeJBir.IL-10−/− and the resistant C57BL/6.IL-10−/− strains and further confirmed by generation of congenic lines (Beckwith et al., 2005). This large region, covering 26 Mb divided into three subloci, has also been implicated in colitis in Gnai2 (G protein α inhibiting 2)-deficient mice (Borm et al., 2005) and, recently, in the T-bet−/− Rag2−/− ulcerative colitis (TRUC) innate model (Ermann et al., 2011).
Here, we identify a major locus for H. hepaticus–induced colitis and associated cancer susceptibility, designated Hiccs, in the same telomeric region of chromosome 3 associated with the IL-10−/−, Gnai2−/−, and TRUC models. We have refined the functional region to a 1.71-Mb interval and show altered expression of three genes as well as differences in nonsynonymous SNPs between resistant and susceptible strains. Importantly, we demonstrate that Hiccs controls CAC through effects on the early innate inflammatory response driven by activation of ILCs.
RESULTS
A major susceptibility locus for H. hepaticus–induced colitis is localized in the telomeric part of chromosome 3
As previously described (Erdman et al., 2003a,b), we found C57BL/6.RAG−/− (B6.RAG) mice to be strongly resistant to H. hepaticus–induced typhlocolitis, showing only limited inflammation of the cecum, reduced splenomegaly, and no colitis compared with the severe inflammation observed in H. hepaticus–infected 129S6.RAG−/− (129.RAG) mice (Fig. 1). The telomeric part of chromosome 3 has been identified in other models of colitis (using IL-10−/− and Gnai2−/− mice) as a major susceptibility locus (Beckwith et al., 2005; Borm et al., 2005). Consequently, we generated a novel congenic line on the 129.RAG background harboring a large interval of B6 origin on chromosome 3 (>40 Mb) delimited by the microsatellite markers D3Mit103 and D3Mit45 (Fig. 1 A). Upon H. hepaticus infection, phenotypic analysis of intercrossed mice from this 129S6.B6-(D3Mit103-D3Mit45).Rag2−/− congenic line (abbreviated 129.C3B.RAG or C3B) revealed that the B6 chromosome 3 interval conferred significant protection from colonic inflammation and splenomegaly but not from cecal inflammation (Fig. 1, B–D). Moreover, the similar protection between heterozygote 129/B6 and homozygote B6/B6 congenic mice indicated a clear dominant effect of the B6 chromosome 3 locus (Fig. 1, B–D). In these mice, intestinal inflammation was strongly reduced in the proximal and distal parts of the colon compared with littermate or 129.RAG controls. Some degree of inflammation could still be found in the mid colon (unpublished data). The differential phenotype in response to chronic H. hepaticus infection was not linked to a change in bacterial burden, as cecal colonization by H. hepaticus was found to be equivalent in infected B6.RAG, 129.RAG, and intercrossed 129.C3B.RAG mice (unpublished data). Using a lymphocyte-replete model of H. hepaticus infection in IL-10−/− mice, we found a marked reduction in colitis in B6.IL10−/− compared with 129.IL10−/− mice as described for the Cdcs 1 locus (Beckwith et al., 2005; Fig. 2). These results indicate that resistance to bacteria-induced colitis in the B6 compared with 129 strain extends to innate and lymphocyte replete models.
Figure 1.
Dominant protection from colitis and splenomegaly conferred by the B6 chromosome 3 interval. (A) Schematic representation of the interval of B6 origin harbored by the congenic line 129.C3B.RAG on chromosome 3, defined by the microsatellite markers D3Mit103 (107.3 Mb from centromere), D3Mit348/319 (126.7/128.0 Mb), and D3Mit45 (147.8 Mb). (B) Cecum and colon inflammation after H. hepaticus infection of parental B6.RAG and 129.RAG strains or of mice generated by intercrossing of the 129.C3B.RAG congenic line (three possible genotypes for the congenic interval: 129/129, B6/129, or B6/B6). Data represent pooled results from two independent experiments. Horizontal lines represent medians. **, P < 0.01, Mann-Whitney nonparametric test (each group compared with 129.RAG mice). (C) Representative photomicrographs of H&E-stained middle colon sections isolated from H. hepaticus–infected parental B6.RAG, 129.RAG, or mice generated by intercrossing of 129.C3B.RAG congenic line. Bars, 0.2 mm. (D) Splenomegaly was assessed in the same experiments by measuring spleen mass and spleen cell numbers. Horizontal lines represent medians. **, P < 0.01, Mann-Whitney nonparametric test (each group compared with 129.RAG mice).
Figure 2.
Differential colitis susceptibility between C57BL/6.IL-10−/− and 129S6.IL-10−/− mice. Evaluation of colon inflammation after H. hepaticus infection for 2 mo. Data represent two pooled independent experiments. Three mice of each strain were also analyzed without H. hepaticus infection. Horizontal lines represent medians. **, P < 0.01, Mann-Whitney nonparametric test between B6 and 129 mice.
Generation, characterization, and fine mapping of subcongenic recombinant lines
The 129.C3BR1.RAG (R1) recombinant was identified during the backcrossing process of the C3B congenic line. It retained the second half of the congenic interval, with a unique recombination event between the close microsatellite markers D3Mit348 and D3Mit319 (Table 1 and Fig. 3 A). Analysis of heterozygote (not depicted) and homozygote R1 mice after H. hepaticus infection showed an equivalent protected phenotype compared with C3B mice, with significantly reduced colitis and splenomegaly (Fig. 3 B). The susceptibility locus was therefore localized in the R1 interval spanning a region of 20 Mb, and the proximal boundary of this locus was delimited by the R1 recombination point.
Table 1.
Genotyping the subcongenic recombinant lines by microsatellite markers and SNPs
| Mbpa | Marker | C3B | R1 | R12 | R12X | R17 | R21 | R6 | R9 |
| 107.273 | D3Mit103 | B6 | 129 | B6 | B6 | 129 | 129 | 129 | 129 |
| 126.744 | D3Mit348 | B6 | 129 | B6 | B6 | 129 | 129 | 129 | 129 |
| 127.086 | rs31622216 | B6 | 129 | B6 | B6 | 129 | 129 | 129 | 129 |
| 127.233 | rs31106293 | B6 | B6 | B6 | B6 | B6 | B6 | 129 | 129 |
| 128.042 | D3Mit319 | B6 | B6 | B6 | B6 | B6 | B6 | 129 | 129 |
| 128.799 | rs31282270 | B6 | B6 | B6 | 129 | B6 | B6 | 129 | 129 |
| 129.011 | rs33862828 | B6 | B6 | B6 | 129 | B6 | B6 | 129 | 129 |
| 129.213 | rs30910985 | B6 | B6 | 129 | 129 | 129 | 129 | B6 | B6 |
| 130.016 | D3Mit357 | B6 | B6 | 129 | 129 | 129 | 129 | B6 | B6 |
| 139.262 | D3Mit351 | B6 | B6 | 129 | 129 | 129 | 129 | B6 | B6 |
| 147.807 | D3Mit45 | B6 | B6 | 129 | 129 | 129 | 129 | 129 | B6 |
Protection or susceptibility to H. hepaticus–induced colitis and splenomegaly is as follows: protected, C3B, R1, R12, R12X, R17, and R21; susceptible, R6 and R9.
rom the chromosome 3 centromere.
Figure 3.
Fine mapping of the Hiccs locus to a 1.71-Mb interval by analysis of subcongenic recombinant lines. (A) Schematic representation of the subcongenic intervals harbored by the respective recombinant lines (compared with the C3B congenic interval). Microsatellite markers used for genotyping are indicated. (B) Evaluation of colon inflammation and splenomegaly after H. hepaticus infection of each recombinant line. Data represent pooled results from five independent experiments. Horizontal lines represent medians. **, P < 0.01, Mann-Whitney nonparametric test (each group compared with R9 recombinant mice). (C) Representative photomicrographs of H&E-stained middle colon sections isolated from H. hepaticus–infected susceptible (R9) or protected (R17 and R12X) mice. Bars, 0.2 mm. (D) Schematic representation of the 1.71-Mb Hiccs interval defined by the recombinant lines R17 and R12X (plain arrow). Phenotype of the recombinant lines is noted as P = protected and S = susceptible. Known genes and microRNAs localized in this interval are indicated (adapted from Ensembl database), as well as markers used for genotyping. Dashed boxes and arrows represent the region containing the recombination point of the indicated line.
Other subcongenic recombinants were generated by intercrossing 129.C3B.RAG and 129.RAG mice. In particular, several recombination events were found between the microsatellite markers D3Mit319 and D3Mit357, as exemplified by the R6, R9, and R12 recombinant lines (Table 1 and Fig. 3 A), indicating a probable recombination hotspot. Mice from the R12 line were found to be protected from colitis and splenomegaly upon H. hepaticus infection, whereas the R6 and R9 lines were found to be as susceptible as the 129.RAG strain (Fig. 3, B and C). The distal boundary of the susceptibility locus was therefore delimited by the recombination point between D3Mit319 and D3Mit357, excluding the large telomeric B6 region harbored by the R6 and R9 lines.
