SUMMARY
Macrophages respond to inflammatory stimuli by modulating the expression of hundreds of genes in a defined temporal cascade, with diverse transcriptional and post-transcriptional mechanisms contributing to the regulatory network. We examined pro-inflammatory gene regulation in activated macrophages by performing RNA-Seq with fractionated chromatin-associated, nucleoplasmic, and cytoplasmic transcripts. This methodological approach allowed us to separate the synthesis of nascent transcripts from transcript processing and the accumulation of mature mRNAs. In addition to documenting the sub-cellular locations of coding and non-coding transcripts, the results provide a high-resolution view of the relationship between defined promoter and chromatin properties and the temporal regulation of diverse classes of co-expressed genes. The data also reveal a striking accumulation of full-length yet incompletely spliced transcripts in the chromatin fraction, suggesting that splicing often occurs after transcription has been completed, with transcripts retained on the chromatin until fully spliced.
INTRODUCTION
Macrophages play an important role in regulating immune responses following exposure to a microbial insult or danger signal. Their response involves widespread changes in gene expression that are often tailored to the stimulus. Some inducible genes encode proteins that help polarize the subsequent innate and adaptive immune responses, while others encode effector molecules that protect the host from the insult. Microbial pathogens have acquired mechanisms to subvert immune responses, and deregulated inflammation has been associated with numerous diseases ranging from inflammatory autoimmune disorders to atherosclerosis and cancer. A major challenge has been to elucidate the molecular circuitry that regulates an inflammatory gene program, with the ultimate aim of selectively altering the response to diminish disease pathology while maintaining anti-microbial immunity (Medzhitov and Horng, 2009; Smale, 2010).
To understand the transcriptional response to an inflammatory stimulus, several studies have focused on the regulatory properties of primary response genes, which are rapidly induced in the absence of new protein synthesis (reviewed in Fowler et al., 2011). Primary response genes induced by Toll-like receptor (TLR) ligands and tumor necrosis factor (TNF) usually contain CpG-island promoters. Such genes are associated with RNA polymerase II in unstimulated cells and possess histone tail modifications commonly found at the promoters of actively transcribed genes (Hargreaves et al., 2009; Ramirez-Carrozzi et al., 2009). These genes can be activated by inducible transcription factors in the absence of SWI/SNF-dependent nucleosome remodeling (Ramirez-Carrozzi et al., 2006, 2009).
Rapid transcriptional induction may also be facilitated by the active release of polymerase molecules that have initiated transcription in unstimulated cells but are paused near the transcription start site (TSS) (Nechaev and Adelman, 2011). Paused polymerase molecules can be released by signal-dependent recruitment of P-TEFb, phosphorylation of RNA polymerase II on serine 2 of the C-terminal domain (CTD), and release of the NELF repressor. Although some genes are regulated by pausing in close proximity to the TSS, others are reported to be efficiently transcribed into precursor transcripts, with inducible P-TEFb recruitment regulating the efficiency of transcript processing (Adelman et al., 2009; Hargreaves et al., 2009).
In contrast to the features described above, many secondary response genes and some primary response genes require SWI/SNF-dependent nucleosome remodeling for their induction (Ramirez-Carrozzi et al., 2006, 2009; Fowler et al., 2011). These genes often lack CpG islands and are inaccessible to nucleases in unstimulated cells. Prior to stimulation, they also lack histone modifications characteristic of active genes. The remodeling requirement may facilitate highly selective regulation by imposing an additional barrier to activation.
A major long-term goal is to elucidate the precise network of events that coordinates the complex, temporally ordered gene expression cascade induced by an inflammatory stimulus. Progress towards this goal has been made primarily through the use of microarrays to examine temporal changes in mRNA levels, followed by efforts to connect transcription factors and signaling pathways to clusters of co-regulated genes (Ramsey et al., 2008; Amit et al., 2009; Litvak et al., 2009). However, temporal changes in mRNA level can diverge considerably from temporal changes in transcription. For example, a lag period can follow the maximal activation of transcription before the peak mRNA level is reached (Hao et al., 2009; Rabani et al., 2011). Microarrays also tend to underestimate the magnitudes of changes in mRNA levels, further limiting efforts to examine transcriptional cascades (Marioni et al., 2008).
RNA-Seq has emerged as an improved method for examining transcriptomes. Although this method is usually used to study mRNAs, three strategies have been described for the analysis of nascent transcripts, thereby allowing dynamic changes in transcription to be distinguished from changes in mRNA levels. First, the genome-wide nuclear run-on (GRO-Seq) involves the incubation of nuclei with labeled nucleoside triphosphates, followed by high-throughput sequencing of cDNA prepared from the labeled nascent transcripts (Core et al., 2008; Hah et al., 2011). In a second method, NET-Seq, ternary complexes containing RNA polymerase, the DNA template, and the nascent transcript are isolated by crosslinking, chromatin fragmentation, and polymerase immunoprecipitation, followed by high-throughput sequencing of cDNAs (Churchman and Weissman, 2011). Finally, metabolic labeling has been used to monitor the fate of nascent transcripts (Friedel and Dölken, 2009, and references therein). This method was recently employed to characterize clusters of co-expressed genes in mouse dendritic cells stimulated with lipopolysaccharide (LPS) (Rabani et al., 2011).
Here, we describe RNA-Seq analysis of biochemically fractionated chromatin-associated, nucleoplasmic, and cytoplasmic transcripts from unstimulated and stimulated macrophages. Like the three methods described above, this method provides basic information about nascent transcription. However, the results have provided several additional insights of broad relevance concerning the dynamic regulation of RNA processing and transport, and have allowed us to define, at high resolution, the properties of specific classes of co-expressed genes activated by an inflammatory stimulus.
EXPERIMENTAL PROCEDURES
Cell Culture, Cell Fractionation, and Library Preparation
Macrophages were prepared from the bone marrow of four week-old C57BL/6 or Ifnar−/− mice (Ramirez-Carrozzi et al., 2009). Adherent macrophages were activated on day six with lipid A (100ng/ml) (Invivogen).
Subcellular fractions were prepared as described (Pandya-Jones and Black, 2009), with minor changes. The cell lysis buffer contained 0.15% NP-40, and the sucrose cushion did not contain detergent. Fraction purity was confirmed by immunoblot analysis of SNRP70, beta-tubulin (Sigma), and histone H3 (Abcam). Cytoplasmic and nucleoplasmic RNA was purified using Qiagen RNeasy columns. Chromatin RNA was isolated using TRI-reagent (MRC), followed by further purification with RNeasy columns. All samples were eluted into 100 ul RNAse-free water. RNA (10 ug) from each fraction was depleted of rRNA using the Mouse/Human Ribominus kit (Invitrogen). Whole cell RNA was purified using TRI-reagent and RNeasy columns. Polyadenylated RNA was purified using the Micro-polyA purist kit (Ambion).
Strand-specific libraries were generated using 500 ng RNA input according to the “dUTP” method (Levin et al., 2010). Librarires from mRNA were not strand-specific. An Illumina HiSeq 2000 was used for sequencing with a single end sequencing length of 50 nt.
