Abstract
Generating neural stem cells and neurons from reprogrammed human astrocytes is a potential strategy for neurological repair. Here we show dedifferentiation of human cortical astrocytes into the neural stem/progenitor phenotype to obtain progenitor and mature cells with a neural fate. Ectopic expression of the reprogramming factors OCT4, SOX2, or NANOG into astrocytes in specific cytokine/culture conditions activated the neural stem gene program and induced generation of cells expressing neural stem/precursor markers. Pure CD44 + mature astrocytes also exhibited this lineage commitment change and did not require passing through a pluripotent state. These astrocyte-derived neural stem cells gave rise to neurons, astrocytes, and oligodendrocytes and showed in vivo engraftment properties. ASCL1 expression further promoted neuronal phenotype acquisition in vitro and in vivo. Methylation analysis showed that epigenetic modifications underlie this process. The restoration of multipotency from human astrocytes has potential in cellular reprogramming of endogenous central nervous system cells in neurological disorders.
Abbreviations: ESC, Embryonic Stem Cell; iPSC, induced Pluripotent Stem Cell; NSC, Neural Stem Cell
Keywords: Astrocytes, Reprogramming, Neural stem cells, Induced pluripotent stem cells
Highlights
► Single expression of OCT4, SOX2, or NANOG reprogram astrocytes into NSCs. ► Astrocyte-derived neural stem cells gave rise to neuroectodermal cells. ► ASCL1 promoted neuronal phenotype acquisition. ► Epigenetic modifications underlie this process.
Introduction
Replacement of neurons after degeneration or damage is absent in the vast majority of the mammalian central nervous system (CNS), and neuronal loss is thought to be definitive in neurological diseases such as stroke or Alzheimer's disease [1] . In apparent contrast, several studies have demonstrated that some cells resembling glial cells function as neural stem cells (NSCs) or progenitors in specific areas of the adult brain, such as the ventricular subependymal zone and the subgranular zone of the hippocampus [1]. These data pose the questions of [1] why glia in the vast majority of the CNS, such as the cortical regions, apparently cannot produce new neurons and [2] if these cells can be redirected towards neurogenesis when supplied with the appropriate transcriptional factors and/or environmental signals.
It has been demonstrated that astrocytes from murine cerebral cortex can be directly differentiated into neurons by the forced expression of a single transcription factor, such as PAX6, Neurog2 or Mash1 and/or Dlx2 [2,3]. These data open the possibility of activation of endogenous astrocytes for neuronal repair of injured brain tissue in neurological disorders. While such studies have evaluated astrocyte conversion in the murine context, a similar possibility has yet to be demonstrated in human cells.
An essential element for developing such applications with therapeutic value is a thorough comprehension of the mechanisms that regulate reprogramming of adult cells into induced pluripotent stem cells (iPSCs) [4] or directly into another committed lineage, such as fibroblasts converted into neurons [5] and also specific neuronal subpopulations like dopaminergic neurons [6]. Even if for the purpose of autologous cell transplantation in neurological disorders, fibroblasts from patients resemble a much more suitable source of neurons than astrocytes from patients, nevertheless, shading light in the mechanisms that make possible to reprogram astrocytes into NSCs is useful for the final goal of using these cells as endogenous cell source for in situ neural repair in the CNS without any invasive cell graft.
Reprogramming towards pluripotency involves the expression of master stem cell factors followed by a series of molecular events that include generation of various intermediate cells and a small fraction of bona fide pluripotent cells [4]. The developmental properties of these intermediate/partially reprogrammed cells are still unclear. The four transcription factors Oct4, Sox2, Klf4, and c-Myc (or substitution of the latter two with Nanog and Lin28) can induce pluripotency in mouse and human fibroblasts [7]. In fact, physiologically, OCT4, SOX2, and NANOG act in concert to promote and maintain embryonic stem cell (ESC) identity [4]. Human astrocytes can be reprogrammed into iPSCs, with similar efficiencies to other cells, using the viral expression of four reprogramming factors (Oct4, Sox2, Klf4, and c-Myc) [8]. Remarkably, overexpression of a single factor like OCT4 in adult cells can induce full reprogramming, as when it is expressed in NSCs (human and mouse) [9,10], or promote the formation of another phenotype, such as the generation of blood cells with its expression in human fibroblasts [11]. These data suggest that the effect of these stem reprogramming factors changes in relationship to the lineage and the differentiation stage of the cells expressing them. These findings also have raised the possibility that in conditions similar to those needed for reprogramming to iPSCs, cells could acquire, as an alternative, a specific well-defined lineage phenotype. Because of their full commitment, these cells could be used for basic and therapeutic applications with a greater safety relative to undifferentiated pluripotent iPSCs, thus overcoming some limitations in the use of the latter. In the current work, using the individual expression of OCT4, SOX2, or NANOG, we demonstrated and characterized the direct neural fate conversion of human astrocytes into multipotent neural progenitors, in vitro and in vivo. These cells were generated in a manner that is independent of iPSC production.
Material and methods
Cell cultures
Human cortical astrocytes (ScienCell) were cultured in astrocyte medium (Science Cell) on poly-l-lysine-coated flasks. Lentiviral vectors (pSIN) containing cDNA of OCT4, SOX2, NANOG, and Lin 28 [12] were obtained from Addgene. Astrocyte transductions were performed at 24 h post-plating on coated dishes as described [12] or with nucleofection [13]. Transfected cells were cultured in ESC/iPSC media (mTERS, Stem Cells) or in human NSC medium (Stem Cell Inc.) supplemented with EGF and FGF and heparin, according to the manufacturers’ instructions. Colonies with human NSC morphology (NSC colonies) were generally picked for expansion on day 21 post-transduction. Control iPSCs were derived as previously described from human fibroblasts with OCT4, SOX2, NANOG, and LIN28 vectors [12]. To select the CD44 + population, primary astrocytes were incubated with PE-labeled anti-CD44 + antibody (BD Pharmingen) followed by FACS selection (FACS Vantage BD). An appropriate isotype Ig control was used. ASCL1 vector (OriGene) was transfected as described above for the other vectors in AstroNSC cells.
Functional/phenotypic analysis
For expansion and differentiation of NSC clones, the cells were cultured in NSC expansion and differentiation medium (Stem Cell Inc.) according to the manufacturer's instructions. Cells were fixed in 4% paraformaldehyde (PFA) for 10 min. Then they were permeabilized with 0.5% Tween-20 in PBS and then exposed to 0.1% Tween-20 with 10% horse serum. We incubated the cells with primary antibodies overnight and with secondary antibodies for 1 h when unconjugated primary antibodies were employed (Alexa Fluor, Invitrogen). We used the following primary antibodies: SSEA-3 (1:100, R&D), SSEA-4 (1:500, DSHB), TRA1-60 (1:500, Chemicon), TRA1-81 (1:500, Chemicon), Pax6 (1:200, Millipore), nestin (1:200, Millipore), Sox2 (Millipore 1:200), TuJ1 (1: 200, Millipore), GFAP (1:300, SIGMA), anti-microtubule-associated protein 2 (MAP2; 1:100, mouse, Sigma-Aldrich), NF (1: 200, SIGMA), synapsin (1:500, Abcam), and GABA (1:200, Abcam). For all images, we used a confocal LEICA LCS2 microscope. Cell counts and measurements were quantified as described previously [14,15] and analyzed using NIH Image Software, and statistical calculations were performed using StatsDirect software. Teratoma formation was investigated by injection into immunocompromised mice as described [13]. Tissues and teratomas were evaluated for the presence of the three germinal layers (mesoderm, endoderm, and ectoderm) through histological examination with hematoxylin and eosin.