Taking advantage of the recombination hotspot between the markers D3Mit319 and D3Mit357, we were able to generate new recombinants from the R1 line, retaining only a small region from B6 origin. The reduced interval was centered on the marker D3Mit319, excluding D3Mit348 and D3Mit357, as defined in the R17 and R21 recombinant lines (Table 1 and Fig. 3 A). Upon H. hepaticus infection, mice from R17 and R21 lines were protected from colitis and splenomegaly (Fig. 3, B and C), confirming the previous definition of the susceptibility locus.
Precise mapping of the recombination events using SNP markers localized the proximal R1 point between the SNPs rs31622216 and rs31106293 in an interval spanning 147 Kb (Table 1 and Fig. 3 D). The distal hotspot of recombination was confirmed by mapping of R6, R9, R12, R17, and R21 recombination points, which revealed a common interval between the SNPs rs33862828 and rs30910985, spanning 202 Kb (Table 1 and Fig. 3 D). The susceptibility interval was then delimited by the SNPs rs31622216 and rs30910985, spanning 2.13 Mb (Fig. 3 D).
In the course of SNP genotyping for the R12 line, a new recombination event was identified and mapped between the microsatellite marker D3Mit319 and the SNP rs31282270 (Table 1 and Fig. 3 D). As mice from this new R12X line were protected upon chronic H. hepaticus infection (Fig. 3, B and C), the distal region containing the genes Pitx2 and Enpep was excluded from the susceptibility locus. The maximal interval was therefore delimited by the SNPs rs31622216 and rs31282270, spanning 1.71 Mb (Fig. 3 D), which define the Hiccs locus. Evaluation of cytokine production by lamina propria leukocytes (LPLs) isolated from colons of protected R17 and susceptible R9 mice 8 wk after H. hepaticus infection demonstrated that protection correlated with reduced secretion of proinflammatory cytokines including IL-17, IL-22, IFN-γ, and MIP-1α (Fig. 4).
Figure 4.
Cytokine production by LPLs. LPLs were isolated from colons of H. hepaticus–infected recombinant mice. Cytokine production after overnight culture of the LPL was evaluated for susceptible R9 (n = 6, filled bar) and protected (n = 5, open bar) R17 mice. Graphs represent mean ± SEM for each indicated cytokine and show a representative experiment out of two independently performed. *, P < 0.05; **, P < 0.01, Mann-Whitney nonparametric test.
Sequencing and expression profile of the genes in the susceptibility interval
Analysis of the 1.71 Mb susceptibility interval using the Ensembl and National Center for Biotechnology Information databases identified a cluster of known genes between the SNP rs31622216 and the microsatellite D3Mit319, as well as an empty region (“gene desert”) between D3Mit319 and the SNP rs31282270 (Fig. 3 D). The cluster included four genes of known function (Neurogenin2, Tifa, Larp7, and Alpk1), three genes identified by mRNA only (4930422G04Rik [abbreviated 493Rik], Ap1ar, and 5730508B09Rik [573Rik]), one hypothetical protein (LOC100043382), and a set of microRNAs (Mir302a-d and Mir367), as shown in Table 2.
Table 2.
Exonic polymorphisms detected in the known genes and microRNAs of the 1.71Mb Hiccs locus
| Starta | Gene symbol | Description | Polymorphisms in exons (total/nonsynonymous) |
| 127239632 | Larp7 | La ribonucleoprotein domain family, member 7 | 8/2 |
| 127248145 | Mir302b | microRNA 302b | 1/- |
| 127248280 | Mir302c | microRNA 302c | 0/- |
| 127248413 | Mir302a | microRNA 302a | 1/- |
| 127248541 | Mir302d | microRNA 302d | 0/- |
| 127248650 | Mir367 | microRNA 367 | 0/- |
| 127256407 | 4930422G04Rik | RIKEN cDNA 4930422G04 gene | 45/25 |
| 127336063 | Neurog2 | Neurogenin 2 | 0/0 |
| 127373228 | Alpk1 | α-Kinase 1 | 31/17 |
| 127492837 | Tifa | Traf2 binding protein | 18/0 |
| 127510182 | Ap1ar | Adaptor-related complex 1 regulatory protein | 9/0 |
| 127573459 | 5730508B09Rik | RIKEN cDNA 5730508B09 gene | 11/1 |
| 127673211 | LOC100043382 | Hypothetical protein LOC100043382 | 27/1 |
Base pair from chromosome 3 centromere.
We took a systematic approach for sequencing of the genes and determining their expression profile in the colon. Sequences for four bacterial artificial chromosome (BAC) clones of 129 origin covering the cluster of known genes in the susceptibility interval were determined using Illumina Solexa sequencing. The sequence of each BAC has been annotated and submitted to GenBank (accession nos. JQ664734, JQ664735, JQ664736, and JQ664737 for 416G21, 225D04, 164I03, and 32B05, respectively). The BAC assembly, covering 530 Kb, revealed >5,000 SNPs compared with the published B6 sequence, indicating that the 129 strain was quite polymorphic for this region compared with the B6 strain (unpublished data). More precisely, we were able to identify nonsynonymous polymorphisms in several of the candidate genes, as well as two SNPs in the microRNA sequences (Table 2). The genes Alpk1 and 493Rik were of particular interest as they showed 17 and 25 coding differences between B6 and 129 backgrounds, respectively (Table 2 and Table S1). We confirmed the exonic SNPs by sequencing PCR products from 129 and B6 cDNA, or genomic DNA in case of the microRNAs. We also found that these nonsynonymous SNPs were identical between the 129 and C3H/HeJ strains (Table S1), the latter being the susceptible background in the IL-10−/− colitis model (Beckwith et al., 2005).
To complement the sequencing approach, extensive expression analysis was performed for all the known genes in the susceptibility interval. Levels of expression were determined by quantitative (Q) PCR on mRNA extracted from colon tissue homogenates, obtained from both uninfected and H. hepaticus–infected mice of resistant (R17) and susceptible (R9) recombinant lines (Fig. 5). We found significantly higher expression of Alpk1 and lower expression of Ap1ar and 573Rik in colon homogenates of uninfected and infected susceptible mice compared with resistant mice (Fig. 5). Of note, we could not detect Neurogenin2 or LOC100043382 mRNAs in the colon or cecum (unpublished data). Finally, no difference in expression of the microRNAs Mir302a-d and Mir367 was found in colon homogenates between resistant (R17) and susceptible (R9) mice, with or without H. hepaticus infection (unpublished data).
Figure 5.
Colon mRNA expression levels of the candidate genes in susceptible (R9) and protected (R17) mice. RNA samples were prepared from colons of uninfected or H. hepaticus–infected susceptible (R9, open circles) or protected (R17, filled diamonds) mice. A representative experiment out of at least two independently performed is shown. Level of expression of each candidate gene was assessed by Q-PCR and calculated relative to expression of Hprt. Horizontal lines represent medians. **, P < 0.01, Mann-Whitney nonparametric test.
Functional effects of the Hiccs locus are contained within the hematopoietic compartment
To gain insight into the mechanism of action of Hiccs and to identify the cells involved, reciprocal BM chimeras were generated using 129.RAG and 129.C3B.RAG mice (Fig. 6 A) or using protected (R17) and susceptible (R9) recombinant lines (Fig. 6 B). After reconstitution of the innate immune system by donor BM cells, chimeric mice were infected with H. hepaticus for 8–10 wk. Both approaches clearly illustrated the transfer of phenotype by the hematopoietic compartment. Indeed, susceptible recipients receiving BM from protected donors and infected with H. hepaticus exhibited significantly decreased colonic inflammation and splenomegaly, equivalent to the protected control groups (Fig. 6, A and B). Conversely, R17 or C3B recipients became susceptible to H. hepaticus–induced colitis when reconstituted with BM from R9 or 129.RAG donors, respectively (Fig. 6, A and B). As in previous experiments, inflammation was abrogated in the proximal and distal colon, whereas a reduced inflammatory infiltrate was observed in the mid colon (unpublished data). Severe inflammation of the cecum was observed in all infected chimeras, confirming that the Hiccs locus was not contributing to this phenotype (unpublished data).
Figure 6.
The hematopoietic compartment genotype determines protection and susceptibility to H. hepaticus–induced colitis. (A and B) Reciprocal BM chimeras: spleen mass and histological evaluation of colon inflammation after H. hepaticus infection for 8 wk, comparing control and chimeric groups. BM donors and recipients are indicated on the X axis, as donor→recipient: (A) 129 = susceptible 129.RAG mice, C3B = protected 129.C3B.RAG mice; (B) R9 = susceptible R9 mice, R17 = protected R17 mice. Horizontal lines represent medians. **, P < 0.01, Mann-Whitney nonparametric test. (C) Expression of candidate genes within the hematopoietic compartment. Pooled colon LPL from H. hepaticus–infected protected or susceptible mice were FACS sorted into CD45+ and CD45− populations. The expression level of the candidate genes was analyzed using Q-PCR, normalized to Hprt mRNA, and is indicated relative to CD45− protected samples. Mean ± SD of triplicates from a representative experiment out of two independently performed is shown. (D) BMDMs from susceptible R9 and protected R17 mice were generated and stimulated for 20 h with 1 µg/ml LPS or live H. hepaticus (20 MOI). Candidate gene expression levels were analyzed as described and are shown relative to R9 unstimulated cells. Graphs represent mean ± SEM of two pooled independent experiments (n = 4). *, P < 0.05, Mann-Whitney nonparametric test.