Sequence Mapping and Analysis
All bioinformatics analyses were conducted using the Galaxy platform except as noted (Goecks et al., 2010). Reads were aligned to the mouse mm9 reference genome with Tophat (Trapnell et al., 2010), using most default parameters. Alignments were restricted to uniquely mapping reads, with two possible mismatches permitted. RPKM values were calculated as described (Mortazavi et al., 2008) for mm9 Refseq genes (Pruitt et al., 2005) using Seqmonk (http://www.bioinformatics.bbsrc.ac.uk/projects/seqmonk/). Because chromatin transcripts were largely unspliced, RPKMs were calculated by counting all locus mapping reads and dividing by the length of the entire locus. RPKMs for nucleoplasmic and cytoplasmic transcripts and mRNAs were calculated by counting exonic reads and dividing by mRNA length. Coexpressed gene classes were generated with Cluster3 by applying k-means clustering to mean-centered log2(RPKM) expression values. Analysis of 5’ to 3’ read distribution trends was conducted using the Seqmonk probe trends tool. Splicing levels were analyzed by comparing the base coverage of exons alone divided by the base coverage of an entire locus; the calculations included exonic and intronic reads, as well as exon:exon and exon:intron junction spanning reads. This ratio was subsequently compared to three data points using the PERCENTRANK function in Excel: 0, X, and 1, where X is the ratio of exon length to locus length. Genome tracks were generated using the genome coverage utility in the BEDTools suite (Quinlan et al., 2010) and visualized in UCSC genome browser. Sequencing data have been submitted to GEO under accession GSE32916.
ChIP-Seq and Motif Analyses
ChIP-Seq datasets were obtained from GEO accession GSE17631 (De Santa et al., 2009). Datasets were converted from mm8 to mm9 genome builds using the liftover utility in Galaxy. Promoter enrichment was calculated as the height of peaks falling within a one kb window centered on the TSS.
Pscan was used to detect DNA motifs over-represented in each class between nucleotides −500 and +250 relative to the TSS (Zambelli et al., 2009). Significance was tested against CpG content matched promoters as background. A binding site was considered significantly over-represented with a p-value<0.01.
RESULTS
RNA-Seq Analysis of Fractionated Transcripts
Inflammatory gene transcription was studied in mouse bone marrow-derived macrophages stimulated with lipid A (the active component of LPS) for 0, 15, 30, 60, and 120 min. We fractionated cells (Wuarin and Schibler, 1994) to isolate chromatin-associated, nucleoplasmic, and cytoplasmic transcripts, as done previously to analyze nascent transcripts from individual genes (Dye et al., 2006; Pawlicki and Steitz, 2008; Pandya-Jones and Black, 2009). In pilot studies, the purity of the chromatin, nucleoplasm, and cytoplasm was assessed by Western blot analysis of β-tubulin, SNRP70, and histone H3, respectively (Figure S1A). To facilitate the analysis of nascent transcripts, an oligo dT purification step was omitted during the isolation of RNA from the three fractions. However, in separate experiments, an oligo dT resin was used to isolate polyadenylated RNA from whole-cell macrophage lysates.
Random primed, strand specific cDNA libraries prepared from the RNA populations were analyzed by high-throughput sequencing. Inspection of the aligned reads for individual genes provided strong support for the notion that the transcripts in the chromatin samples were newly transcribed. First, as illustrated for the inducible Nfkb1 gene (Figure 1A), the reads from the chromatin samples were broadly distributed across both the exons and introns of many genes, whereas in nucleoplasmic and cytoplasmic samples exon reads were strongly enriched. Reads were not observed at genomic regions flanking transcription units, demonstrating that the reads were derived from RNA rather than DNA. Reads from oligo dT-selected RNA showed a similar pattern of exon enrichment (Figure S1B). Second, transcript levels for inducible genes increased at earlier time points in the chromatin samples than in the nucleoplasmic and cytoplasmic samples (see Figure 1B for Nfkb1), or the mRNA (Figure S1C). This finding suggests that an analysis of chromatin transcripts provides a more accurate view of the kinetics of transcriptional induction than is provided by mRNA analyses.
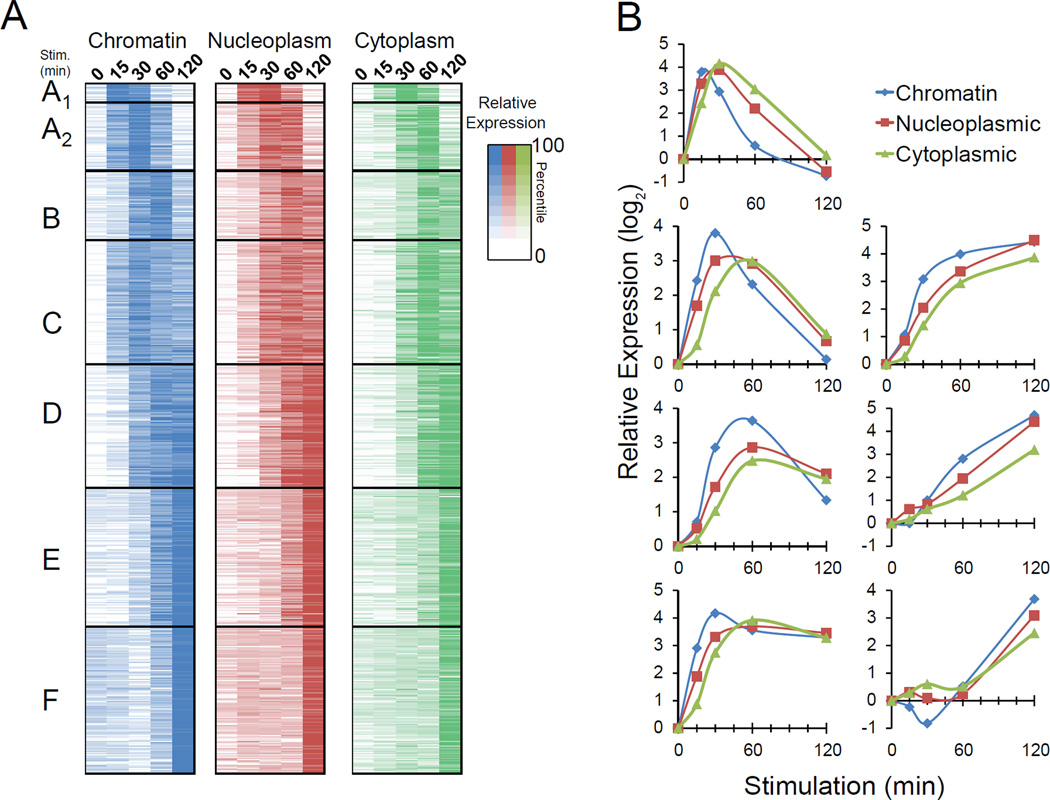
Figure 1. RNA-Seq Read Distributions and Induction Kinetics for Nfkb1 Transcripts.
(A) The distribution of RNA-Seq reads at the Nfkb1 locus is shown for libraries prepared from cytoplasmic, nucleoplasmic and chromatin-associated RNA. Time points and scale are indicated at the left. Exons and introns are shown at the bottom, and the TSS is indicated (bent arrow).
(B) Nfkb1 transcript levels in each of the three fractions through the time-course are shown as fold-induction values relative to the unstimulated transcript values (determined from RPKMs). See also Figure S1.