Microarray analysis of gene expression: Astro vs AstroNANOG
Microarray expression analysis of AstroNANOG and parental astrocytes was performed with human genome GeneChip Gene 1.0 ST Array (Affymetrix, Santa Clara, CA). This analysis was performed by Imagenes Laboratory (Germany). The GeneChip IVT Express Kit and the GeneChip Hybridization wash and stain kit were used with 100 ng of total RNA that was mixed with 2 μl of 1/500,000 diluted spikes. Synthesis of the first strand and the second strand of cDNA was performed according to the kit instructions at 42 °C, transcribed, and amplified into cRNA. The cRNA was quantified by NanoDrop. Then fragmentation of 12.5 μg cRNA was performed and hybridized onto the GeneChip according to the Affymetrix protocol. An Affymetrix RUO platform was used and data processed using the Affymetrix GCOS program. Quality control was performed based on Affymetrix quality-control metrics. All chips were normalized by the quantile method and background corrected using robust multi-array analysis. After normalization, the probe-level data were then summarized using median polish to obtain the probe set level measurement. Hierarchical cluster analyses were performed with 1-PCC (Pearson correlation coefficient) as the distance parameter. The maximum distance between cluster members was used as the basis to merge lower-level clusters (complete linkage) into higher-level clusters. To assess the certainty of the existence of a cluster, we applied multiscale bootstrap resampling (10,000 bootstraps) to the hierarchical clustering of the six samples and calculated P values of the hypotheses as well as bootstrap probabilities for each cluster. Microarray data are available in the GEO database.
Cell transplantation
As donor cells for in vivo experiments, we used AstroNSCs and CD44-NSCs ± ASCL1 that were genetically modified to express the GFP gene reporter. Cells were transplanted into the lateral ventricles of anesthetized 1-to-2-day-old mice pups as described previously [16]; 2 μl of cell suspension (100,000 viable cells/μl) was slowly injected. As control, mice received human fibroblasts or freeze-killed cells. The immunosuppressor FK506 administered i.p. at 1.0 mg/kg body weight was administered to all animal groups for the entire length of the experiment. At least 12 animals for each experimental group were analyzed. All animal experiments were performed according to institutional guidelines in compliance with national and international law and policies.
Tissue analysis
Two months after transplantation, the animals were killed, perfused, and fixed with 4% PFA in PBS (pH 7.4). The brain was isolated, immersed in PFA solution for 1 h, and then in sucrose 20% solution in PBS (pH 7.4) overnight and frozen. The tissues were cryosectioned and mounted on gelatinized glass slides. Cerebral tissue was cut in the coronal plane from the frontal lobes (10 or 50 μm for stereological cell count). All CNS sections were blocked with 1% fetal calf serum in PBS and permeabilized with 0.25% Triton X-100. Sections were processed for multiple markers to determine the cellular phenotype of GFP-human donor cells. Immunohistochemistry was performed as described previously [14–16] for the following proteins: nestin, TuJ1 (1:200; Chemicon), NeuN (1:200; Chemicon), MAP2 (1:200; Sigma), neuronal nuclei (NeuN), NF (1: 200, SIGMA), GAD67 (1:500, DSHB), oligodendrocyte marker O4 (1:200; Chemicon), and GFAP (mouse monoclonal, 1:200; Sigma). Mouse and rabbit antibodies conjugated with RPE, CY3, or biotin (1:200; Jackson ImmunoResearch and Dako) were employed for 1 h at room temperature as secondary antibodies when unconjugated primary antibodies were used. Co-expression of GFP/YFP and tissue-specific markers was evaluated using a conventional fluorescence microscope (Zeiss Axiophot) and laser confocal scanning (Leica TCS SP2 AOBS) microscopic analysis. A stereological count of total GFP-positive cells and double-labeled cells with neural and glial markers was conducted using a modified version of the optical fractionator method on every fifth section in selected areas with three ROIs (0.3 × 0.3 mm). Resulting numbers were tallied and multiplied by tissue thickness (50 μm) and the number of intervening sections (n = 5) as described previously [16]. Repeated measures ANOVA were used for statistical analyses.
Sample labeling and hybridization to NimbleGen array for methylation analysis
The labeling of dsDNA, microarray hybridization, and scanning were performed by the Imagenes Laboratory according to the manufacturer's standard protocol. The NimbleGen 3x720K CpG Island Plus RefSeq Promoter Arrays were used. Each array covers important regulatory elements including RefSeq gene promoters and 27,728 annotated CpG islands with 100 bp probe spacing throughout all tiled regions. Tiling of each RefSeq gene promoter (~ 22,532 total) begins 2.4 kb upstream of the transcription start site and extends downstream 0.6 kb for a total of 3 kb of promoter coverage per gene. This array also tiles through various positive, negative, and non-CpG control regions to facilitate assessment of experimental performance. Data were extracted from scanned images by using NimbleScan extraction software. Microarray data are available in the GEO database.
DNA methylation find-peaks analysis
DNA Methylation Find-Peaks analysis identifies regions of significant positive enrichment in IP-based methylation microarray data, using a modified ACME algorithm for peak identification [17]. The analysis accepts normalized, scaled log2-ratio data in GFF format. Map-Peaks Analysis associates peaks with genomic features, such as transcription start sites or CpG islands. Per peak position, the peak scores of the Astro and AstroNANOG experiments were subject to a t-test evaluation, which resulted in a P value for differential peak observation. The ratio between the group peak scores (= fold change) was calculated by the ratio of the respective group median peak scores.
Results
Reprogramming of human cortical astrocytes by the ectopic expression of individual stem transcription factors
We investigated the effects of the individual expression of the key reprogramming factors OCT4, SOX2, and NANOG in human cortical astrocytes. Transduced versus untransduced astrocytes were examined between 14 and 21 days post-transduction (D14–D21) in human ESC/iPSC culture conditions. Fig. 1a summarizes the experimental design. The starting population was positive for astrocytic markers like CD44 and GFAP and completely negative for neural stem cells markers like CD133, CD15, SOX2 and PAX6. No neural stem cells clones both as adherent cell clusters or neurospheres were detected both in bulk and clonal condition in neural stem cells medium.
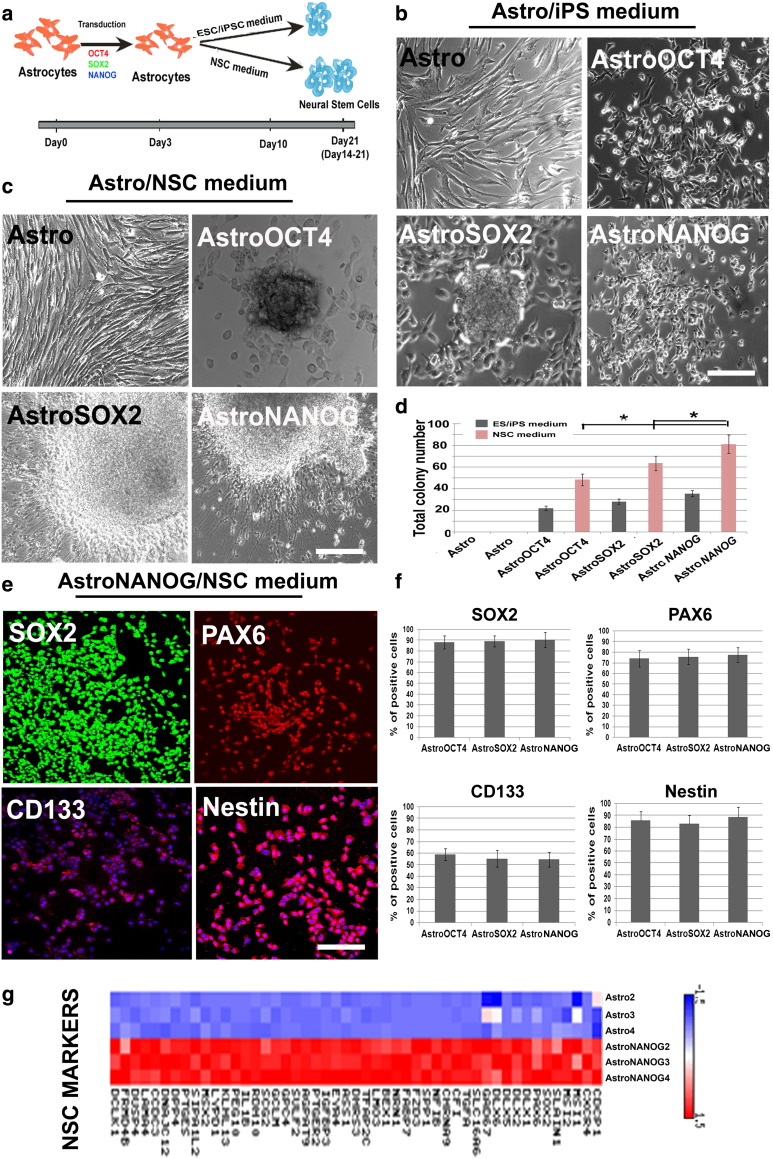
Fig. 1.