As we identified cells of the BM compartment as the site of action of Hiccs, we investigated the expression of the candidate genes by leukocytes in the intestine. Thus, we sorted lamina propria cells isolated from the colon of H. hepaticus–infected recombinant mice using the pan-leukocyte marker CD45, isolated RNA, and assessed expression of each candidate gene by Q-PCR. Increased expression of Alpk1 and 573Rik mRNA was observed in CD45+ intestinal leukocytes compared with CD45− nonhematopoietic cells (Fig. 6 C). Moreover, within the CD45+ leukocyte population there was increased expression of Alpk1 and 573Rik in cells isolated from susceptible (R9) compared with protected (R17) mice (Fig. 6 C).
The expression profile for the candidate genes was also evaluated in BM-derived macrophages (BMDMs) from both susceptible (R9) and protected (R17) recombinant lines. Alpk1, but not 573Rik, was expressed at higher levels in unstimulated BMDM derived from susceptible versus protected mice (Fig. 6 D). Furthermore, we observed increased expression of Alpk1 and 573Rik in LPS or H. hepaticus–stimulated BMDM compared with unstimulated regardless of their origin (R17 or R9 lines). Tifa and the other candidate genes showed no significant change of their mRNA expression in BMDM, with or without stimulation (Fig. 6 D and not depicted). Collectively these results indicate that susceptibility is determined by genes expressed in hematopoietic cells.
The Hiccs locus acts on early inflammation and cytokine expression
To gain further insight into how Hiccs controls the early inflammatory response, we performed a kinetic analysis of the innate response after H. hepaticus infection in protected and susceptible mice. As early as 3 to 8 d after H. hepaticus infection of susceptible 129.RAG mice, we observed a small increase in number of colonic LPL, as well as signs of systemic inflammation, identified by an increased spleen weight (Fig. 7 A). By day 14 after infection, clear differences in spleen weight and LPL numbers were apparent between susceptible 129.RAG and protected R17 mice (Fig. 7 A).
Figure 7.
The Hiccs locus controls the early inflammatory response through ILCs. (A–D) Time course of early H. hepaticus infection in susceptible 129.RAG (gray circles) and protected R17 (open diamonds) mice. (A) Spleen weight and total colonic LPL count. (B) Frequencies and absolute numbers of granulocytes (CD11bhi Gr1hi), DC (CD11chi CD11b+/−), and monocytes/macrophages (CD11bhi Gr1−/int CD11c−) in colon LPL. For ILC (lin neg, Thy1hi, Sca1hi), absolute numbers are shown. For each time point and genotype, the mean and SEM of five to seven mice pooled from two independent experiments are shown. (C) Representative FACS plot of 129.RAG and R17 colonic LPL, showing granulocyte gate (CD11bhi Gr1hi) and frequency. (D) TNF, IL-1β, IFN-γ, IL-17A, and IL-22 were detected by FlowCytomix in the supernatants of overnight cultured LPL. Each dot represents the mean and SEM of two pools with two mice each. IL-23p19 mRNA expression was analyzed by Q-PCR of RNA isolated from fresh LPL. Each dot represents the mean and SEM of two to four mice. One representative experiment out of two with similar results is shown. (E and F) 129.RAG and R17 mice were treated with isotype control or anti-Thy1 antibody to deplete ILC at days 6 and 0 before H. hepaticus infection. Mice were analyzed at day 6 after infection. One of two independent experiments with similar results is shown. (E) Total LPL number, granulocyte frequency, and absolute number. Horizontal lines represent means. (F) TNF, IL-1β, and IFN-γ were detected by FlowCytomix in the supernatants of overnight cultured LPL. Mean and SEM are shown for three mice per condition. **, P < 0.01; *, P < 0.05, Mann-Whitney nonparametric test for A, B, and E, unpaired Student’s t test for F.
To monitor the composition of the colon lamina propria during the course of early inflammation, we performed FACS staining for different leukocyte populations: DCs, granulocytes, monocytes/macrophages, and ILC (Fig. 7 B). As early as 3 d after infection, a marked increase in both the frequency and absolute numbers of recruited granulocytes was found in LPL of susceptible 129.RAG mice compared with protected R17 mice (Fig. 7, B and 7C). In contrast, increased infiltration by DC and monocytes/macrophages was detected in susceptible mice only at later time points (Fig. 7 B).
These acute inflammatory changes were accompanied by an early peak of proinflammatory cytokines. This response was apparent by day 3 after infection, as LPL from 129.RAG mice showed increased production of TNF, IL-1β, and IFN-γ compared with LPL from R17 mice (Fig. 7 D). Somewhat unexpectedly, IL-17 levels were higher in R17 mice throughout the early inflammatory response. The reduction in IL-17A production observed in susceptible 129.RAG mice in the early inflammatory response was reversed at later time points (8 wk after infection) where higher levels of these cytokines correlated with disease susceptibility (Fig. 4). Lower levels of proinflammatory cytokines in R17 mice did not seem to reflect an overall hyporesponsiveness of the R17 line toward H. hepaticus, as BMDM of both protected (R17) and susceptible (R9) lines reacted similarly to LPS or H. hepaticus stimulation in terms of cytokine production and IκB phosphorylation (unpublished data).
The IL-23 axis is highly relevant for IBD development and has been shown to be a key pathway in mouse models of colitis (Hue et al., 2006; Kullberg et al., 2006). Early during H. hepaticus infection, we found that levels of IL-23p19 mRNA gradually increased and were also more elevated in LPL from 129.RAG compared with R17 mice (Fig. 7 D).
A novel population of ILCs that produce IFN-γ and IL-17 in response to IL-23 was recently identified to be crucially involved in the H. hepaticus innate colitis model (Buonocore et al., 2010). To examine whether ILC played a major role in the early inflammatory response dependent on Hiccs, 129.RAG and R17 mice were treated with ILC-depleting anti-Thy1 antibody and infected with H. hepaticus for 6 d (Buonocore et al., 2010). The depletion of ILC resulted in an abrogation of early granulocyte recruitment in the susceptible 129.RAG strain (Fig. 7 E). ILCs were found at a similar level in LPL of infected 129.RAG and R17 mice and were depleted equally well by the anti-Thy1 antibody (Fig. 7 B and not depicted). Furthermore, the production of TNF, IL-1β, and IFN-γ was not elevated in LPL from anti-Thy1–treated 129.RAG mice above levels observed in LPL from protected R17 mice (Fig. 7 F). Interestingly, MIP-1α, a chemokine involved in the recruitment of myeloid cells, was also increased in LPL from 129.RAG mice compared with R17 mice early after H. hepaticus infection and was abrogated by depletion of ILC (Fig. 7 F). Together, these results suggested a role for the Hiccs locus in early inflammatory events, such as production of inflammatory cytokines and granulocyte recruitment, that are driven by ILC.
The congenic interval on chromosome 3 determines susceptibility to CAC
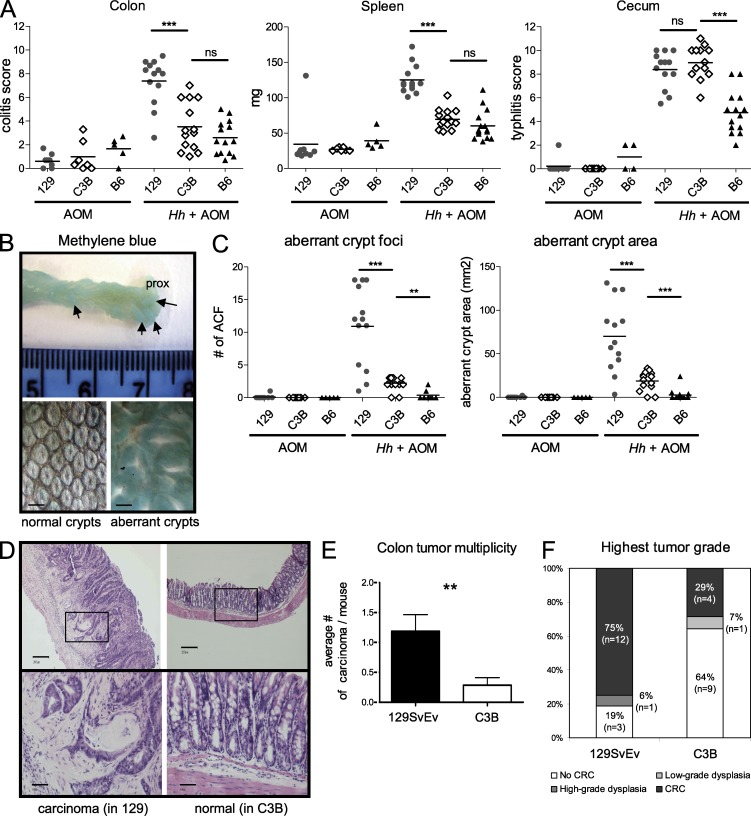
To study if the Hiccs locus was also playing a role in the development of CAC, we tested the chromosome 3 congenic and recombinant lines in a H. hepaticus–driven innate tumor model. The development of epithelial dysplasia and invasive cancer in the colon of H. hepaticus–infected 129.RAG mice has been described to be evident 2 mo after infection (Erdman et al., 2003a,b). However we experienced a later development and lower frequency of neoplasia (unpublished data); therefore, we used a new tumor-promoting protocol that combined H. hepaticus infection and administration of the carcinogen azoxymethane (AOM; Nagamine et al., 2008). Even after 5 mo of H. hepaticus infection, the protected C3B group (pooled C3B and R17 mice) showed reduced susceptibility to colitis and systemic inflammation but not cecal inflammation, compared with the susceptible 129 group (pooled 129.RAG and R9 mice; Fig. 8 A). To assess colonic inflammation and tumor formation after H. hepaticus plus AOM treatment, dissected colons were stained with methylene blue. In mice from the susceptible 129 group, an increased thickening of the mid colon could be observed, as well as protruding lesions in the proximal part, indicative of aberrant crypt foci (Fig. 8 B). These aberrant crypt lesions were significantly decreased in mice from the C3B group and were absent in mice treated with AOM alone (Fig. 8 C), indicating the requirement for inflammation and H. hepaticus infection for the development of the lesions.