Distinct Sub-Cellular Transcript Profiles
We divided 13,199 expressed RefSeq genes (RPKM >1 in at least one of the three fractions) into twelve clusters based on their transcript profiles in the three subcellular fractions and five time points (Figure 2A). Most lipid A-induced genes were found in Clusters 1–3 (Figures 2A and 2B). Transcriptional downregulation was detected in Clusters 4–6 (Figures 2A and 2B). Surprisingly, far more genes were downregulated than upregulated (Figure 2C). However, the average magnitude of downregulation in two clusters (Clusters 4 and 6) was only slightly greater than 2-fold in the chromatin sample (Figure 2B). It is noteworthy that reduced transcript levels were observed in the chromatin samples at earlier time points than in the nucleoplasmic and cytoplasmic samples (Figures 2A and 2C). This difference is presumably due to the time required for pre-existing mRNAs to decay. The remaining clusters showed little change in transcript levels during the time course (Figures 2A and 2B, Clusters 7–12).
Figure 2. Distributions of Transcripts in the Three Subcellular Fractions.
(A) RefSeq genes exceeding 400-bp in length and exhibiting an RPKM of at least one in one of the samples were divided into 12 clusters on the basis of their pattern of transcript levels in the three subcellular fractions and five time points. Colors indicate the percentile of the relative expression level.
(B) The average relative transcript levels within representative clusters are shown. Although relative cluster-to-cluster differences are of interest, absolute transcript abundances in each sub-cellular compartment cannot be determined because of possible variability in RNA isolation efficiencies from the three fractions.
(C) The number and percentage (parenthesis) of genes within each of the 12 clusters in panel A are shown. See also Figure S2.
The cluster analysis also reveals extensive variability in the distribution of transcripts between the chromatin, nucleoplasm, and cytoplasm (Figure 2A). Although we cannot accurately calculate the percentage of transcripts for a gene in each of the three fractions, relative differences are apparent. Cluster 7, for example, contains abundant transcripts in the chromatin, with much lower transcript levels in the nucleoplasm and cytoplasm relative to the other clusters. This cluster is dominated by annotated and unannotated non-coding transcripts and miRNA precursors. (The analysis excluded transcripts shorter than 400 nucleotides and therefore excluded mature miRNAs and many miRNA precursors.) Some non-coding RNAs, such as Xist, are highly enriched in the chromatin because they function at this location (Figure S2). Examples of other non-coding RNAs in Cluster 7 are Neat1 and Malat1/Neat2 (Figure S2). Further mining of the data sets may provide insights into the subcellular locations at which many non-coding RNAs function. Cluster 7 also includes transcripts from constitutive protein-coding genes that may be highly unstable and therefore present at low levels in the cytoplasm (e.g. Leng8 in Figure S2).
Cluster 11, by contrast, contains abundant cytoplasmic transcripts, with fewer transcripts in the chromatin and nucleoplasm. This cluster includes genes that are likely to encode highly stable mRNAs, such as Actb (β-actin) and Tuba (α–tubulin).
Clusters 8 and 9 contain genes whose transcripts are more abundant in the nucleoplasmic fraction than the chromatin or cytoplasm. Many genes in these clusters encode proteins that would be expected to be translated by ribosomes associated with the endoplasmic reticulum (ER; see Chen et al. 2011). Thus, their enrichment in this fraction is likely due to the co-purification of ER-associated transcripts with the nuclear envelope.
Further analysis of individual transcripts or clusters of transcripts enriched in different subcellular fractions is likely to provide important insights into the role of RNA stability in gene regulation at a genome-wide scale, the functions of specific non-coding RNAs, and possibly other facets of RNA dynamics.
Fundamental Properties of the Lipid A-Induced Transcriptome
For this analysis, we focused on lipid A-induced genes. An examination of reads for approximately 26,000 Refseq genes (normalized as RPKM) revealed changes in chromatin-associated transcript abundance through the time course (Figure S3A). Examining genes with a read coverage of at least 1 RPKM at one of the stimulated time points, we found 560 genes that were induced in the chromatin samples at least 5-fold during the 2-hr stimulation period. 246 genes were induced between 5 and 10 fold, 247 between 10 and 100 fold, and 67 more than 100-fold (Figure S3B). Peak chromatin transcript values were reached at the 15-min time point for only 16 genes, with 145, 94, and 304 genes reaching their highest levels at 30, 60, and 120 min, respectively (Figure S3C). We also determined the time-point at which each gene exhibited its maximum fold-increase, or “ramp,” relative to the preceding time-point. Although only 16 genes exhibited their peak chromatin transcript level at the 15-min time point, 154 genes showed maximum ramp values within 15 min of stimulation (Figure S3C).
To examine the reproducibility of the data sets, Pearson correlation values (R values) were determined for chromatin-associated transcripts from two independent experiments performed several months apart with macrophages from different mice (Figure S4). R values derived from a comparison of RPKMs for inducible genes equaled or exceeded 0.94 at each time point (Figure S4A). Hierarchical clustering showed that each time point correlated more closely with the corresponding time point from the second experiment than with any of the other time points (Figure S4B). Similar results were obtained when all expressed genes or all RefSeq genes were examined rather than all induced genes (Figure S4C–F).
The 560 lipid A-induced genes were clustered into six classes, A through F, on the basis of the temporal profiles of their chromatin-associated transcripts (Figures 3A and 3B). We further subdivided class A into Classes A1 and A2 to highlight, in class A1, the 16 genes that exhibited peak chromatin transcript levels at the 15-min time-point in the two experiments. Interestingly, ten of these sixteen genes encode proteins that directly regulate gene expression: transcription factors (Fos, FosB, Jun, Atf3, Egr1, Egr3, and Nr4a1 [NUR77]), transcriptional co-regulators (Nfkbid [IκBNS] and Btg2), and an RNA-binding protein (Zfp36 [TTP]). The other six participate in diverse functions (Ppp1r15a, Plk3, Pmaip1, Ier2, Gdf15, Fabp4). Class A2 is largely composed of transiently induced genes that exhibit maximal chromatin transcripts 30 min post-stimulation. Most Class B genes peak at 60 min, with substantial decreases by 120 min. Class C genes, like Class A2 genes, are induced early and usually peak at the 30 min time point, but unlike the transient induction of Class A2 genes, transcription of Class C genes is sustained. Classes D, E, and F exhibit increasingly later induction of chromatin-associated transcripts (Figure 3B).
Figure 3. Kinetic Analysis of Lipid A-Induced Transcripts.
(A) Lipid A-induced genes were divided into six classes (A–F) on the basis of their chromatin-associated transcript profiles. Class A was divided into Classes A1 and A2 to highlight 16 genes that exhibit peak or near-peak transcript levels at the 15-min time point. Nucleoplasmic and cytoplasmic transcript levels were aligned on the basis of the clustering of chromatin transcripts. Transcript values are normalized to the average RPKM for each fraction. Colors indicate percentile values.
(B) The average fold induction within each class (y-axis) is shown for each time point (x-axis). Chromatin (blue), nucleoplasmic (red), and cytoplasmic (green) transcripts were analyzed separately. See also Figures S3, S4, and S5.