OCT4/SOX2/NANOG-transduced human cortical astrocytes give rise to neural stem cell (NSC) colonies.
a) Schematic illustration of NSC generation from human cortical astrocytes (Astro). Human astrocytes were transfected with OCT4, SOX2, or NANOG. The cells were cultured in ESC/iPSC or NSC media. NSC colonies were observed between days 14 and 21 post-transfection. b, c) Bright-field images of human untransduced astrocytes (Astro) and OCT4- (AstroOCT4), SOX2- (AstroSOX2), or NANOG- (AstroNANOG) transduced astrocyte colonies (dashed lines indicate the spheres) in ESC/iPSC medium or NSC medium (n = 12/condition). Scale bar b: 150 μm; scale bar c, Astro and AstroOCT4: 150 μm; AstroSOX2: 75 μm; AstroNANOG: 100 μm. d) Quantification of colonies in human astrocytes after transduction in ESC/iPSC medium (gray bar) or in neuronal medium (pink bar) at 21 days (12 biological replicates; error bars, s.d.; *P < .00001). e) Immunocytochemical expression of SOX2, PAX6, CD133, and nestin in AstroNANOG colonies. Similar results were obtained with AstroOCT4 and AstroSOX2 colonies. Scale bar: 150 μm. f) Quantification of NSC marker expression as a percentage of positive cells in AstroOCT4, AstroSOX2, and AstroNANOG colonies. g) Global gene analysis of NSC marker expression of Astro vs AstroNANOG cells. For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.
Unlike untransduced astrocytes, astrocytes transduced with OCT4 (AstroOCT4), SOX2 (AstroSOX2), or NANOG (AstroNANOG) gave rise to colonies of small cells resembling NSCs (Fig. 1b). The vast majority of these cells were adherent colonies composed of small round/spindle cells, while the other cell populations took the structure of spheroids similar to neurospheres.
To enhance the emergence of NSCs, we used human NSC culture conditions that included neuronal medium supplemented with EGF and FGF (Fig. 1c). This treatment increased the frequency of NSC colony emergence from astrocytes by 2- to 3-fold, compared with untreated astrocytes, while no effect was detected from control untransfected astrocytes (Fig. 1d). The number of colonies obtained was significantly greater with NANOG than with SOX2 or OCT4 (P < .00001) (Fig. 1d). NSCs were then clonally selected and further expanded in the neuronal medium. Supplementary Video 1 shows the typical contrast-phase aspect of these cells. These neural stem colonies were positive for neuronal precursor-specific proteins like SOX2, PAX6, CD133, and nestin (Fig. 1e), and they were negative for astrocytic markers like CD44 and for pluripotency factors such as SSEA-3 and Tra-1–60. We evaluated the self-renewal AstroNSCs in vitro by growing them at clonal density. Clones can be passaged and generate secondary and tertiary NSCs clones. These data indicate that single reprogramming factors are sufficient to initiate the emergence of NSCs from astrocytes and that these cells are responsive to stimulation by NSC growth factors.
To further characterize these cells molecularly, we performed genome-wide expression profiling of AstroNANOG-derived NSCs in comparison with parental astrocytes. AstroNANOG cells showed increased expression of NSC/precursor markers with a parallel reduction in astrocyte-specific gene expression, resulting in a shift towards the NSC identity (Supplementary Table 1). Overall, the gene expression pattern of AstroNANOG cells was significantly different relative to untransfected astrocytes as demonstrated by hierarchical clustering of signals, the heat map, and the volcano-plot t-test (Supplementary Fig. 1). To confirm that cortical astrocytes were converted and that our results were not linked to the presence of other cells such as NSC/precursors, we selected a population of CD44 + astrocytes by FACS (Supplementary Fig. 2). CD44 + cells mark a population of fully committed astrocytes that do not differentiate into neuronal or oligodendrocytic lineages [18]. After sorting the purified cells are all positive for CD44 + as shown in Supplementary Fig. 2c and are negative for neural stem cells markers like CD133, CD15, SOX2 and PAX6 (data not shown). We then transduced the purified CD44 + cells using reprogramming factors (Supplementary Fig. 2). We demonstrated that these cells also generated NSCs with a similar efficiency observed with the whole primary culture. The untrasfected CD44 + never generated clones resembling NSCs. The isolated CD44-NSCs clones were negative for CD44 and positive for SOX2, PAX6, CD133, and nestin (Supplementary Fig. 2). These cells are expanded in vitro in NSC medium and were grown at clonal density. They generated secondary and tertiary NSC clones demonstrating that they are self-renewing cells. Thus, fully committed human cortical astrocytes can be reprogrammed into NSCs.
Direct reprogramming into NSCs does not require passage through a pluripotent state
Expression of OCT4 alone has been demonstrated as sufficient to reprogram NSCs into iPSCs [11]. We wanted to investigate whether the individual expression of OCT4/SOX2/NANOG could reprogram astrocytes into iPSCs or if the generation of NSCs was obtained through passage into a pluripotent state. We observed no formation of typical iPSCs in culture, and the pluripotency markers SSEA3, SSEA4, TRA-1–60, and TRA-1–81 were not detectable in the transduced condition, whereas both features increased during establishment of iPSCs from human fibroblasts transduced contemporaneously with four factors (OCT4/SOX2/NANOG/Lin28) as previously described [12] (Fig. 2 and Supplementary Fig. 3).