Figure 8.
The B6 congenic interval confers a lower frequency of colorectal cancer. 129 (pooled 129.RAG and R9 mice), C3B (pooled 129.C3B.RAG and R17 mice), or B6 (B6.RAG) groups were infected with H. hepaticus and injected 6 wk later with AOM (one time per week) for 5 wk. Colons were analyzed for tumor formation 5 mo after H. hepaticus infection. (A) Evaluation of colitis, typhlitis, and spleen weight in mice treated with AOM alone or with H. hepaticus + AOM for 5 mo. Data shown are pooled results from two independent experiments. Horizontal lines represent medians. ***, P < 0.001, Mann-Whitney non-parametric test. (B) Changes in crypt structure were analyzed by methylene blue staining. Top picture: characteristic formation of inflammatory lesions in the colon of an Hh+AOM-treated 129.RAG mouse. Microscopic examination revealed the formation of aberrant crypts (bottom right) compared with the structure of normal crypts in an Hh+AOM treated C3B mouse (bottom left). Bars, 0.4 mm. (C) Methylene blue–stained colons were assessed for aberrant crypt foci. Number of foci and area are shown from two pooled independent experiments. Horizontal lines represent medians. ***, P < 0.001, Mann-Whitney nonparametric test. (D–F) Histological evaluation of complete sequential sections of colon from 129 and C3B mice for dysplasia and carcinoma (pooled results from three independent experiments). (D) Photomicrograph of an invasive carcinoma with dysplasia in a 129.RAG mouse (left) compared with the normal histological appearance of a C3B mouse (right). Top bars, 0.2 mm; bottom bars, 0.05 mm. (E) Tumor multiplicity: number of invasive adenocarcinomas per mouse in each group (n = 14 and 16). Graphs represent mean ± SEM. **, P = 0.0086, Mann-Whitney nonparametric test. (F) Tumor incidence: percentage of mice with the indicated tumor grade (highest tumor grade found) within each group.
Further histological assessment of sequentially cut and hematoxylin/eosin-stained sections revealed the presence of H. hepaticus–induced adenocarcinomas that seem to follow the inflammation-dysplasia-carcinoma sequence as described for human CAC (Ullman and Itzkowitz, 2011). Carcinoma location correlated with the sites of highest inflammation, as illustrated in Fig. 8 D, which shows hyperplastic and heavily inflamed colonic mucosa with a focus of moderately differentiated adenocarcinoma (top left). This comprises several architecturally and cytologically abnormal crypts, which have invaded beyond the muscularis mucosa into the submucosa and have elicited a mild desmoplastic response (Fig. 8 D, bottom left). Carcinomas were found penetrating through the muscularis mucosa but not through the muscularis propria and would be staged in the TNM scoring scheme as pT1 (Sobin and Wittekind, 1997). Tumors were present throughout the colon with predominance in the rectum and correlated with foci of inflammation. Immunohistochemical staining of areas of CAC showed an aberrant expression of proliferation markers, such as Ki-67 and Cyclin D1, in the upper part of the crypts as well as an increased expression of COX2 (unpublished data).
Invasive colon carcinomas were found in 75% (12/16) of mice from the susceptible 129 group (pooled 129.RAG and R9 mice) treated with H. hepaticus plus AOM, with a mean of 1.2 carcinoma (range 0–4) per mouse (Fig. 8, E and F). Invading foci were small, with a diameter between 0.1 and 3 mm (mean size 0.81 ± 0.18 mm SEM). In contrast, in the protected C3B group (pooled 129.C3B.RAG and R17 mice) the incidence rate of colon tumors was reduced to 29% (4/14) and the mean tumor number was decreased to 0.3 carcinoma/mouse (Fig. 8, E and F). However, the mean tumor size (0.57 ± 0.21 mm SEM) in the protected C3B group was not significantly different from the 129 group and the severity of the residual tumors was similar, still proceeding to invasive adenocarcinoma (Fig. 8 F). Within the protected C3B group, the tumor incidence of the R17 recombinant mice was 14% (1/7) and therefore comparable or even lower than in the 129.C3B.RAG mice carrying the longer congenic interval at 42% (3/7). In summary, the B6 chromosome 3 congenic interval defining the Hiccs locus leads to a lower incidence rate and frequency of carcinoma after H. hepaticus infection and AOM administration.
DISCUSSION
Tumorigenesis induced by chronic inflammation has been associated with several human inflammatory diseases, including IBD, but the genetic factors that drive CAC are still largely unknown (Mantovani et al., 2008). Herein, we identify a 1.71-Mb interval on mouse chromosome 3 as a major susceptibility locus for H. hepaticus–induced innate colitis and associated colon cancer on the 129S6.RAG−/− genetic background, designated Hiccs. This interval encompasses eight known genes and five microRNAs, of which several contain nonsynonymous SNPs and show expression variations in innate cells making them promising candidate genes. Hiccs acts in innate hematopoietic cells and controls ILC-driven initiation of inflammation after H. hepaticus infection. Importantly, the congenic interval is capable of reducing the frequency of colitis-associated invasive adenocarcinoma from 75 to 30%.
Host genetic factors that predispose to a dysregulated interplay between gut microbiota and the innate immune system are believed to play a key role in development of chronic intestinal inflammation and associated colon cancer (Saleh and Trinchieri, 2011). Although a defined bacterial etiology for CAC in humans has not yet been identified, recent GWASs have linked genes involved in bacterial sensing and handling, such as NOD2 or autophagy genes, with susceptibility to IBD (Khor et al., 2011). Similarly, in many mouse models of IBD, chronic inflammation and cancer development are dependent on the genetic background. A decrease in CAC after H. hepaticus infection was previously reported for the B6 background (Erdman et al., 2003a) and similar observations were made for innate immune colitis in the TRUC model between the BALB/c.TRUC and B6.TRUC strains (Ermann et al., 2011). However, to our knowledge, no systematic genome-wide analysis has been conducted in bacteria or chemically induced CAC models. Using a novel tumor induction protocol that combined AOM administration and H. hepaticus infection, we now identify Hiccs, a colitis susceptibility locus on chromosome 3 which contributes to colon tumorigenesis.
Hiccs is part of a larger region previously identified in two other models of colitis, using IL-10−/− and Gnai2−/− mice (Beckwith et al., 2005; Borm et al., 2005). The original Cdsc1 locus described in the IL-10−/− model has been further subdivided into three distinct intervals covering a total of 26 Mb (Bleich et al., 2010). Recently, a study in TRUC mice also highlighted a role for Cdcs1 in an innate colitis model (Ermann et al., 2011). The complex genetic structure of the Cdcs1 locus is not seen in our H. hepaticus innate model, where we found that Hiccs, the reduced 1.7-Mb congenic interval, is sufficient to confer protection. However, Hiccs overlaps with the most telomeric sublocus Cdcs1.3, which covers 2.4 Mb (Bleich et al., 2010), suggesting that this region may represent a strong and common colitogenic determinant in several models of colitis and CAC. Similar to results in H. hepaticus–induced innate colitis, we found that H. hepaticus–induced colitis in lymphocyte-replete mice was less severe in B6.IL10−/− mice than 129.IL10−/− mice. Further experiments are required to determine whether the B6 Hiccs locus will be sufficient to confer resistance in this lymphocyte-replete setting. Interestingly, there is a clear synteny between the 1.7-Mb Hiccs interval on mouse chromosome 3 and a region of human chromosome 4 localized in 4q25, with clustering of the same genes in reverse orientation (as indicated by Ensembl database). Although an association of the 4q25 region with IBD has not been identified by recent GWAS (The Wellcome Trust Case Control Consortium, 2007; Anderson et al., 2011; Thompson and Lees, 2011), a disease-relevant role is still possible as GWAS based on SNPs analysis cannot detect all loci, particularly those involving rare variants (Manolio et al., 2009). Further studies are required to assess the potential contribution of the human syntenic interval in IBD and CAC.