Progression of Transcripts from Chromatin to Nucleoplasm to Cytoplasm
To monitor the progression of transcripts from the chromatin to the nucleoplasm and cytoplasm, we visualized the profiles of individual genes and analyzed complete co-expression classes. Nucleoplasmic and cytoplasmic transcripts (and mRNAs) largely followed the same temporal profile as the chromatin transcripts, but with a clearly detectable delay (Figures 3B, 3B, and S5). These data are consistent with those obtained by metabolic labeling (Rabani et al., 2011). Thus, transcription appears to be a dominant regulator of mRNA expression kinetics. We have been unable to identify genes that exhibit large changes in mRNA levels without corresponding changes in nascent transcripts, which would be suggestive of regulation primarily at the level of mRNA stability.
Promoter and Chromatin Properties within Co-Expression Classes
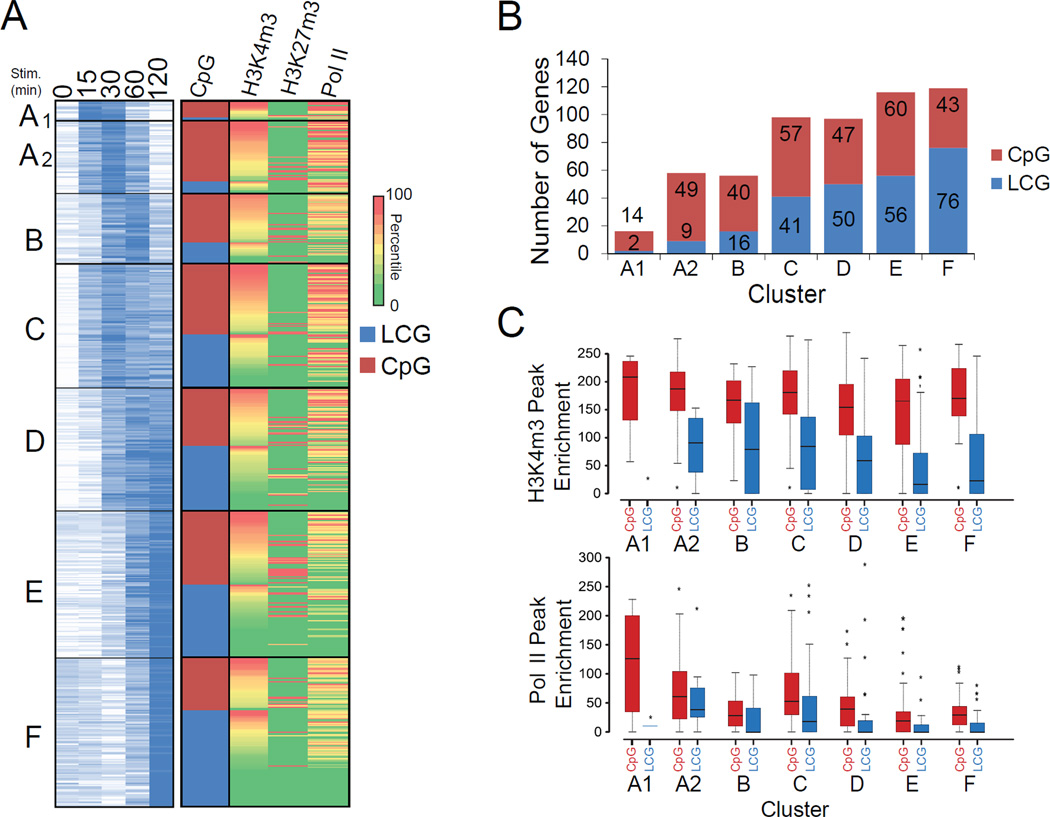
Our ability to define gene classes from the temporal profiles of nascent transcripts allowed us to examine promoter and chromatin properties within each class. Previous studies suggested that rapidly induced genes frequently contain CpG-island promoters, whereas secondary response genes and primary response genes induced with delayed kinetics usually contain low CpG (LCG) promoters (Hargreaves et al., 2009; Ramirez-Carrozzi et al., 2009). Consistent with these studies, 88, 84, and 71 percent of genes in Classes A1, A2, and B contain CpG-island promoters (Figures 4A and 4B). Interestingly, however, 36–58% of genes in the remaining classes also contain CpG-island promoters, which is higher than predicted from the previous studies of limited numbers of genes. It is noteworthy that a high prevalence of CpG-island promoters correlated more closely with transient induction than with rapid induction. That is, Classes A1, A2, and B contain a much higher percentage of CpG island promoters than Class C (Figure 4B); Class C is induced as rapidly as Class A2 and more rapidly than Class B, but Class C transcription is sustained, in contrast to the transient induction of Classes A1, A2, and B.
Figure 4. Promoter and Chromatin Properties of Co-Expressed Genes.
(A) The distribution of CpG-island promoters was determined for each class from Figure 3 on the basis of expression kinetics of chromatin-associated transcripts. Levels of histone H3K4me3, H3K27me3, and RNA polymerase II at the promoters in unstimulated cells is shown. ChIP-Seq values represent the signal within a 1 kb window centered on the TSS. Each column is normalized to the average enrichment at all inducible promoters, and color-coded according to percentile.
(B) The number of genes that contain CpG island (red) or LCG (blue) promoters within each co-expressed class is shown.
(C) The quantitative distributions of promoter-associated histone H3K4me3 (top) and RNA polymerase II (bottom) in unstimulated cells is shown for each class subdivided by promoter CpG content. The scale of the boxplots represents the intensity of ChIP-Seq peaks as determined in De Santa et al. (2009).
It has been suggested that CpG-island promoters facilitate rapid induction due to their assembly, in unstimulated cells, into chromatin with features commonly associated with transcriptionally active genes (Hargreaves et al., 2009; Ramirez-Carrozzi et al., 2009). Consistent with this hypothesis, histone H3 lysine 4 trimethylation (H3K4me3), a mark found at the promoters of active genes, was generally higher at CpG-island promoters than at LCG promoters in unstimulated cells (Figures 4A and 4C; ChIP-Seq data from De Santa et al., 2009). It is noteworthy, however, that this active chromatin mark was enriched at CpG-island promoters in all classes. Similarly, RNA polymerase II association was more prevalent in unstimulated cells at CpG island promoters than at LCG promoters in all of the classes (Figures 4A and 4C).
An examination of the repressive histone H3K27me3 modification revealed association with a small fraction of promoters within all classes (Figure 4A). Interestingly, this repressive mark appears to be most prevalent in CpG-island promoters that exhibit relatively low levels of the H3K4me3 mark.
Together, these results confirm that rapidly induced genes frequently contain CpG-island promoters, high levels of H3K4me3, and RNA polymerase II association in unstimulated cells. However, these features were also found in many genes with delayed induction kinetics. A more detailed examination of the distinct regulatory capabilities conferred by CpG-island and LCG promoters is presented below.
Dynamic Range of Inducible Transcription
It has been proposed that rapidly induced genes with active chromatin features in unstimulated cells are transcribed at relatively high levels prior to cell stimulation, but that the elevated transcription does not lead to elevated mRNA levels because transcription elongation and RNA processing are inefficient prior to stimulation (Hargreaves et al., 2009). The RNA-Seq analysis of chromatin-associated transcripts allowed us to test this hypothesis.