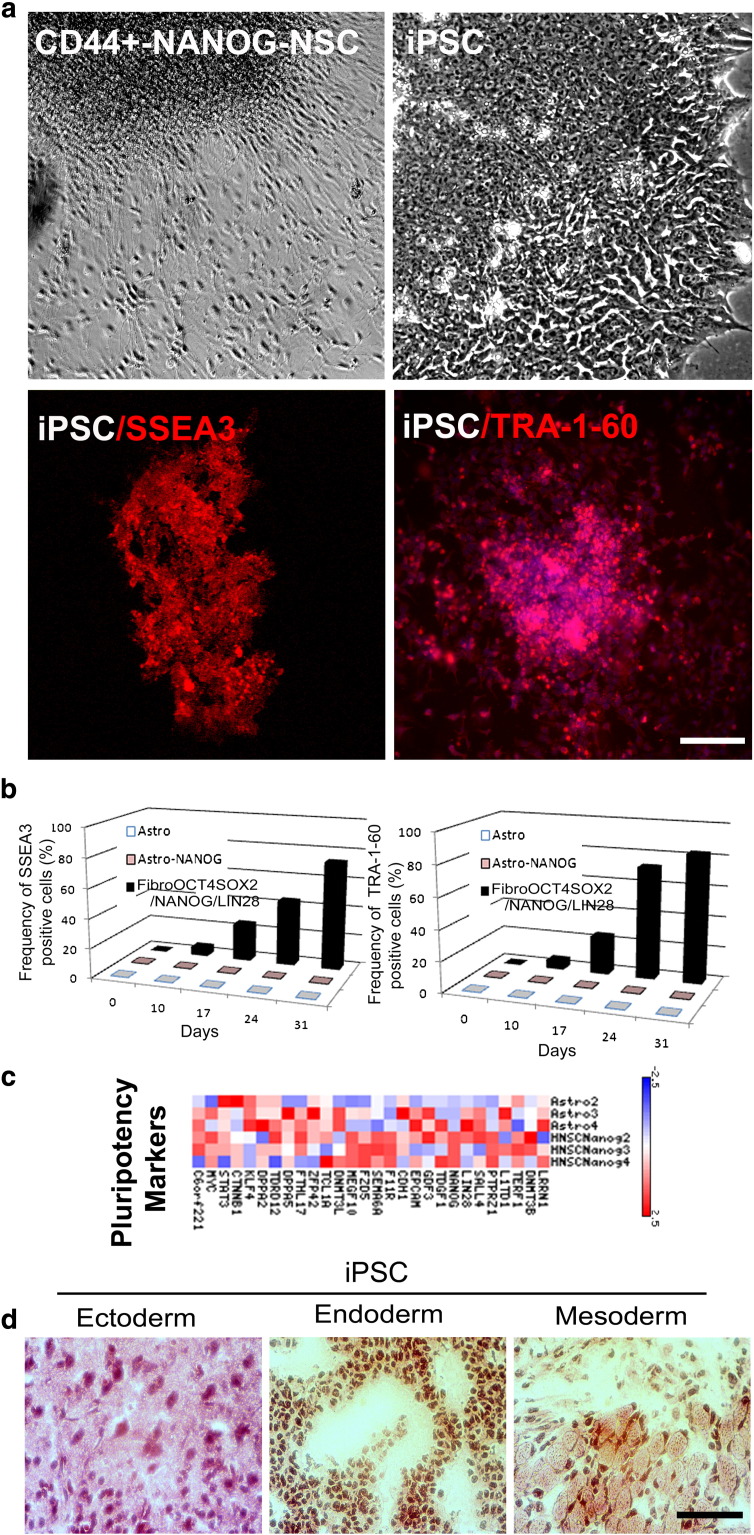
Fig. 2.
Conversion of astrocytes into NSCs by OCT4/SOX2 and NANOG does not require the pluripotent state.
a) A colony of CD44 + NANOG-NSCs presented a different morphological aspect under contrast-phase microscopic observation relative to the iPSC colony obtained from fibroblasts by contemporaneous transfection of OCT4/SOX2/NANOG/LIN28. iPSCs were positive for pluripotency markers SSEA3 and TRA-1-60. Scale bar: 150 μm. b) Quantitative analysis of SSEA3 and Tra-1-60 in Astro, AstroNANOG, and human fibroblasts transduced with OCT4, SOX2, NANOG, and LIN28, over the human iPSC derivation timeline. c) Pluripotency gene profile in parental astrocytes and in AstroNANOG. d) Teratomas derived from human iPSCs and testicular sections from mice injected with astrocytes, AstroNANOG, and saline (CTR). Scale bar: 300 μm.
We analyzed the expression in AstroNANOG cells of a specific gene subset known to be crucial for induction and maintenance of pluripotency in NSCs derived from astrocytes. Apart from the up-regulation of NANOG itself, NANOG expression did not modify the pluripotency gene expression pattern as evaluated by microarray analysis (Fig. 2). In contrast to the fully reprogrammed iPSCs, transplantation of AstroNANOG-transduced or untransduced astrocytes into immunodeficient mice did not generate teratomas composed of all three germ layers or any overgrowth resembling a tumor (Fig. 2). Overall, our results indicated that the Astro-derived NSCs were directly diverted towards an NSC fate without passing through a detectable phenotype or functional status of pluripotent cells.
Neuronal differentiation of NSCs derived from astrocytes in vitro
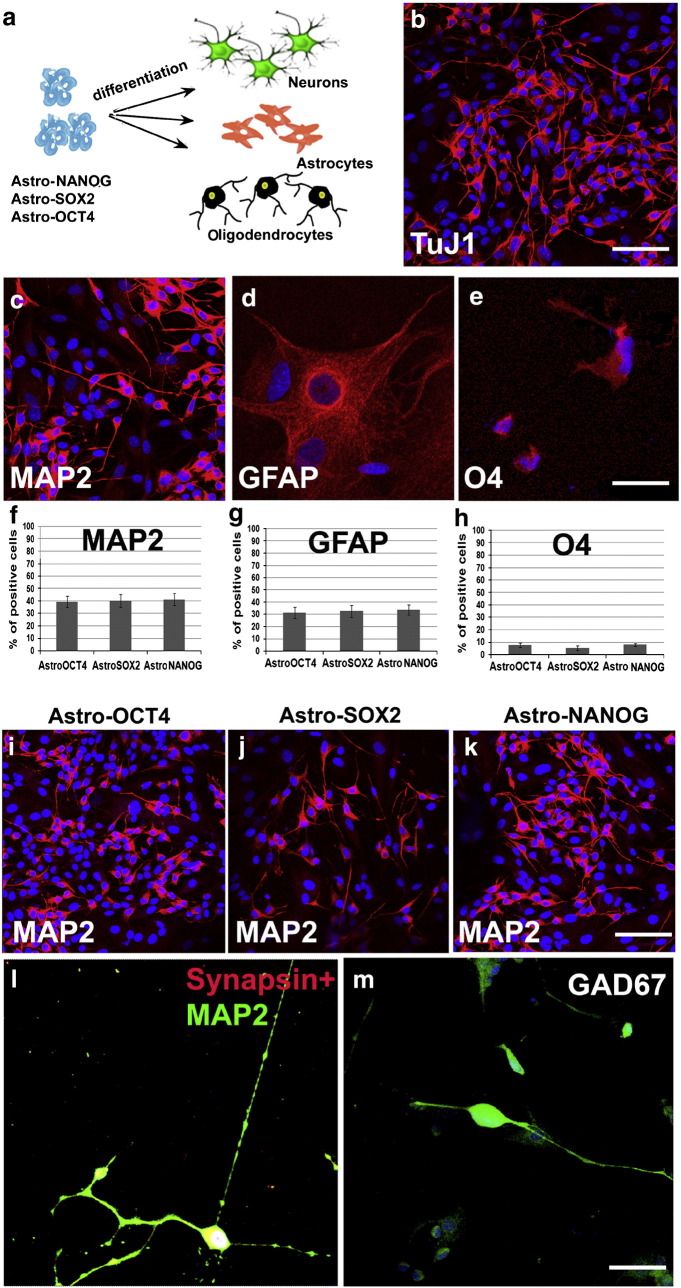
Global gene expression analysis showed that the AstroNANOG-derived cells exhibited a gene expression pattern consistent with that of neuronal stem/precursor cells, supporting the hypothesis that they may have functional neurogenic properties of multiple neuroectodermal phenotypes. To functionally characterize neuronal differentiation capacity, AstroNSC clones (for each condition: OCT4, SOX2, NANOG) were cultured in human neuronal differentiation medium. The resulting progeny presented features consistent with differentiation into the three neuroectodermal lineages (Fig. 3). After 15 days of differentiation, we observed 41.4 ± 4.9% neurons (positive for neuronal markers TuJ1, MAP2, and NF; quantification performed with MAP2), 33.6 ± 4.2% astrocytes (positive for GFAP), and 7.9 ± 1.3% oligodendrocytes (positive for O4 in AstroNANOG-NSCs). The remaining cells had features of neuronal precursors positive for SOX2. Similar results were obtained with AstroNSCs transfected with OCT4 or SOX2 and also with CD44 +-reprogrammed cells (Supplementary Fig. 4). The AstroNSC-derived neurons presented typical neuronal morphology and not only were positive for pan-neuronal markers but also for synapsin, supporting the presence of synaptic contact, and for the neurotransmitter GABA, both suggesting a functionally correct competence of these cells (Supplementary Fig. 4).