Systematic analyses of sequence and expression of the eight known genes and five microRNAs localized in the 1.7-Mb interval identified several candidate genes. So far, the best candidate is Alpk1, as this gene shows 17 nonsynonymous nucleotide variations between 129 and B6 strains, 2 of which are located in the kinase domain, as well as a significantly higher expression level in susceptible mice. Alpk1 encodes an α kinase present in polarized epithelial cell lines that is essential for transport of lipid rafts carrying vesicles from the Golgi apparatus to the plasma membrane (Heine et al., 2005). Alpk1 was also recently identified as a gout-susceptibility factor and shown to play a role in the activation of proinflammatory cytokines in monocytes upon challenge with urate crystals (Wang et al., 2011). However, other genetic elements in the susceptibility interval also need to be considered, for example, 4930422G04Rik, which shows numerous amino acid differences but is not differentially expressed, as well as Ap1ar, which is differentially regulated. Tifa, through its function in TLR downstream signaling by TRAF6 binding would be an interesting candidate (Ea et al., 2004) but does not show strain-specific nonsynonymous polymorphisms or differential expression. As further reduction of the susceptibility interval using congenic recombinant lines is not feasible, a knockin or BAC transgenic approach will be required to conclusively identify the responsible gene. It is notable that upon H. hepaticus infection, B6.RAG mice showed a reduced inflammation of the cecum compared with 129.RAG mice. However, this phenotype was not found in infected 129.C3B.RAG mice or any of the subcongenic recombinants, which displayed a level of typhlitis equivalent to 129.RAG mice. This indicates that genetic loci outside the chromosome 3 interval control typhlitis susceptibility and act independently of Hiccs. Further investigation of these loci requires a genome-wide analysis focused on cecal inflammation.
Nevertheless, we were able to gain insight into the cellular mechanism underlying the susceptibility imparted by the Hiccs locus. In particular, BM chimera experiments demonstrated that colitis protection or susceptibility is transmissible with the genotype of the donor BM. This result parallels recent findings in the TRUC model of innate colitis, where the genetic background was also shown to control colitis severity through hematopoietic cells (Ermann et al., 2011). Innate inflammation in the absence of appropriate immune regulation seems to be a key driver of inflammation-induced cancer. Mediators of this process are proinflammatory cytokines and chemokines that shape a pro-neoplastic microenvironment (Mantovani et al., 2008; Balkwill and Mantovani, 2010; Saleh and Trinchieri, 2011). Recent studies exemplifying procarcinogenic effects of cytokines in the development of CAC showed that antiapoptotic effects of IL-6 on intestinal epithelial cells were mediated through STAT3 signaling (Bollrath et al., 2009; Grivennikov et al., 2009). Interestingly we see a clear effect of Hiccs on early innate immune responses, which could pave the way for later cancer development through protumorigenic inflammatory reactions. As early as 3 d after H. hepaticus infection, we observed a marked increase in neutrophil recruitment and in the production of the proinflammatory cytokines TNF, IL-1β, and IFN-γ in LPL from susceptible 129.RAG mice compared with protected R17 mice. Depletion of TNF was previously shown to abrogate colitis development in the H. hepaticus innate model (Maloy et al., 2003), and more recent studies demonstrated that the presence of Gr1+ neutrophils and elevated TNF and nitric oxide in the gut were prerequisites for tumor development (Erdman et al., 2009). TNF is a well known protumorigenic factor in the development of colon cancer in humans, and in mice and anti-TNF therapy has been successfully applied in IBD patients (Popivanova et al., 2008; de Silva et al., 2010). However, in the TRUC model, where dysregulated TNF expression by DC was suggested to be driving colitis and tumor development, late intervention with anti-TNF antibody could not cure colitis (Garrett et al., 2009). Together, these findings suggest an early role for TNF as a mediator that reshapes the microenvironment toward dysplasia and cancer.
Depletion of ILC by anti-Thy1 antibody treatment at the time of H. hepaticus infection blocked early neutrophil recruitment and expression of TNF, IL-1β, and IFN-γ by LPL from susceptible mice, and therefore, ILCs seem to be crucial in the chain of early inflammatory events. Our previous work has shown that both IL-17 and IFN-γ contribute to immune pathology in H. hepaticus–induced innate colitis (Buonocore et al., 2010). Strikingly, analysis of these ILC-derived cytokines very early in the response showed differential kinetics. Thus, IFN-γ was increased at day 3 in susceptible mice and remained high at 8 wk after infection. In contrast, IL-17 levels were low in susceptible mice in the first 14 d after infection but had risen by the chronic phase of disease. The situation was reversed in resistant mice that exhibited low IFN-γ throughout the time course but high early levels of IL-17 that had fallen by day 14. These results raise the interesting possibility that an acute IL-17 response is host protective and that later chronic production may be deleterious. Although ILCs clearly contribute to the amplification of the genetically controlled inflammatory response in susceptible mice, it remains to be determined whether the susceptibility locus acts directly on ILC or indirectly through other innate cells. Our earlier studies identified IL-23 as a pivotal player in chronic innate colitis (Hue et al., 2006). However, resistant and susceptible strains displayed similar early H. hepaticus–induced expression of IL-23 up to day 8, after which IL-23 levels continued to rise in susceptible mice. These results suggest IL-23 may drive the chronicity of the response and that differential production of other mediators, such as TNF or IL-1β, contribute to the Hiccs controlled early innate response.
Recently, several studies identified a role for the microbial flora and flora-derived virulence factors in the development of intestinal cancer in different mouse models (Garrett et al., 2009; Uronis et al., 2009; Wu et al., 2009). Interestingly, H. hepaticus–driven colitis has been shown to be strictly dependent on MyD88 expression by innate hematopoietic cells (Asquith et al., 2010). In contrast, colitis and CAC in the TRUC model seem to be MyD88 independent (Garrett et al., 2009). This illustrates the complexity of the interactions between the innate immune system and the gut microbiota in models where the same susceptibility locus may be involved. Preliminary studies failed to show differences in the frequency of major gut microbial phyla/families in 129.RAG versus congenic R17 mice (unpublished data), suggesting that it is the genetically controlled response to H. hepaticus infection itself that is the trigger for colitis and associated tumorigenesis in this model. In contrast, CAC in TRUC mice is associated with altered bacterial community structure and the presence of distinct colitogenic agents (Garrett et al., 2010). Why H. hepaticus, which is a commensal in many animal facilities, turns into a carcinogen in a particular host genetic background is not clear. An H. hepaticus virulence factor possibly involved in CAC development could be the toxin Cdt, which was shown to induce dysplasia in the liver and is thought to generate double-strand DNA breaks, thereby triggering the DNA damage response via Myc and ATM (Ge et al., 2007; Guerra et al., 2010). Therefore, it is tempting to speculate that besides the deleterious effects of chronic inflammation, H. hepaticus could actively be involved in transformation events leading to invasive adenocarcinoma.
Collectively, the Hiccs locus promotes through its actions in innate immune cells a procarcinogenic environment, where the accumulation of inflammatory cytokines and neutrophils could lead to increased DNA damage and eventually to cancer. Further characterization of the causal genetic element and the relevant molecular pathways that increase susceptibility to colitis and CAC in our mouse model could lead to the discovery of novel interplays between the innate immune system, the colonic epithelium, and the gut microbiota. This, in turn, could offer insights into human IBD and more generally into inflammation-driven cancer, as well as providing novel therapeutic targets for the treatment of these diseases.
MATERIALS AND METHODS
Mice and genotyping.
All inbred mice used in this study were maintained in an accredited specific pathogen-free facility and experiments conducted in accordance with the UK Scientific Procedures Act (1986) under a Project License (PPL) authorized by the UK Home Office Animal Procedures Committee and approved by the Sir William Dunn School Ethical Review Committee. C57BL/6.IL10−/− and 129S6.IL10−/− mice were bred in our facility. Congenic 129S6.B6-(D3Mit103-D3Mit45).Rag2−/− (abbreviated 129.C3B.RAG) mice were generated by iterative backcrossing to the 129S6.Rag2−/− (129.RAG) background, using a marker-assisted speed congenic method (Markel et al., 1997). For the iterative backcrossing procedure, a total of 63 polymorphic microsatellite markers, evenly spaced on each chromosome, were used to screen the offspring at each generation. The final 129.C3B.RAG congenic line was established after the eighth generation of backcross. Primer sequences for the microsatellite markers were obtained from the Ensembl or MGI database. Mice intercrossed at seventh and eighth generation of backcross were infected with H. hepaticus and analyzed.