An examination of RPKMs revealed that basal nascent transcript levels span more than three orders of magnitude within most classes (Figure 5A). The median basal RPKMs were similar for Classes A2 through F, whereas the median RPKM for the 16 genes in Class A1 was moderately higher. The significance of this higher median value is difficult to determine due to the small size of Class A1. Little difference was observed between genes containing CpG-island promoters and LCG promoters (Figure 5A).
Figure 5. Dynamic Ranges of Expression within Co-Expression Classes.
(A) Chromatin-associated transcript levels (RPKMs) in unstimulated macrophages are shown for all genes in each class. Genes containing CpG-island promoters (diamonds) and LCG promoters (stars) are shown separately. The median transcript level for each group is indicated by a black bar.
(B) Chromatin-associated transcript levels (RPKMs) at their peak time point are shown for all genes in each class, with CpG-island and LCG promoters shown separately.
(C) The maximum fold-induction of chromatin transcripts for each gene is shown, by dividing the peak RPKM by the RPKM in unstimulated cells. Representative genes with LCG promoters that exhibit strong activation are indicated.
(D) The numbers of genes in each class activated by more than 100-fold and by only 5–10-fold are shown.
(E) Over-represented transcription factor binding sites are shown for each class. Data are presented after hierarchical clustering. CpG-island and LCG promoters were analyzed separately. Transcription factor families are shown at the right (see Figure S6A for details). Color intensity is proportional to the negative log(p-value). Single and double stars indicate p-values < 0.01 and < 0.001, respectively. See also Figure S6.
An examination of RPKMs at the nascent transcript peak for each gene (i.e. the RPKM at the time point with the highest nascent transcript level) also revealed similarities between all classes (Figure 5B). However, the median peak RPKMs for LCG genes in Classes D, E, and F were approximately 3-fold higher than the RPKMs for CpG-island genes in the same classes.
Most interestingly, the median fold-induction values for LCG genes in Classes D and E were substantially higher than for CpG-island genes (Figure 5C). Of particular relevance, LCG promoters are highly prevalent among genes induced by more than 100-fold, whereas CpG-island promoters are more prevalent among genes that are weakly induced (5–10-fold) (Figures 5C and 5D). Similar profiles were observed when cytoplasmic RNA or mRNA were examined (data not shown), demonstrating that these differences in nascent transcripts lead to similar distributions in mature mRNA.
These findings explain why LCG promoters were found to be more prevalent during the secondary response in previous studies (Ramirez-Carrozzi et al., 2009; Hargreaves et al., 2009). The previous studies focused on limited numbers of genes that encode critical regulators of immunity, such as Il6, Nos2, and Il12b. Nascent transcripts for these genes were induced in this study by 700-, 1300-, and 1600-fold, respectively (see Figure 5C), placing them in a fold-induction range dominated by genes with LCG promoters. The unusually potent induction of these and other LCG genes may be related to their key roles in regulating innate and adaptive immune responses.
Together, these results reveal a broad range of basal and peak nascent transcript levels, with LCG promoters strongly enriched among genes that are most potently induced. We propose that the absence of H3K4me3 and the absence of associated RNA polymerase II at LCG promoters in unstimulated cells contributes to potent induction by limiting basal transcription from promoters that have evolved to support particularly high levels of transcription following stimulation. CpG-island promoters can support either high or low levels of transcription, but they rarely need to support the potent induction observed at many LCG genes.
Importantly, the quantitative RNA-Seq results fail to support models which proposed that rapidly induced genes with CpG island promoters are transcribed at high levels in unstimulated cells, with mRNA induction due primarily to stimulus-dependent enhancement of RNA processing. Additional results presented below (Figure 6) provide further support for the view that inducible genes are transcribed at variable basal levels, with increased transcription initiation/promoter release playing a prominent role in the induced expression of all or almost all genes. Our findings are not incompatible with models in which cell stimulation enhances transcription elongation and/or the rate of RNA processing. However, we propose that any enhancement of elongation and processing that occurs following stimulation primarily serves the purpose of promoting the efficient expression of genes that are also strongly upregulated at the level of transcription initiation and/or promoter release.
Figure 6. High Prevalence of Completed Transcripts Cleaved at the Polyadenylation Site in the Chromatin Samples.
(A) Normalized read distributions from the 5’ to 3’ end of all genes within each class are shown for each time point. Data from chromatin-associated transcripts were used and genes were normalized for length. Relative gene position is shown on the x-axis and cumulative reads at each relative position are shown on the y-axis.
(B) Cumulative read distributions spanning the polyadenylation site are shown for lipid A-induced genes. The x-axis shows the gene position relative to the polyadenylation cleavage site and the y-axis shows cumulative reads at each position.
(C) Read profiles are shown for chromatin-associated transcripts at the Il12b locus. Time points and scale are shown at the left and exon-intron structure of the locus is at the bottom. In the nucleoplasmic and cytoplasmic fractions, abundant exonic reads accumulated by the 60-minute time point (not shown).
Class-Specific Differences in Transcriptional Regulation
The high-resolution temporal profiles of nascent transcripts provide a step towards identifying signaling pathways and transcription factors that regulate genes within each co-expression class. To gain insight into factors that may regulate specific classes, a motif enrichment analysis was carried out with promoter sequences (Figures 5E and S6A). NF-κB motifs were over-represented in most classes, consistent with the expectation that this factor contributes to the regulation of many lipid A-induced genes. In addition, binding sites for factors within the ATF, JUN, and CREB families were strongly enriched in Class A1 and A2 promoters. These motifs were more strongly enriched than NF-κB motifs in these classes, whereas NF-κB motifs were strongly over-represented in Class C. Genes in all three classes were potently induced at the 15-min time point, but Classes A1 and A2 were transiently induced whereas Class C transcription was sustained (see Figure 3). These results suggest that different sets of factors may coordinate the transient and sustained responses to TLR4 signaling.
The motif analysis also uncovered a high prevalence of STAT and IRF binding sites within the promoters of genes in Classes E and F (Figure 5E). Genes containing these motifs may be direct targets of interferon signaling. IFN-β is potently induced by lipid A and can induce IFN response genes through the Type 1 interferon receptor, IFNAR (Vaidya and Cheng, 2003). To identify IFN-dependent genes, we performed RNA-Seq analysis with mRNA from wild-type and Ifnar−/− macrophages stimulated with lipid A. Figure S6C highlights genes whose mRNA levels were reduced by 3–10-fold (orange) or greater than 10-fold (red) in the Ifnar−/− macrophages compared to wild-type. Consistent with the results of the motif analysis, 94% of the genes that exhibit at least 3-fold dependence on IFNAR were located in classes E and F. Also consistent with the motif analysis, IFNAR-dependence was biased towards genes with LCG promoters (Figures S6B and S6C). These results are consistent with previous suggestions that, unlike the primary responses to LPS and TNF, the primary response to IFN favors LCG promoters (Ramirez-Carrozzi et al., 2009).