Fig. 3.
In vitro generation of three neuroectodermal lineages in particular in neurons from AstroOCT4/SOX2 or NANOG.
Protocol of AstroNSC differentiation into neurons, astrocytes, and oligodendrocytes. AstroNSCs were cultured in human neuronal differentiation medium. The cell phenotype was determined after 15 days of differentiation. b–e) Differentiation of AstroNANOG cells into neurons positive for TuJ1 (b) and MAP2 (c), astrocytes positive for GFAP (d), and oligodendrocytes positive for O4 (e). f–h) Quantification of the differentiation efficiency into neurons (MAP2), astrocytes (GFAP), and O4 for the three conditions (NANOG, OCT4, SOX2). i–k) AstroOCT4/SOX2/NANOG had similar neuronal differentiation abilities as demonstrated here with immunocytochemistry for MAP2. l–m) The AstroNSC-derived neurons acquired a complex mature phenotype and were positive for synapsin (l, shown here in red as double staining with MAP2, green) and neurotransmitters like GAD67 positivity, enzyme for the biosynthesis of the neurotransmitter GABA suggests. Scale bar: b, c, i, j, k: 150 μm; d, e: 50 μm; l, m: 50 μm.
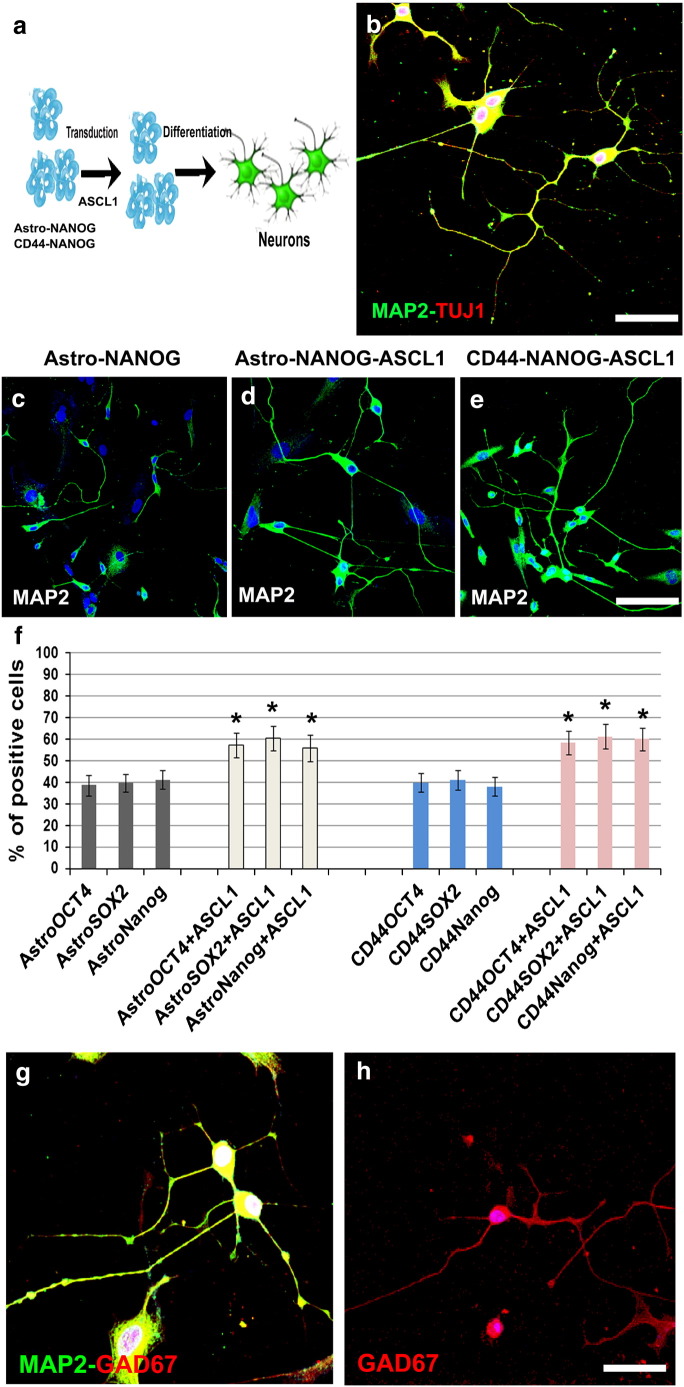
Overexpression of the helix–loop–helix ASCL1 can induce neurogenesis in NSCs [19] and rodent astrocytes [2] and, in association with other factors, in murine and human fibroblasts [5,20]. Thus, we transduced AstroNSCs with ASCL1 (Fig. 4, Supplementary Fig. 5). Transfected cells had a significantly augmented number of neurons compared with untreated cells (transfected cells 47.4 vs 36.3%, P < .00001). The ASCL1 neurons presented highly complex morphologies and expressed the pan-neuronal markers TuJ1, MAP2, and NF. Indeed, they were positive for synapsin and also for neurotransmitters like GAD67 positivity, the enzyme for the biosynthesis of the neurotransmitter GABA suggests (Fig. 4, Supplementary Fig. 5).
Fig. 4.
ASCL1 promotes neuronal differentiation of Astro and CD44 + NANOG cells.
a) Schematic illustration of Astro and CD44 + NANOG-ASCL1 differentiation into neurons. Astro/CD44 + NSCs were cultured in human neuronal differentiation medium. The cell phenotype was analyzed after 15 days. b) AstroNANOG-ASCL neurons presented long dendritic arborization and axons and were positive for pan-neuronal markers MAP2 (green) and TuJ1 (red), shown here as merged signal. c–e) Neurons differentiated from AstroNANOG-ASCL and CD44-NANOG-ASCL1 were present at higher rates and with a complex mature morphology in culture relative to AstroNANOG. The neurons are shown here with immunocytochemistry for MAP2. f) Quantification of the differentiation efficiency into neurons (MAP2), demonstrating that efficiency was significantly higher with AstroNANOG-ASCL1 and CD44-NANOG-ASCL1 vs AstroCD44-NANOG. g, h) The AstroNSC-derived neurons were positive for GAD67 (shown here in red as double staining with MAP2, green (g) or in single staining (h)).
Scale bar: b: 50 μm; c, d: 150 μm; g: 50 μm; h: 70 μm.
Neuronal differentiation of NSCs from astrocytes in vivo
To assess the ability of AstroNSC clones cells to engraft and differentiate into neurons in vivo in mammalian CNS, Astro/CD44NANOG cells were transplanted intracerebroventricularly in immunosuppressed mice (n = 12) to characterize its in vivo potential (Fig. 5a). Animals transplanted with fibroblasts and freeze-killed cells were used as control. Human donor cells ultimately engrafted in the brains of all transplanted animals. In particular, donor-transplanted cells incorporated into the gray and white matter of the brain, migrating from the cerebral ventricles within 2 months after delivery. These cells were observed also in cortical areas.
Fig. 5.