Subcongenic recombinant mice were generated by backcrossing the 129.C3B.RAG line with 129.RAG mice and by extensive genotyping using microsatellite markers and SNPs. Homozygote recombinant lines were established using the initial recombinant founder. To characterize precisely the location of the recombination point, SNPs were chosen in the various intervals based on their position on the chromosome 3 sequence (taken from Ensembl database). The identified SNPs were genotyped in recombinant lines by sequencing of PCR product (Source Bioscience Geneservice) using the following primer pairs: rs31622216, 5′-ATAGCCACCTCTCTAGAGTC-3′ and 5′-GGCAATCCTAAAAGCGTGAC-3′; rs31106293, 5′-GTTAATTGCAGAGCCACATAGC-3′ and 5′-TCTCAGTCCTTATCTCTCAGTGC-3′; rs31282270, 5′-TGGATGTGAGGAGATACTGTAGG-3′ and 5′-ACCTATGGTTACACTCATTCTGC-3′; rs33862828, 5′-TGCATATATTCAGCCAGAGACG-3′ and 5′-CAGATGCACAGCAGGAAGC-3′; and rs30910985, 5′-TAGAGTGGCTGCACCGTAGG-3′ and 5′-TGTGTGGAACCAGATGAAGC-3′.
Induction of H. hepaticus colitis.
H. hepaticus NCI-Frederick isolate 1A (strain 51449) was grown on blood agar plates containing trimethoprim, vancomycin, and polymyxin B (Oxoid) under microaerophilic conditions as described previously (Maloy et al., 2003). Mice were fed three times on alternate days with H. hepaticus (0.5–1.0 × 108 CFU) by oral gavage. For the early time course experiments, mice were infected with a single dose of H. hepaticus. To minimize cage effects, susceptible mice (R9 or 129.RAG) were systematically co-housed with protected mice (R17). Recombinant congenic lines were also initially analyzed as heterozygotes with littermate controls. For quantitation of H. hepaticus colonization, cecal contents were collected upon sacrifice and DNA extracted using a DNA Stool kit (QIAGEN). Q-PCR was performed with primers specific for H. hepaticus cdtB gene as previously described (Ge et al., 2001), using the Chromo4 detection system.
Histological assessment of inflammation.
Colonic and cecal inflammation in H. hepaticus–infected mice was assessed at 6–8 wk (if not otherwise stated). Samples of proximal, mid, and distal colon, as well as cecum, were fixed in 10% formalin, embedded in paraffin, and cut into 5-µm sections for H&E staining. Scoring of inflammation was performed using previously published criteria (Izcue et al., 2008). In brief, each sample was graded semiquantitatively from 0 to 3 for the four following criteria: degree of epithelial hyperplasia and goblet cell depletion, leukocyte infiltration in the lamina propria, area of tissue affected, and presence of markers of severe inflammation (such as crypt abscesses, submucosal inflammation, and ulcers). Scores for each criterion were added to give a final quantification for intestinal inflammation ranging from 0 to 12. For colitis scores, a mean value of the three sections (proximal, mid, and distal colon) was calculated. Photomicrographs of H&E-stained colon sections were taken with a Coolscope Slide Scanner (Nikon).
Cell preparation, isolation, and FACS staining.
LPLs were isolated as previously described (Maloy et al., 2003). In brief, colons were longitudinally opened, cut into 1-cm pieces, and incubated (three times) in RPMI 1640 with 10% FCS and 5 mM EDTA at 37°C to remove epithelial cells. Tissue was then digested with 100 U/ml of type VIII collagenase in two cycles of 1 h at 37°C. The isolated cells were layered on a 30/40/75% Percoll gradient (GE Healthcare), which was centrifuged for 20 min at 600 g, and the 40/75% interface, containing mostly leukocytes, was recovered. For FACS sorting, LPL preparations were stained with an antibody against CD45.2 (BD). CD45-negative and -positive cell populations were sorted on a MoFlow cell sorter (Dako). To identify different cell populations during early colitis, we stained LPL by FACS using antibodies against F4/80 (BM8), CD11b (M1/70), Ly6G (Gr1, RB6-8C5), CD11c (N418), B220 (RA3-6B2), Thy1 (53–2.1), and Sca1 (D7; all from eBioscience).
BAC contig and sequence.
BACs of 129S7 origin, from the SANGER library bMQ, were identified based on mapping of their extremities by Ensembl database. Four BAC clones, bMQ416G21, bMQ225D4, bMQ164I3, and bMQ32B5 (Source Bioscience Geneservice), were purified using a long-construct Maxiprep kit (QIAGEN). Gene content was confirmed by PCR amplification of the corresponding candidate gene. The BAC preparation was checked for integrity by NotI digestion and migration on agarose gel. The four BACs were pooled and send to the Gene Pool for Illumina Solexa high-throughput sequencing. Sequences of the four BACs were established by the Gene Pool using the published C57BL/6 sequence as framework and were integrated for analysis into Gbrowse by the Computational Biology Research Group. The mRNA sequence of each candidate gene was confirmed in C57BL/6, 129S6, and C3H/HeJ strains by de novo sequencing of PCR products amplified from the corresponding cDNA (or genomic DNA in the case of the candidate microRNAs).
Cytokine detection.
For detection of cytokines, 105 isolated LPL cells were cultured overnight in 100 µl RPMI 1640 with 10% FCS at 37°C. Cytokines in the supernatant were detected using the FlowCytomix Multiplex system for IL-17, IL-22, IFN-γ, IL-1β, IL-6, TNF, and MIP-1α (eBioscience). Samples were analyzed on a FACSCalibur (BD) according to the manufacturer’s instructions.
Quantification of mRNA levels by real-time PCR.
Isolated tissues (from colon or cecum) or cell preparations were snap-frozen in liquid nitrogen. Tissue material was homogenized in RLT buffer (QIAGEN) with β-mercaptoethanol using a homogenizer (MP Biomedicals). RNA isolation was performed using the RNeasy kit (QIAGEN) including a DNase I digest step. Content and purity of RNA was controlled with a NanoDrop spectrophotometer (Thermo Fisher Scientific). cDNA synthesis was performed using the Superscript III reverse transcription kit from Invitrogen. Quantitative real-time PCR for the candidate genes was done using SYBR green chemistry (SensiMix; Bioline) or the TaqMan system. cDNA samples were analyzed in triplicate using the Chromo4 detection system (Bio-Rad Laboratories) and values were normalized to Hprt. Analysis was performed according to the Δ-Ct method or, to show relative expression, the ΔΔCt method (Pfaffl, 2001).
The following primer pairs were used for the detection of candidate genes with SYBR green: QuantiTect Primer Assay for Hprt (QIAGEN): Larp7, 5′-GAAGATTGTGAGCGGAGAGC-3′ and 5′-CTCGAGGTTCCAACTGTGC-3′; 4930422G04Rik, 5′-TGTTAGGAAGATCGCCAAGC-3′ and 5′-AGCAGGACTCTGTTGGTTCC-3′; Alpk1, 5′-GCTATGCAGTGAAGCTGTGG-3′ and 5′-TGTCCATGCAAGATGAGTCG-3′; Mm_Alpk1_2_SG QuantiTect Primer Assay (QIAGEN): Tifa, 5′-GCCACTGGAAGACTCTCAGG-3′ and 5′-AACGTATACTGGCACATGTTGG-3′; Ap1ar, 5′-CGACAGCACCTCCTTAGACC-3′ and 5′-ACTCAGAGTTGCCGTTGTCG-3′; and 5730508B09Rik, 5′-GGCGAGCAGGATATTGAAGA-3 and 5′-ATCCTTGTGAAGCCCATGTT-3′. Primer pairs and probes for TaqMan were the following: Hprt, 5′-GACCGGTCCCGTCATGC-3′ and 5′-TCATAACCTGGTTCATCATCGC-3′, VIC/TAMRA probe 5′-ACCCGCAGTCCCAGCGTCGTC-3′; Neurog2, 5′-GCGTAGGATGTTCGTCAAATC-3′ and 5′-CAGCAGCATCAGTACCTCCTC-3′, FAM/TAMRA probe 5′-TTCCTCCTTCAACTCCAGAGTC-3; LOC100043382, 5′-TCTGAACATGGTTGTCCCAA-3′ and 5′-GTGAATCAGCCTGCCTTAGC-3′, FAM/TAMRA probe 5′-CCCTGCCTCCTGTCTTTCTT-3′; and IL-23p19, 5′-AGCGGGACATATGAATCTACTAAGAGA-3′ and 5′-GTCCTAGTAGGGAGGTGTGAAGTTG-3′, FAM/TAMRA probe 5′-GCCAGTTCTGCTTGCAAAGGATCCGC-3′.
BM chimeras.
For BM chimera experiments, recipient mice were lethally irradiated (2× 550 rads/5.5 Gys) before reconstitution with BM from the indicated donor line. BM was aseptically harvested from tibia and femur of the respective donor strain, and 5 × 106 cells were injected into the tail vein of the irradiated recipients. Mice were allowed to reconstitute for 8 wk before being infected with H. hepaticus. Susceptible recipient mice (129.RAG or R9) were mixed with protected recipient (129.C3B.RAG or R17) before irradiation to minimize cage effects and maintained for the duration of the experiment.
BMDM stimulation.