Accumulation of Full-Length, Incompletely Spliced Transcripts in the Chromatin Fraction
As discussed above, the contributions of promoter-proximal pausing, transcription elongation, and transcript processing to the regulation of inducible transcription are of considerable interest. The RNA-Seq data from chromatin-associated transcripts allowed an examination of transcript dynamics, by analyzing the read density across inducible transcription units over time. An initial expectation was that read density would exhibit a decreasing 5’ to 3’ gradient across actively transcribed genes. This gradient was expected because 5’ end reads will be generated by both partial and complete transcripts, whereas 3’ reads will be generated only by complete or nearly complete transcripts. We also expected to observe read peaks at the 5’ end due to promoter-proximal pausing.
As shown in Figure 1 for the Nfkb1 gene, a clear 5’-to-3’ gradient was detected at the 15- and 30-min time points in the chromatin fraction. However, a gradient was not observed in the unstimulated cells, despite substantial basal transcription prior to stimulation (see Figure S1B). The gradient was also absent at the 60- and 120-min time points, after transcription had reached its peak. These results reveal that, during steady-state transcription (prior to induction or after transcription had reached its peak), full-length transcripts accumulated on the chromatin and that release from the chromatin was slow relative to the completion of transcription. Moreover, many of these transcripts remained incompletely spliced (see below).
The features described above for Nfkb1 were apparent when genes of each class were analyzed together, after normalizing the lengths of all genes (Figure 6A). For example, in Class A2, a 5’-to-3’ gradient was observed at the 15-min time point, which is the point at which transcripts were beginning to accumulate. By the 30-min time point, the gradient had disappeared. The nascent transcript levels declined at the 60 and 120-min time points, but without a 5’-to-3’ gradient. The only class that failed to show evidence of a gradient is Class A1. This class is notable for its extremely rapid induction and the short length of its genes (1–4 kb, data not shown). Genes of this length would be predicted to be fully transcribed well before the 15-min time point.
These results suggest that, following the completion of transcription, a lag period exists prior to transcript release. Splicing may be completed during this lag period, as introns are abundant on the chromatin but scarce in the nucleoplasm and cytoplasm. Consistent with the hypothesis that completed transcripts accumulate on the chromatin, we found that sequence reads for most genes in the chromatin fraction declined dramatically at the polyadenylation site. This is apparent in Figure 6B, which shows the read distributions across the RefSeq polyadenylation sites for all inducible genes. Similar profiles were observed when polyadenylated mRNA was analyzed, providing additional evidence that the decline observed in the chromatin fraction corresponds to cleavage of completed transcripts at the polyadenylation site (data not shown). RNA-Seq analysis of oligo dT-primed cDNA libraries revealed that polyadenylated transcripts are abundant on the chromatin (data not shown). However, only a fraction of the unspliced transcripts cleaved at the polyadenylation site appeared to be polyadenylated (data not shown), with the precise percentage of polyadenylated transcripts difficult to determine.
The data described above suggest that completed transcripts accumulated at many genes in the chromatin fraction and that these transcripts were usually cleaved at the polyadenylation site but incompletely spliced. Additional insight into transcript dynamics was obtained from an examination of transiently induced genes after transcription had subsided. As shown for the Il12b gene in Figure 6C, intronic reads were lost more rapidly than exonic reads from the chromatin fraction after transcription subsided. At the 60-minute time point, abundant Il12b transcripts were present on the chromatin (Figure 6C), and also in the nucleoplasm and cytoplasm (data not shown); in the chromatin fraction, reads were abundant in both the exons and introns. Il12b transcription peaked between 60 and 120 minutes and then subsided by the 120-minute time point. At this latter time point, exon reads were far more abundant than intron reads (Figure 6C), suggesting that excised introns were released from the chromatin and/or degraded more rapidly than spliced transcripts were released. These results demonstrate that excised introns are not stably maintained on the chromatin and suggest that the incompletely spliced transcripts observed at Il12b and many other genes are precursors of fully spliced transcripts (see Discussion).
To examine splicing efficiency of chromatin-associated transcripts in greater depth, we determined the ratio of exon reads to total reads for genes within each constitutive and inducible class from Figures 2A and 3A. Using constitutively expressed Cluster 11 as an example (Figure 7A, blue), we found that, in the chromatin fraction, most genes were incompletely spliced. In Figure 7A, the genes are ordered on the basis of splicing level. Genes at the far left correspond to those whose chromatin-associated transcripts are efficiently spliced, as they primarily have exon reads in the chromatin fraction. Genes at the far right correspond to those with greater numbers of reads in introns than exons; examining several of these genes revealed that they contain aberrant intron read spikes that are likely to be derived from additional transcripts from the locus (data not shown). Between these extremes, the large majority of genes were comparable to Nfkb1 in that only a moderate fraction of introns had been excised from the pool of transcripts. In contrast to the incomplete splicing observed in the chromatin fraction, efficient splicing was observed at almost all genes in the nucleoplasm and cytoplasm (Figure 7A). Similar results were obtained with each constitutive and inducible class (data not shown).
Figure 7. Analysis of Exon:Intron Ratios and a Comparison of Nascent Transcript Methods.
(A) Reads mapping to approximately 1,450 genes from Cluster 11 (Figure 2) were analyzed for relative exon and intron coverage. The genes were rank-ordered according to splicing levels (length-normalized exon reads over total reads) in the chromatin fraction (blue), and were compared to splicing levels in the nucleoplasm (red) and cytoplasm (green). Similar results were obtained with all gene clusters (data not shown).
(B) RNA-Seq read distributions at Nfkb1 are shown comparing reads from the cytoplasmic (green), nucleoplasmic (red), and chromatin (blue) fractions to data obtained by 4sU-Seq and GRO-Seq (Rabani et al. 2011; Escoubet-Lozach et al. 2011). GRO-Seq was performed with LPS-stimulated bone marrow-derived macrophages. 4sU-Seq was performed with LPS-stimulated bone marrow-derived dendritic cells. The fifth track displays the 4sU-Seq data with a scale of 15 reads to show the reduced number of reads after the polyadenylation site relative to the last intron. The cleavage site (red arrow) and reads representing transcription past this site (green bracket; abundant only in the GRO-Seq data set) are indicated. See also Figure S7.
As mentioned above (Figure 6B), the decline in reads at the polyadenylation site suggests that most transcripts in the chromatin fraction are full-length and cleaved at the polyadenylation site, despite the incomplete splicing. Additional evidence that incompletely spliced transcripts are frequently cleaved at the polyadenylation site is provided by published RNA-Seq data obtained using the metabolic labeling (4sU-Seq) and global nuclear run-on (GRO-Seq) methods (Rabani et al., 2011; Escoubet-Lozach et al., 2011). Figure 7B shows the read profiles obtained with these methods at Nfkb1 in dendritic cells (4sU-Seq) or macrophages (GRO-Seq) stimulated with LPS for 1 hr. The 4sU profile was similar to the profile observed with chromatin-associated transcripts; a modest 5’ to 3’ gradient was observed, similar to the gradient observed in Figure 1A at the 15 and 30 min time points with the chromatin transcripts. The 4sU profile also shows a dramatic decline in reads at the polyadenylation site, despite abundant intronic reads throughout the gene. This decrease is most apparent in the fifth track of Figure 7B, in which the 15-read scale revealed greatly reduced read numbers after the polyadenylation site in comparison to the final intron. Thus, both the 4sU method and the analysis of chromatin-associated transcripts revealed a high abundance of incompletely spliced transcripts that had been cleaved at the polyadenylation site.