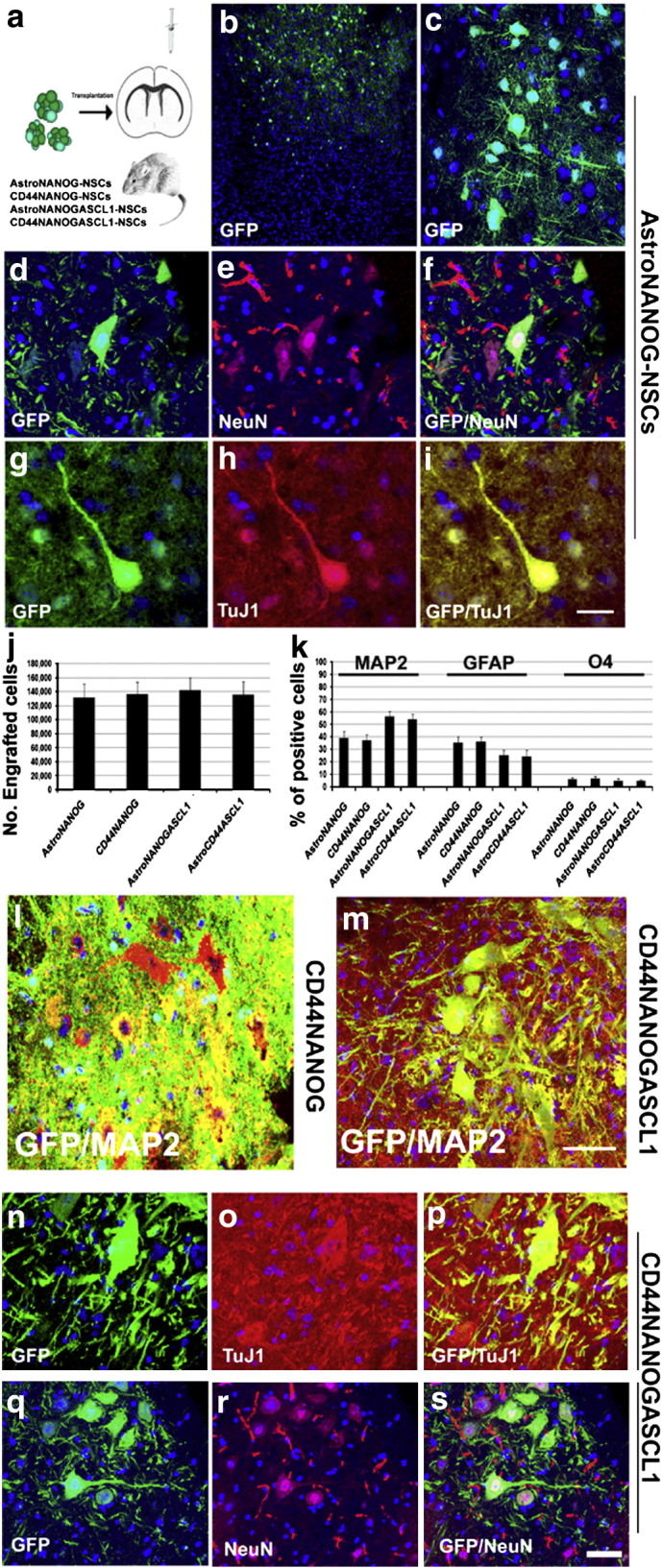
AstroNSCs transplanted in rodent brain survive and generate neurons.
a) For in vivo experiments, we transplanted AstroNSCs (AstroNANOG, CD44NANOG ± ASCL1) intracerebroventricularly in the mouse brain. To trace the fate of transplanted cells, cells were modified to express the gene reporter GFP. b, c) After transplantation of AstroNANOG cells, we demonstrated the presence of integrated donor cells in brain cortical areas. d–i) A significant proportion of GFP cells (d, g: green) acquired a neuronal phenotype as demonstrated here by the expression of the neuronal markers NeuN (e: red) and TuJ1 (h: red). f, i: merged images. j, k) Quantification of donor-engrafted cell number (j) and of different phenotypes acquired (k, expressed as percentage of positive cells: neurons (MAP2), astrocytes (GFAP), and oligodendrocytes (O4)), for each condition. l, m) Transplantation of CD44-NANOG-ASCL1 cells gave rise to a higher proportion of mature neurons relative to the CD44-NANOG population as shown here by immunohistochemistry for MAP2 (red) merged (yellow) with GFP (green). n–s) The neurons derived from CD44-NANOG-ASCL1-GFP cells (n, q) presented a complex phenotype and were positive for neuronal markers like TuJ1 (o: red) and NeuN (r: red). p, s: merged images.
Scale bar: b: 300 μm; c: 80 μm; d–i: 75 μm; l, m: 75 μm; n–p: 50 μm; q–s: 80 μm.
The total number of GFP-positive human donor AstroNSCs in the brain sections from treated animals was determined by stereological estimation at 2 months post-transplantation. The results indicate that cell number was 131,764 ± 19,546 cells (65.88 ± 14.38% of the initial population of 200,000 cells) in the case of AstroNANOG cells, and 136,872 ± 16,845 cells (68.44 ± 12.31% of the initial population of 200,000 cells). Donor-derived cells exhibited a predominantly neuronal phenotype (MAP2: 39.3 ± 5.2% AstroNANOG; 37.2 ± 4.8% CD44NANOG). These cells presented highly mature, morphologically differentiated cells with complex and long neuritic extensions. A proportion of the engrafted cells retained NSC features, while the remaining cells acquired a glial phenotype (GFAP: 35.4 ± 4.9% AstroNANOG, 36.3 ± 3.8% CD44NANOG; O4: 6.1 ± 1.6% AstroNANOG, 6.6 ± 1.9% CD44NANOG) (Fig. 5). Donor-derived neuronal cells express also neurotransmitter proteins like GABA (Supplementary Fig. 6).
The ability to generate neuronal progenitors and the presence of the three neuroectodermal lineages supports the in vivo functional capacity of Astro-derived cells.
The transplantation of AstroNANOG/ASCL1 + cells gave rise to a similar number of engrafted cells and a larger proportion of a neuronal phenotype, confirming the pro-neurogenic effect of ASCL1 expression (Fig. 5).
Taken together, our data demonstrate that AstroNSCs can give rise to functional neuronal progenitor-like cells that can differentiate into neuronal lineages in vitro and in vivo.