For BMDMs, BM cells were isolated from femur and tibia of R9 or R17 recombinant mice and were cultured in L929 fibroblast-conditioned RPMI 1640 medium (containing 15% L929 supernatant and 10% FCS) for 7 d. The resulting macrophages were stimulated with 1 µg/ml LPS (Sigma-Aldrich) or 20 MOI of live H. hepaticus.
In vivo treatment with anti-Thy1 antibody.
To deplete Thy1+ ILC, mice were treated with 1 mg anti-Thy1 antibody (YTS 154.7.7.10) or rat isotype control 6 d before and on the day of H. hepaticus infection. Depletion of ILC was controlled at day 6 after infection by FACS staining for lineage negative (CD11b, Gr1, and B220), Thy1hi, Sca1hi cells (Buonocore et al., 2010).
Inflammation-associated colon cancer.
Mice were infected with H. hepaticus, and 6 wk after infection, AOM (Sigma-Aldrich) was administered i.p. once a week at a dose of 10 mg/kg for 5 wk according to a previously published protocol (Nagamine et al., 2008). 21 wk after H. hepaticus infection, mice were sacrificed and colons were removed. Whole colons were fixed in 10% formalin, stained in 2% methylene blue, and aberrant crypt area was analyzed using a dissection microscope and sliding caliper. Colons were then either cut in several pieces (covering the whole length) or swiss-rolled, and 5-µm paraffin sections were prepared. Randomly selected colon ribbons (200–300 sections per mouse) were prepared and every 10th section was H&E stained and graded by a pathologist. Low-grade dysplasia, high-grade dysplasia, and adenocarcinoma were scored blindly using criteria for grading dysplasia in human IBD, as well as the TNM classification of colorectal tumors. Criteria for dysplasia in human IBD: low grade dysplasia, requires intact to mild alteration of gland architecture and nuclear alterations that extend to mucosal surface without appreciable loss of nuclear polarity; high grade dysplasia, requires intact to severe alterations of gland architecture with clear evidence of a loss of nuclear polarity. TNM classification of colorectal tumors (Sobin and Wittekind, 1997): (pT = primary tumor) pTX, primary tumor cannot be assessed; pT0, no evidence of primary tumor; pT1, tumor invades submucosa; pT2, tumor invades muscularis propria; pT3, tumor invades through muscularis propria into subserosa or nonperitonealized pericolic or perirectal tissues; pT4, tumor directly invades other organs (pT4a) and/or involves the visceral peritoneum (pT4b).
Statistical analysis.
Statistical analysis and graphical representations were done using Prism5 (GraphPad Software). For comparison of Q-PCR results and histological analysis, the nonparametric Mann-Whitney test was applied.
Online supplemental material.
Table S1 gives details on the nonsynonymous SNPs found in the candidate genes (and microRNAs) of the susceptibility Hiccs locus, in particular their dbSNP ID number and genotype for C57BL/6, 129S6, and C3H/HeJ strains. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20120239/DC1.
Supplementary Material
Acknowledgments
We would like to thank Carolina Arancibia and Julia Bollrath for critical reading of this manuscript, Richard Stillion for histology, Nigel Rust and Helen Ferry for cell sorting, B. Warren for advice on cancer pathology, N. Saunders for help with BAC sequencing, and S. Cobbold and H. Waldman for the Thy1 hybridoma. We wish to acknowledge the Computational Biology Research Group (Medical Sciences Division, Oxford UK) for use of their services in this project.
O. Boulard was supported by Cancer Research UK grants A7297 and A11663; S. Kirchberger by the European Commission research program Infla-Care (EC contract No. 223151) and an Austrian FWF Schroedinger Fellowship; D.J. Royston was supported by The Royal College of Pathologists and the Jean Shanks Foundation; and K.J. Maloy and F.M. Powrie were supported by the Wellcome Trust.
The authors declare no competing financial interest.
Footnotes
Abbreviations used:
- AOM
- azoxymethane
- BAC
- bacterial artificial chromosome
- BMDM
- BM-derived macrophage
- CAC
- colitis-associated cancer
- GWAS
- genome-wide association study
- IBD
- inflammatory bowel disease
- ILC
- innate lymphoid cell
- LPL
- lamina propria leukocyte
- SNP
- single nucleotide polymorphism
- TRUC
- T-bet−/− Rag2−/− ulcerative colitis
References
- Ahmadi A., Polyak S., Draganov P.V. 2009. Colorectal cancer surveillance in inflammatory bowel disease: the search continues. World J. Gastroenterol. 15:61–66 10.3748/wjg.15.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C.A., Boucher G., Lees C.W., Franke A., D’Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A., et al. 2011. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 43:246–252 10.1038/ng.764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asquith M.J., Boulard O., Powrie F., Maloy K.J. 2010. Pathogenic and protective roles of MyD88 in leukocytes and epithelial cells in mouse models of inflammatory bowel disease. Gastroenterology. 139:519–529: 529: e1–e2 10.1053/j.gastro.2010.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F., Coussens L.M. 2004. Cancer: an inflammatory link. Nature. 431:405–406 10.1038/431405a [DOI] [PubMed] [Google Scholar]
- Balkwill F., Mantovani A. 2010. Cancer and inflammation: implications for pharmacology and therapeutics. Clin. Pharmacol. Ther. 87:401–406 10.1038/clpt.2009.312 [DOI] [PubMed] [Google Scholar]
- Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. NIDDK IBD Genetics Consortium. Belgian-French IBD Consortium. Wellcome Trust Case Control Consortium 2008. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat. Genet. 40:955–962 10.1038/ng.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J.C., Lee J.C., Lees C.W., Prescott N.J., Anderson C.A., Phillips A., Wesley E., Parnell K., Zhang H., Drummond H., et al. UK IBD Genetics Consortium. Wellcome Trust Case Control Consortium 2 2009. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat. Genet. 41:1330–1334 10.1038/ng.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith J., Cong Y., Sundberg J.P., Elson C.O., Leiter E.H. 2005. Cdcs1, a major colitogenic locus in mice, regulates innate and adaptive immune response to enteric bacterial antigens. Gastroenterology. 129:1473–1484 10.1053/j.gastro.2005.07.057 [DOI] [PubMed] [Google Scholar]
- Bleich A., Büchler G., Beckwith J., Petell L.M., Affourtit J.P., King B.L., Shaffer D.J., Roopenian D.C., Hedrich H.J., Sundberg J.P., Leiter E.H. 2010. Cdcs1 a major colitis susceptibility locus in mice; subcongenic analysis reveals genetic complexity. Inflamm. Bowel Dis. 16:765–775 10.1002/ibd.21146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollrath J., Phesse T.J., von Burstin V.A., Putoczki T., Bennecke M., Bateman T., Nebelsiek T., Lundgren-May T., Canli O., Schwitalla S., et al. 2009. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 15:91–102 10.1016/j.ccr.2009.01.002 [DOI] [PubMed] [Google Scholar]
- Borm M.E., He J., Kelsall B., Peña A.S., Strober W., Bouma G. 2005. A major quantitative trait locus on mouse chromosome 3 is involved in disease susceptibility in different colitis models. Gastroenterology. 128:74–85 10.1053/j.gastro.2004.10.044 [DOI] [PubMed] [Google Scholar]
- Buonocore S., Ahern P.P., Uhlig H.H., Ivanov I.I., Littman D.R., Maloy K.J., Powrie F. 2010. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 464:1371–1375 10.1038/nature08949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill R.J., Foltz C.J., Fox J.G., Dangler C.A., Powrie F., Schauer D.B. 1997. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect. Immun. 65:3126–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe S.E. 2005. Helicobacter infection, chronic inflammation, and the development of malignancy. Curr. Opin. Gastroenterol. 21:32–38 [PubMed] [Google Scholar]
- de Silva S., Devlin S., Panaccione R. 2010. Optimizing the safety of biologic therapy for IBD. Nat. Rev. Gastroenterol. Hepatol. 7:93–101 10.1038/nrgastro.2009.221 [DOI] [PubMed] [Google Scholar]
- Duerr R.H., Taylor K.D., Brant S.R., Rioux J.D., Silverberg M.S., Daly M.J., Steinhart A.H., Abraham C., Regueiro M., Griffiths A., et al. 2006. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 314:1461–1463 10.1126/science.1135245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ea C.K., Sun L., Inoue J., Chen Z.J. 2004. TIFA activates IkappaB kinase (IKK) by promoting oligomerization and ubiquitination of TRAF6. Proc. Natl. Acad. Sci. USA. 101:15318–15323 10.1073/pnas.0404132101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaden J.A., Abrams K.R., Mayberry J.F. 2001. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut. 48:526–535 10.1136/gut.48.4.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S.E., Poutahidis T., Tomczak M., Rogers A.B., Cormier K., Plank B., Horwitz B.H., Fox J.G. 2003a. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am. J. Pathol. 162:691–702 10.1016/S0002-9440(10)63863-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S.E., Rao V.P., Poutahidis T., Ihrig M.M., Ge Z., Feng Y., Tomczak M., Rogers A.B., Horwitz B.H., Fox J.G. 2003b. CD4(+)CD25(+) regulatory lymphocytes require interleukin 10 to interrupt colon carcinogenesis in mice. Cancer Res. 63:6042–6050 [PubMed] [Google Scholar]