In contrast to the read profiles obtained with the 4sU-Seq and chromatin-associated transcript methods, abundant reads extending past the polyadenylation site were observed by GRO-Seq (Figure 7B). This finding is consistent with the knowledge that polymerase molecules continue transcription past the polyadenylation site and that these transcripts can be captured in the short labeling time of the GRO-Seq experiment.
Finally, although reads in our chromatin fraction declined at the polyadenylation site on most genes, exceptions were apparent. One example is the Mapkapk2 gene, where reads can be detected more than 20 kb downstream of the gene 30 min after induction (Figure S7). Reads from apparent Mapkapk2 transcripts decline gradually with distance from the gene and extend into the neighboring Idl10 (IL-10) gene; Il10 is transcribed on the opposite strand and is also induced by many stimuli (Figure S7). It is difficult to predict at this time whether the extensive read-through transcription observed at this gene has functional relevance or simply reflects the fortuitous absence of transcription termination sequences.
DISCUSSION
We describe the use of RNA-Seq to examine chromatin-associated, nucleoplasmic, and cytoplasmic transcripts in macrophages exposed to an inflammatory stimulus. This approach allowed us to obtain novel insights into transcription and RNA dynamics, and to examine the inflammatory gene transcription program at high resolution. Although other methods have been developed for genome-wide analysis of nascent transcripts, the experimental strategy described here is unique in its ability to provide information about the properties of transcripts that remain associated with chromatin and the changes that occur as the transcripts proceed to the nucleoplasm/ER or cytoplasm.
At most protein-coding genes, full-length transcripts appear to accumulate on the chromatin during steady state transcription, and these transcripts are often cleaved at the polyadenylation site but incompletely spliced. The retention of polyadenylated yet incompletely spliced transcripts at a gene locus has been observed previously in an analysis of integrated DNA constructs (Brody et al., 2011). Our results demonstrate that such transcripts accumulate at most endogenous genes in mammalian cells. Fully spliced transcripts were predominant within the nucleoplasm and cytoplasm, suggesting that the temporal delay in transcript release could allow the completion of splicing. Such a delay may be indicative of a quality control step ensuring full processing prior to release (Schmid and Jensen, 2010). It is noteworthy that the splicing delay is not indicative of when spliceosome assembly occurs. Spliceosome assembly may be linked to active transcription, with elongating transcripts assembling factors at exon-intron junctions as the RNA polymerase proceeds along the transcription unit. (Perales and Bentley, 2009).
Previous studies have provided strong evidence of co-transcriptional splicing (Singh and Padgett, 2009; Perales and Bentley, 2009). Other studies have indicated that splicing can follow the completion of transcription (Nevins and Darnell, 1978; Vargas et al., 2011). Our results demonstrate that, in mammalian cells, the completion of splicing often follows the completion of transcription, although some genes accumulate chromatin-associated transcripts that have been efficiently spliced, consistent with co-transcriptional splicing. One recent study analyzed chromatin-associated transcripts in Drosophila at steady-state and reported evidence of more extensive co-transcriptional splicing (Khodor et al., 2011). It is possible that co-transcriptional splicing is more prevalent in some species than others. However, differences in the experimental procedures and analysis methods may also have influenced the interpretation of the results. In particular, our finding that full-length transcripts frequently accumulate on the chromatin was strongly dependent on the analysis of inducible transcription; the dynamic transition from a read distribution lacking a 5’ to 3’ gradient to one exhibiting a gradient during transcriptional induction to one in which the gradient is again lost (Figure 6A) provided clear evidence of the accumulation of full-length transcripts. Furthermore, Khodor et al. (2011) depleted polyadenylated transcripts from their chromatin fraction prior to RNA-Seq analysis, which may have obscured the abundance of full-length transcripts that are incompletely spliced.
One important question that must be considered when interpreting our RNA-Seq data is whether the abundant full-length, incompletely spliced transcripts that accumulate on the chromatin are precursors to productive fully-spliced mRNA, or whether they might instead by non-productive “dead-end” transcripts. The rapid increase and decrease in chromatin transcripts at transiently induced genes (Classes A1, A2, and B in Figure 3) argues against the existence of a large pool of non-productive chromatin-associated transcripts. The hypothetical non-productive transcripts would need to be eliminated from the chromatin at the same rate as the release of productive transcripts. The fact that the increase in chromatin transcripts is followed by an increase in nucleoplasmic and cytoplasmic transcripts, with a defined temporal delay, further supports the notion that the chromatin transcripts are precursors to mRNA. In fact, since chromatin transcripts are not maintained on transiently induced genes for a prolonged period, the hypothetical full-length non-productive transcripts would need to be far more abundant than the pool of nascent productive transcripts to obscure the 5’ to 3’ read gradient generated by these nascent transcripts. Finally, the results in Figure 6C suggest that the full-length incompletely spliced transcripts are competent for the completion of splicing, as the incompletely spliced transcripts resolve into transcripts containing only exons after transcription has subsided.
It was also necessary to consider the possibility that the broad read coverage of both exons and introns on the chromatin may be due to the stable maintenance of introns that were excised immediately after transcription. However, our data suggest that excised introns are short-lived on the chromatin. For example, at the Fos gene, intron reads peaked at the 15-minute time point and decreased by 8-fold at the 30-minute time point (data not shown), suggesting that introns were released from the chromatin and/or degraded with a half-life of approximately five minutes. Fos exon reads decreased by only 3-fold during these 15 minutes, indicating that introns were lost from the chromatin more rapidly than exons. Introns were also lost more rapidly than the fully spliced RNA at the Il12b gene (Figure 6C) and at many other genes (data not shown) after transcription had declined, possibly because the spliced transcripts were not released until all introns had been excised. A detailed analysis of the kinetics of intron and exon accumulation and loss relative to the kinetics of splice exon-exon junction accumulation provides additional evidence of a splicing delay following transcription through an intron (A.P.J. and D.L.B., unpublished results).
Our results and the results of Rabani et al. (2011) highlight the importance of transcriptional induction kinetics in shaping the temporal mRNA profiles of lipid A-induced genes. Although genes could not be identified that are induced primarily via enhanced mRNA stability, mRNA stability must act in concert with transcriptional control to shape the temporal expression profile of each gene. Since the transient induction of nascent transcripts (i.e. transcription) for genes in Classes A and B is mirrored by the transient induction of the mRNA, these transcripts must either be intrinsically unstable or their stability must be tightly regulated (Caput et al., 1986; Hao and Baltimore, 2009). Conversely, mRNAs from genes that exhibit sustained induction are likely to be intrinsically stable or may be stabilized in stimulated cells. Thus, our results support the hypothesis that the temporal dynamics of gene expression required the co-evolution of mechanisms regulating transcription and mRNA stability.
In previous studies, we defined chromatin and promoter properties of 67 genes by qRT-PCR (Ramirez-Carrozzi et al., 2009). By RNA-Seq, we can now discern 560 genes whose nascent transcripts are activated by lipid A at least 5-fold within 2 hrs of stimulation. By classifying genes on the basis of nascent transcript kinetics, we found that binding sites for transcription factors in the ATF, JUN, and CREB families are abundant in the most rapidly and transiently induced genes and that IFN-dependent genes are tightly clustered in two gene classes whose nascent transcripts are induced at later times. The framework provided by classifying genes on the basis of their nascent transcript profiles will greatly aid future efforts to understand how a broad range of signaling pathways, transcription factors, and chromatin events shape the inflammatory gene transcription program.