Epigenetic modifications underlie the reprogramming of astrocytes into NSCs
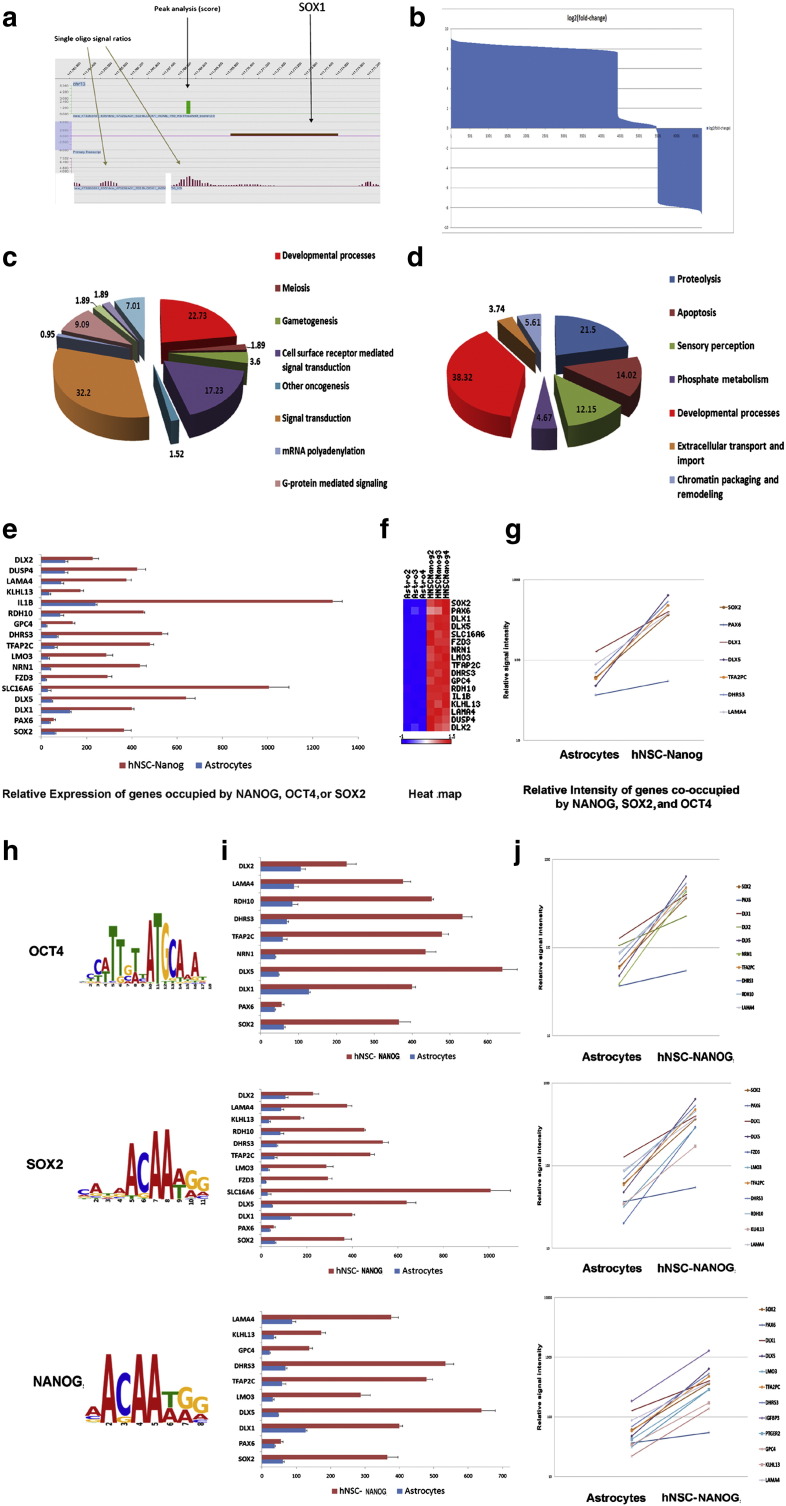
Astro-derived NSCs are likely derived by epigenetic reprogramming, and we analyzed their DNA methylation patterns relative to untransfected astrocytes. We compared three human AstroNANOG-NSC clones to the astrocytes from which they were derived using NimbleGen 3x720K CpG Island Plus RefSeq Promoter Arrays. This approach allowed the interrogation of promoter regions and CpG sites. DNA Methylation Find-Peaks analysis identified regions of significant positive enrichment in microarray data, using a modified algorithm for capturing microarray enrichment (ACME) for peak identification [17] (Fig. 6a). Per peak position, the peak scores of the AstroNANOG and astrocyte experiments were subject to a t-test evaluation. A total of 6667 of 67,652 regions analyzed were found to significantly differ (Supplementary file 1). We termed the differentially methylated regions R-DMRs. The number of R-DMRs that were hypermethylated in astrocytes significantly predominated over hypomethylated, as shown in Fig. 6b.
Fig. 6.
Epigenetic and gene expression modifications associated with astrocyte reprogramming.
a) We compared the methylation pattern of three human AstroNANOG-NSC clones to the astrocytes from which they were derived. DNA Methylation Find-Peaks analysis identified regions of significant positive enrichment in microarray data as shown here in the case of SOX1 as an example. b) A total of 6667 of 67,652 regions analyzed were found to significantly differ. The number of differentially methylated regions, R-DMRs, that were hypermethylated in astrocytes significantly predominated over hypomethylated. This diagram shows the log2 values of the fold changes (Astro/NANOG). The distribution shows a clear majority of positive values, i.e., the respective fold changes were mostly > 1. c, d) We then performed a gene ontology (GO) annotation analysis of the genes near these regions (within 1000 bp of the transcriptional start site (TSS) of a gene). The genes that were hypermethylated in astrocytes compared to AstroNSCs are shown in c, while the genes that were hypomethylated in astrocytes compared to AstroNANOG are shown in d. e–j: Correlations for the global gene expression profile of astrocytes and AstroNANOG and the OCT4, SOX2, and NANOG occupancy. e) Relative expression of genes occupied by OCT4, SOX2, and NANOG and their heat map representation (f) as well as relative intensity (g). Binding site sequences of OCT4, SOX2, and NANOG (h). Relative expression of genes occupied by OCT, SOX2, or NANOG, individually considered, and their relative intensity.
We then performed a gene ontology annotation analysis of the genes near these regions (within 1000 bp of the transcriptional start site (TSS) of a gene) that revealed significant enrichment for genes involved in developmental and regulatory processes (Figs. 6c, d). For example, 22% of the genes that were hypermethylated in astrocytes compared to AstroNSCs and 38% of the genes that were hypomethylated in astrocytes compared to AstroNANOG were involved in developmental processes.
When we observed the overlap of the R-DMRs with known binding sites for OCT4, SOX2, and NANOG [21], we saw that some of the R-DMRs belong to these regions (Supplementary File 3). We then correlated the global gene expression profile of astrocytes and AstroNANOG and the known OCT4, SOX2, and occupancy. NSC genes that were up-regulated in AstroNANOG cells were significantly related with predicted OCT4, SOX2, and NANOG binding sites (Figs. 6e–j). These observations indicated that the sites of epigenetic and gene expression changes during reprogramming of astrocytes to NSCs are tightly linked to genes that are functionally important for pluripotency.
Discussion
Our present study demonstrates for the first time that expression of a single key stem transcription factor is sufficient to rapidly and efficiently convert human cortical astrocytes into neural stem/precursor cells. OCT4/SOX2/NANOG genes were selected because of their demonstrated roles in the maintenance of pluripotency and reprogramming [4]. Astro-derived NSCs are self-renewing and multipotent, present specific NSCs morphology and marker expression, and respond to NSC growth factor stimuli. To unequivocally demonstrate that terminally differentiated astrocytes can be directly converted into NSCs using this strategy, we also employed CD44 +-selected fully mature astrocytes, ruling out the possibility that the presence of rare stem/precursor cells can account for our observations. Furthermore, no spontaneous conversion into NSCs or neurons in control untransfected or mock transfected astrocytes was observed in our culture conditions.
The change in lineage fate occurred without the need for passage through a pluripotent state similar to the iPSCs. This direct conversion approach may allow avoidance of some complications linked to the undifferentiated state of iPSCs, including the risk of teratoma formation. In the context of the production of cells for clinical application for regenerative medicine in neurological diseases, further studies are warranted to investigate if, with a similar strategy, it is possible to directly convert into NSCs other cells that are of a more distant embryonic lineage, like human skin fibroblasts.
The functional differentiation ability of Astro-derived NSCs in neurons and glia in vitro and in vivo confirmed the acquisition of NSC fate by human astrocytes. A remarkable tendency of these cells to differentiate into neurons was observed. These cells are positive not only for pan-neuronal antigens but also for synaptic proteins and neurotransmitters, supporting their functionally correct features.
By inserting the transcription factor ASCL1, we further modulated and promoted the neuronal differentiation ability of reprogrammed cells. ASCL1, also known as Mash1, belongs to the bHLH transcription factors of the achaete-scute family and is a telencephalic transcription factor upstream of Dlx2 during neuronal specification in development [22,23]. This factor, in combination with other neurogenic genes, has been described to promote the neuronal phenotype in murine and human fibroblasts [5,20] and murine astrocytes [3,4]. Also in our experiments, the expression of ASCL1 was neurogenic and associated with the generation of a larger number of neurons with a more complex and mature phenotype. Overall, our data suggest that astrocytes can be induced to acquire a specific desired phenotype also following multiple sequential genetic modifications. Other combinations of multiple neurogenic transcription factors might produce a wider spectrum of neuronal phenotypes.