- Erdman S.E., Rao V.P., Poutahidis T., Rogers A.B., Taylor C.L., Jackson E.A., Ge Z., Lee C.W., Schauer D.B., Wogan G.N., et al. 2009. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc. Natl. Acad. Sci. USA. 106:1027–1032 10.1073/pnas.0812347106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermann J., Garrett W.S., Kuchroo J., Rourida K., Glickman J.N., Bleich A., Glimcher L.H. 2011. Severity of innate immune-mediated colitis is controlled by the cytokine deficiency-induced colitis susceptibility-1 (Cdcs1) locus. Proc. Natl. Acad. Sci. USA. 108:7137–7141 10.1073/pnas.1104234108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagins L.A., Souza R.F., Spechler S.J. 2009. Carcinogenesis in IBD: potential targets for the prevention of colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 6:297–305 10.1038/nrgastro.2009.44 [DOI] [PubMed] [Google Scholar]
- Garrett W.S., Punit S., Gallini C.A., Michaud M., Zhang D., Sigrist K.S., Lord G.M., Glickman J.N., Glimcher L.H. 2009. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 16:208–219 10.1016/j.ccr.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett W.S., Gallini C.A., Yatsunenko T., Michaud M., DuBois A., Delaney M.L., Punit S., Karlsson M., Bry L., Glickman J.N., et al. 2010. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 8:292–300 10.1016/j.chom.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity-Park M.M., Loftus E.V., Jr, Sandborn W.J., Bryant S.C., Smyrk T.C. 2009. MHC Class II alleles in ulcerative colitis-associated colorectal cancer. Gut. 58:1226–1233 10.1136/gut.2008.166686 [DOI] [PubMed] [Google Scholar]
- Ge Z., White D.A., Whary M.T., Fox J.G. 2001. Fluorogenic PCR-based quantitative detection of a murine pathogen, Helicobacter hepaticus. J. Clin. Microbiol. 39:2598–2602 10.1128/JCM.39.7.2598-2602.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z., Rogers A.B., Feng Y., Lee A., Xu S., Taylor N.S., Fox J.G. 2007. Bacterial cytolethal distending toxin promotes the development of dysplasia in a model of microbially induced hepatocarcinogenesis. Cell. Microbiol. 9:2070–2080 10.1111/j.1462-5822.2007.00939.x [DOI] [PubMed] [Google Scholar]
- Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., Karin M. 2009. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 15:103–113 10.1016/j.ccr.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra L., Albihn A., Tronnersjö S., Yan Q., Guidi R., Stenerlöw B., Sterzenbach T., Josenhans C., Fox J.G., Schauer D.B., et al. 2010. Myc is required for activation of the ATM-dependent checkpoints in response to DNA damage. PLoS ONE. 5:e8924 10.1371/journal.pone.0008924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F.M., Briggs J., et al. 2007. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39:207–211 10.1038/ng1954 [DOI] [PubMed] [Google Scholar]
- Hatakeyama M. 2004. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer. 4:688–694 10.1038/nrc1433 [DOI] [PubMed] [Google Scholar]
- Heine M., Cramm-Behrens C.I., Ansari A., Chu H.P., Ryazanov A.G., Naim H.Y., Jacob R. 2005. Alpha-kinase 1, a new component in apical protein transport. J. Biol. Chem. 280:25637–25643 10.1074/jbc.M502265200 [DOI] [PubMed] [Google Scholar]
- Houlston R.S., Webb E., Broderick P., Pittman A.M., Di Bernardo M.C., Lubbe S., Chandler I., Vijayakrishnan J., Sullivan K., Penegar S., et al. Colorectal Cancer Association Study Consortium. CoRGI Consortium. International Colorectal Cancer Genetic Association Consortium 2008. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat. Genet. 40:1426–1435 10.1038/ng.262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hue S., Ahern P., Buonocore S., Kullberg M.C., Cua D.J., McKenzie B.S., Powrie F., Maloy K.J. 2006. Interleukin-23 drives innate and T cell–mediated intestinal inflammation. J. Exp. Med. 203:2473–2483 10.1084/jem.20061099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugot J.P., Chamaillard M., Zouali H., Lesage S., Cézard J.P., Belaiche J., Almer S., Tysk C., O’Morain C.A., Gassull M., et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 411:599–603 10.1038/35079107 [DOI] [PubMed] [Google Scholar]
- Izcue A., Hue S., Buonocore S., Arancibia-Cárcamo C.V., Ahern P.P., Iwakura Y., Maloy K.J., Powrie F. 2008. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 28:559–570 10.1016/j.immuni.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti M., Mino-Kenudson M., Mizoguchi E. 2011. Animal models of colitis-associated carcinogenesis. J. Biomed. Biotechnol. 2011:342637 10.1155/2011/342637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M. 2006. Nuclear factor-kappaB in cancer development and progression. Nature. 441:431–436 10.1038/nature04870 [DOI] [PubMed] [Google Scholar]
- Khor B., Gardet A., Xavier R.J. 2011. Genetics and pathogenesis of inflammatory bowel disease. Nature. 474:307–317 10.1038/nature10209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg M.C., Ward J.M., Gorelick P.L., Caspar P., Hieny S., Cheever A., Jankovic D., Sher A. 1998. Helicobacter hepaticus triggers colitis in specific-pathogen-free interleukin-10 (IL-10)-deficient mice through an IL-12- and gamma interferon-dependent mechanism. Infect. Immun. 66:5157–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg M.C., Jankovic D., Feng C.G., Hue S., Gorelick P.L., McKenzie B.S., Cua D.J., Powrie F., Cheever A.W., Maloy K.J., Sher A. 2006. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J. Exp. Med. 203:2485–2494 10.1084/jem.20061082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy K.J., Salaun L., Cahill R., Dougan G., Saunders N.J., Powrie F. 2003. CD4+CD25+ T(R) cells suppress innate immune pathology through cytokine-dependent mechanisms. J. Exp. Med. 197:111–119 10.1084/jem.20021345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. 2009. Finding the missing heritability of complex diseases. Nature. 461:747–753 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Allavena P., Sica A., Balkwill F. 2008. Cancer-related inflammation. Nature. 454:436–444 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- Markel P., Shu P., Ebeling C., Carlson G.A., Nagle D.L., Smutko J.S., Moore K.J. 1997. Theoretical and empirical issues for marker-assisted breeding of congenic mouse strains. Nat. Genet. 17:280–284 10.1038/ng1197-280 [DOI] [PubMed] [Google Scholar]
- Nagamine C.M., Rogers A.B., Fox J.G., Schauer D.B. 2008. Helicobacter hepaticus promotes azoxymethane-initiated colon tumorigenesis in BALB/c-IL10-deficient mice. Int. J. Cancer. 122:832–838 10.1002/ijc.23175 [DOI] [PubMed] [Google Scholar]
- Ogura Y., Bonen D.K., Inohara N., Nicolae D.L., Chen F.F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R.H., et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 411:603–606 10.1038/35079114 [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popivanova B.K., Kitamura K., Wu Y., Kondo T., Kagaya T., Kaneko S., Oshima M., Fujii C., Mukaida N. 2008. Blocking TNF-alpha in mice reduces colorectal carcinogenesis associated with chronic colitis. J. Clin. Invest. 118:560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh M., Trinchieri G. 2011. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat. Rev. Immunol. 11:9–20 10.1038/nri2891 [DOI] [PubMed] [Google Scholar]
- Sobin L.H., Wittekind C. 1997. UICC TNM classification of malignant tumors [5th edition]. Wiley-Liss, New York; 227 pp [Google Scholar]
- Thompson A.I., Lees C.W. 2011. Genetics of ulcerative colitis. Inflamm. Bowel Dis. 17:831–848 10.1002/ibd.21375 [DOI] [PubMed] [Google Scholar]
- Ullman T.A., Itzkowitz S.H. 2011. Intestinal inflammation and cancer. Gastroenterology. 140:1807–1816 10.1053/j.gastro.2011.01.057 [DOI] [PubMed] [Google Scholar]
- Uronis J.M., Mühlbauer M., Herfarth H.H., Rubinas T.C., Jones G.S., Jobin C. 2009. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE. 4:e6026 10.1371/journal.pone.0006026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.J., Tu H.P., Ko A.M., Chiang S.L., Chiou S.J., Lee S.S., Tsai Y.S., Lee C.P., Ko Y.C. 2011. Lymphocyte α-kinase is a gout-susceptible gene involved in monosodium urate monohydrate-induced inflammatory responses. J. Mol. Med. 89:1241–1251 10.1007/s00109-011-0796-5 [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium 2007. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 447:661–678 10.1038/nature05911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Rhee K.J., Albesiano E., Rabizadeh S., Wu X., Yen H.R., Huso D.L., Brancati F.L., Wick E., McAllister F., et al. 2009. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15:1016–1022 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.