Finally, the current analysis provides genome-scale confirmation of previous models of inflammatory gene activation, while suggesting revisions of other models. The results confirm that a high percentage of genes that are rapidly induced by a TLR stimulus contain CpG-island promoters. In unstimulated cells, these promoters exhibit chromatin features of active genes and can be activated in a nucleosome remodeling-independent manner (Ramirez-Carrozzi et al., 2009). The results also support the previous finding that IFN-induced genes are biased towards LCG promoters, which are not assembled into constitutively active chromatin, consistent with the view that IFN-induced transcription factors promote nucleosome remodeling (Huang et al., 2002; Liu et al., 2002).
However, in contrast to previous suggestions, our results reveal that both CpG-island and LCG promoters are abundant among genes induced at late times. One important distinction is that LCG promoters are highly enriched among the most potently induced genes, with CpG-island promoters more prevalent among weakly induced genes. The CpG-island promoters associated with late genes possess features of active chromatin in unstimulated cells, and are thus similar to CpG-island promoters associated with early genes. We propose that late genes containing CpG-island promoters are poised for induction just like the early genes, but that transcriptional induction of the late genes requires transcription factors or signaling pathways that are expressed following the primary response to the stimulus. These genes may tolerate constitutively active chromatin because the required dynamic range of expression between the unstimulated and stimulated states is relatively small. In contrast, genes that require a large dynamic range of expression often contain LCG promoters. An LCG promoter may help limit basal transcription and, at the same time, facilitate tight regulation by conferring a requirement for an inducible nucleosome remodeling event. The ability to perform high-resolution genome-scale analysis of nascent transcripts should facilitate future efforts to further dissect and understand inflammatory gene regulation networks.
Supplementary Material
Highlights.
Coding and non-coding transcripts exhibit characteristic subcellular distributions
The most potently induced genes favor promoters with low CpG content
Full-length, incompletely spliced transcripts accumulate on the chromatin
Delayed transcript release may reflect a requirement for the completion of splicing
ACKNOWLEDGMENTS
We thank Christopher Glass for sharing results prior to their publication, Alexander Hoffmann, Steve Horvath, Justin Langerman, Steve Ley, Chia-Ho Lin, and Matteo Pellegrini for helpful discussions, and Owen Witte for invaluable support. This work was supported by a UCLA Dissertation-Year Fellowship (to A.P.J.), by NIH grants R01GM086372 and R01CA127279 (to S.T.S.), and by the Broad Stem Cell Research Center at UCLA. D.L.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc. Natl. Acad. Sci. USA. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science. 2009;326:257–263. doi: 10.1126/science.1179050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody Y, Neufeld N, Bieberstein N, Causse SZ, Bohnlein EM, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Identification of a common nucleotide sequence in the 3’-untranslated region of mRNA molecules specifying inflammatory mediators. Proc. Natl. Acad. Sci. USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Jagannathan S, Reid DW, Zheng T, Nicchitta CV. Hierarchical regulation of mRNA partitioning between the cytoplasm and the endoplasmic reticulum of mammalian cells. Mol. Biol. Cell. 2011;22:2646–2658. doi: 10.1091/mbc.E11-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–373. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola S, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Gromak N, Proudfoot NJ. Exon tethering in transcription by RNA polymerase II. Mol. Cell. 2006;21:849–859. doi: 10.1016/j.molcel.2006.01.032. [DOI] [PubMed] [Google Scholar]
- Escoubet-Lozach L, Benner C, Kaikkonen MU, Lozach J, Heinz S, Spann NJ, Crotti A, Stender J, Ghisletti S, Reichart D, et al. Mechanisms Establishing TLR4-Responsive Activation States of Inflammatory Response Genes. PLoS Genet. 2011;7:e1002401. doi: 10.1371/journal.pgen.1002401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler T, Sen R, Roy AL. Regulation of primary response genes. Mol. Cell. 2011;44:348–360. doi: 10.1016/j.molcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel CC, Dölken L. Metabolic tagging and purification of nascent RNA: implications for transcriptomics. Mol. Biosyst. 2009;5:1271–1278. doi: 10.1039/b911233b. [DOI] [PubMed] [Google Scholar]
- Goecks J, Nekrutenko A, Taylor J. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biology. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hah N, Danko CG, Core LJ, Waterfall JJ, Siepel A, Lis JT, Kraus WL. A rapid, extensive, and transient transcriptional response to estrogen signaling in breast cancer cells. Cell. 2011;145:622–634. doi: 10.1016/j.cell.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Qian F, Hu Y, Ang C, Li Z, Wen Z. Chromatin-remodeling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat. Cell Biol. 2002;4:774–781. doi: 10.1038/ncb855. [DOI] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25:2502–2012. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A, Regev A. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods. 2010;7:709–715. doi: 10.1038/nmeth.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V, Ramsey SA, Rust AG, Zak DE, Kennedy K, Lampano AE, Nykter M, Shmulevich I, Aderem A. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat. Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Kang H, Liu R, Chen X, Zhao K. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 2002;22:6471–6479. doi: 10.1128/MCB.22.18.6471-6479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins JR, Darnell JE., Jr. Steps in processing of Ad2 mRNA: poly(A)+ nuclear sequences are conserved and poly(A) addition precedes splicing. Cell. 1978;15:1477–1493. doi: 10.1016/0092-8674(78)90071-5. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–1908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Bentley D. “Cotranscriptionality”: the transcription elongation complex as a nexus for nuclear transactions. Mol. Cell. 2009;36:178–191. doi: 10.1016/j.molcel.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, Amit I, Regev A. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 2011;29:436–442. doi: 10.1038/nbt.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST. Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey SA, Klemm SL, Zak DE, Kennedy KA, Thorsson V, Li B, Gilchrist M, Gold ES, Johnson CD, Litvak V, et al. Uncovering a macrophage transcriptional program by integrating evidence from motif scanning and expression dynamics. PLoS Comput. Bio. 2008;4:e1000021. doi: 10.1371/journal.pcbi.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Jensen TH. Nuclear quality control of RNA polymerase II transcripts. Wiley Interdiscip. Rev. RNA. 2010;1:474–485. doi: 10.1002/wrna.24. [DOI] [PubMed] [Google Scholar]
- Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST. Selective transcription in response to an inflammatory stimulus. Cell. 2010;140:833–844. doi: 10.1016/j.cell.2010.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya SA, Cheng G. Toll-like receptors and innate antiviral responses. Curr. Opin. Immunol. 2003;15:402–407. doi: 10.1016/s0952-7915(03)00070-0. [DOI] [PubMed] [Google Scholar]
- Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell. 2011;147:1054–1065. doi: 10.1016/j.cell.2011.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambelli F, Pesole G, Pavesi G. Pscan: finding over-represented transcription factor binding site motifs in sequences from co-regulated or co-expressed genes. Nucleic Acids Res. 2009;37:W247–W252. doi: 10.1093/nar/gkp464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.