Remarkably the acquisition of a neuronal phenotype was also obtained in vivo after grafting into the rodent brain. This finding is promising ground for future attempts to convert endogenous astrocytes directly in vivo with transcription factor expression.
To investigate the molecular mechanisms underlying this astrocyte-to-NSC conversion, we performed genome-wide expression and methylation analysis. The results demonstrated that the presence of key stem factors activates specific features of the NSC-fate transcription program. It is likely that auto-regulation and positive feedback loops involving reprogramming factors as well as feed-forward regulation of common target genes [24] maintain the expression of key neural stem-determining genes and stabilize the phenotype.
In complementary findings, we showed that the same genes undergo extensive epigenetic modifications. Significant modification of the transcriptional profile could also lead to genome-wide changes in epigenetic features such as DNA methylation and chromatin remodeling that further promote the stability of the new transcriptional network. The analysis of differentially methylated regions revealed a significantly higher methylation with regard to NSCs. Integrating these data with known mapped biding sites of OCT4/SOX2/NANOG [25], we conclude that the reprogramming is likely associated with the direct binding of reprogramming factors to the regulatory loci of neuronal-specific genes.
Overall, our results support the relevance of investigating partial reprogramming as a possible strategy for autologous cell therapy not only ex vivo but also prospectively in vivo by direct conversion of endogenous astrocytes in the CNS for the treatment of neurological diseases.
Funding
The financial support of the following research grants to S.C. and G.P.C. is gratefully acknowledged: Telethon grant: GGP09107, “Neuroprotection in Spinal Muscular Atrophy (SMA) using neural stem cells as a therapeutic approach,” Telethon grant GGP10062: “Development of a therapeutic approach for Spinal Muscular Atrophy with Respiratory Distress (SMARD1) using human induced pluripotent stem cell-derived neural stem cells and motor neurons”; and FIRB RBFR08RV86: “Development of a stem cell approach for motor neuron diseases.”
The following are the supplementary materials related to this article.
Supplementary data.
Differential expressed genes from Astro vs AstroNANOG, ordered by absolute values of logfold changes.
Contrast-phase time-lapse acquisition of AstroNSCs in culture (24 h of recording). The movie shows the small round morphology of the cells and their proliferative capacity.
Conflict of interest
The authors declare that there are no conflicts of interest.
Acknowledgment
We wish to thank the Associazione Amici del Centro Dino Ferrari for their support.
References
- 1.Kriegstein A., Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinrich C., Blum R., Gascón S., Masserdotti G., Tripathi P., Sánchez R., Tiedt S., Schroeder T., Götz M., Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinrich C., Gascón S., Masserdotti G., Lepier A., Sanchez R., Simon-Ebert T., Schroeder T., Götz M., Berninger B. Generation of subtype-specific neurons from postnatal astroglia of the mouse cerebral cortex. Nat. Protoc. 2011;6:214–228. doi: 10.1038/nprot.2010.188. [DOI] [PubMed] [Google Scholar]
- 4.Hanna J.H., Saha K., Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pang Z.P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D.R., Yang T.Q., Citri A., Sebastiano V., Marro S., Südhof T.C., Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caiazzo M., Dell'Anno M.T., Dvoretskova E. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 7.Jopling C., Boue S., Izpisua Belmonte J.C. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz S., Brennand K., Panopoulos A.D., Herrerías A., Gage F.H., Izpisua-Belmonte J.C. High-efficient generation of induced pluripotent stem cells from human astrocytes. PLoS One. 2010;5:e15526. doi: 10.1371/journal.pone.0015526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim J.B., Sebastiano V., Wu G., Araúzo-Bravo M.J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., Meyer J., Hübner K., Bernemann C., Ortmeier C., Zenke M., Fleischmann B.K., Zaehres H., Schöler H.R. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Kim J.B., Greber B., Araúzo-Bravo M.J., Meyer J., Park K.I., Zaehres H., Schöler H.R. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009;461:649–653. doi: 10.1038/nature08436. [DOI] [PubMed] [Google Scholar]
- 11.Szabo E., Rampalli S., Risueño R.M., Schnerch A., Mitchell R., Fiebig-Comyn A., Levadoux-Martin M., Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 12.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., Slukvin I.I., Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 13.Yu J., Hu K., Smuga-Otto K. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corti S., Nizzardo M., Nardini M., Donadoni C., Salani S., Ronchi D., Saladino F., Bordoni A., Fortunato F., Del Bo R., Papadimitriou D., Locatelli F., Menozzi G., Strazzer S., Bresolin N., Comi G.P. Neural stem cell transplantation can ameliorate the phenotype of a mouse model of spinal muscular atrophy. J. Clin. Invest. 2008;118:316–3330. doi: 10.1172/JCI35432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corti S., Nizzardo M., Nardini M., Donadoni C., Salani S., Ronchi D., Simone C., Falcone M., Papadimitriou D., Locatelli F., Mezzina N., Gianni F., Bresolin N., Comi G.P. Embryonic stem cell-derived neural stem cells improve spinal muscular atrophy phenotype in mice. Brain. 2010;133:465–481. doi: 10.1093/brain/awp318. [DOI] [PubMed] [Google Scholar]
- 16.Corti S., Locatelli F., Papadimitriou D., Donadoni C., Del Bo R., Fortunato F., Strazzer S., Salani S., Bresolin N., Comi G.P. Multipotentiality, homing properties, and pyramidal neurogenesis of CNS-derived LeX(ssea-1)+/CXCR4 + stem cells. FASEB J. 2005;19:1860–1862. doi: 10.1096/fj.05-4170fje. [DOI] [PubMed] [Google Scholar]
- 17.Scacheri P.C., Crawford G.E., Davis S. Statistics for ChIP-chip and DNase hypersensitivity experiments on NimbleGen arrays. Methods Enzymol. 2006;411:270–282. doi: 10.1016/S0076-6879(06)11014-9. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y., Wu Y., Lee J.C., Xue H., Pevny L.H., Kaprielian Z., Rao M.S. Oligodendrocyte and astrocyte development in rodents: an in situ and immunohistological analysis during embryonic development. Glia. 2002;40:25–43. doi: 10.1002/glia.10111. [DOI] [PubMed] [Google Scholar]
- 19.Kim H.J., McMillan E., Han F., Svendsen C. Regionally specified human neural progenitor cells derived from the mesencephalon and forebrain undergo increased neurogenesis following overexpression of ASCL1. Stem Cells. 2009;27:390–398. doi: 10.1634/stemcells.2007-1047. [DOI] [PubMed] [Google Scholar]
- 20.Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Südhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doi A., Park I.H., Wen B., Murakami P., Aryee M.J., Irizarry R., Herb B., Ladd-Acosta C., Rho J., Loewer S., Miller J., Schlaeger T., Daley G.Q., Feinberg A.P. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillemot F., Lo L.C., Johnson J.E., Auerbach A., Anderson D.J., Joyner A.L. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 23.Pattyn A., Guillemot F., Brunet J.F. Delays in neuronal differentiation in Mash1/Ascl1 mutants. Dev. Biol. 2006;295:67–75. doi: 10.1016/j.ydbio.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Tam W.L., Lim B. StemBook [Internet] Harvard Stem Cell Institute; Cambridge (MA: 2008. Genome-wide transcription factor localization and function in stem cells. Sep 15. [PubMed] [Google Scholar]
- 25.Loh Y.H., Wu Q., Chew J.L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., Wong K.Y., Sung K.W., Lee C.W., Zhao X.D., Chiu K.P., Lipovich L., Kuznetsov V.A., Robson P., Stanton L.W., Wei C.L., Ruan Y., Lim B., Ng H.H. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;8:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data.
Differential expressed genes from Astro vs AstroNANOG, ordered by absolute values of logfold changes.
Contrast-phase time-lapse acquisition of AstroNSCs in culture (24 h of recording). The movie shows the small round morphology of the cells and their proliferative capacity.