Abstract
This study determined the survivability of methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) for extended periods of time and temperatures using a standard swab for assessment. Our study showed that transportation in Liquid Amies medium could be performed at room temperature or 4°C for up to 14 days without a decrease in recovery of MRSA or VRE.
TEXT
Consolidation of laboratories is a cost-containment measure that has made available to physicians a wider variety of laboratory tests (9). However, with consolidation, the time of transport of microbiological specimens to clinical laboratories within the institution or to other institutions has increased. It is essential to transport these specimens in a fashion that does not adversely affect the number of viable organisms or their morphology.
Appropriate collection and optimal transportation of specimens are important to achieving accurate clinical laboratory results. Swab transport systems have often filled this need when specimens are not expected to be processed quickly (6, 14). The swab transport system must be able to keep organisms viable as well as maintain organisms at a relatively high proportion of what was initially obtained (8, 10, 11, 13, 14). Optimal time and the method of transportation differ for each organism due to the differences in survivability of each bacterial species.
The objective of this study was to determine the survivability of clinical strains of methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) for variable periods of time and temperatures that might be encountered during transport using a standard microbiological specimen swab and Clinical Laboratory and Standards Institute (CLSI) guidelines for assessment.
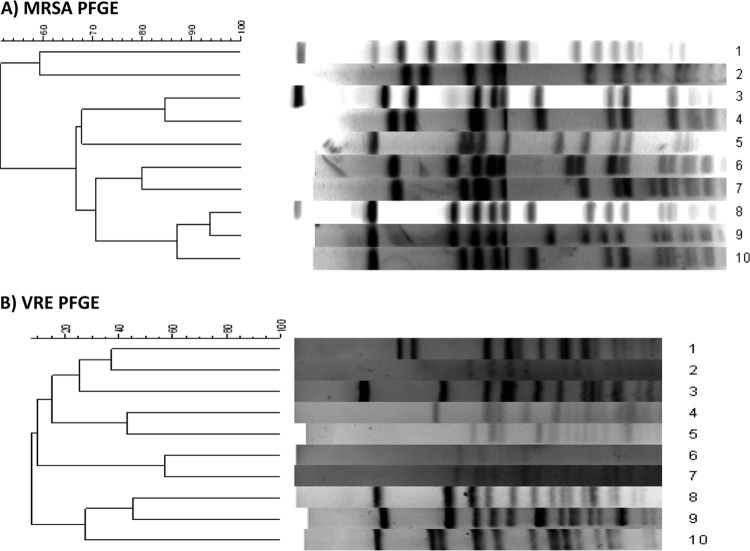
This study was approved by the University of Maryland, Baltimore Institutional Review Board. Isolates were collected from the nares (MRSA) and perirectal area (VRE) of patients in intensive care units at the University of Maryland Center and Kernan Hospital. Isolates were identified using standard laboratory protocol, and susceptibilities were determined using CLSI guidelines (1). Pulsed-field gel electrophoresis (PFGE) was performed to identify clinically unique isolates of MRSA (5) and VRE (2). Isolates were characterized as unique if PFGE patterns of isolates had a band difference greater than 2 (see Fig. 1) (12). In this study, 10 unique isolates of MRSA and 10 unique isolates of VRE were used to inoculate swabs for assessment of survivability.
Fig 1.
Pulsed-field gel electrophoresis to confirm that the strains of MRSA and VRE selected for the study were unique (all had a >2-band difference by Tenover criteria).
The swab elution method described in the M40-A NCCLS guidelines (7) was used to inoculate BD BBL CultureSwab Liquid Amies, Single Swab (Becton, Dickinson, Sparks, MD). Swabs were inoculated with 100 μl of a 1:10 dilution of a 0.5 MacFarland inoculum made for each organism, giving a total of 45 samples for each isolate. The 45 samples included 3 for baseline culture as well as 3 cultures for each of the 2 temperatures (room temperature and 4°C) on days 2, 4, 6, 8, 10, 12, and 14. The swabs were then stored at either room temperature or 4°C for up to 14 days to mimic conditions encountered during transport.
Swabs were vortexed in 1 ml of 0.85% saline before successive 10-fold dilutions were made, and 100 μl was plated in duplicate onto tryptic soy agar (Sigma-Aldrich, Switzerland). Colony counts were performed on each plate after overnight incubation. Growth at each temperature and time point was compared to the baseline performance of the swab for that particular organism to assess whether there was an increase or decrease in CFU. According to CLSI guidelines, there should be no more than a 3 log10 decrease or a 1 log10 increase in CFU between baseline counts and counts of the swabs stored at different temperatures (7).
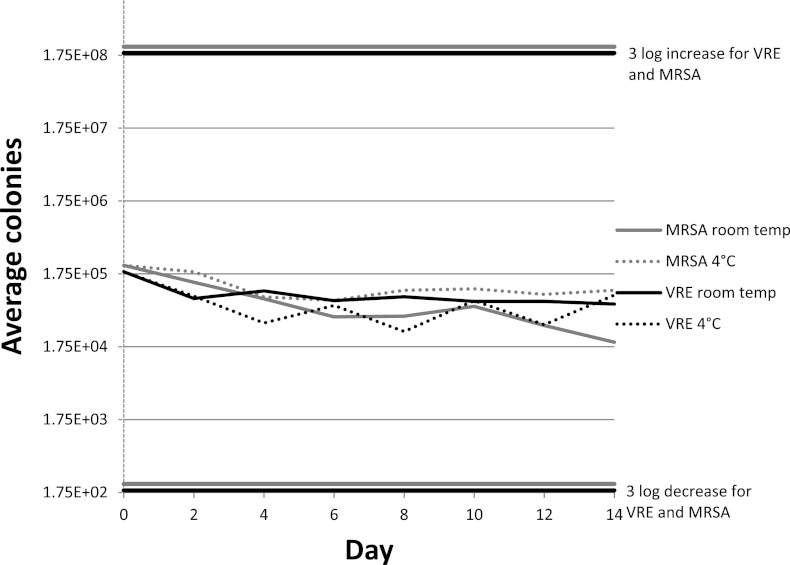
At baseline, 2.29 × 105 CFU/ml for MRSA and 1.87 × 105 CFU/ml for VRE were extracted from the swabs. The swabs containing unique strains of MRSA and VRE remained stable, as demonstrated by a change of less than 3 log10 compared to baseline counts (see Fig. 2). At the day 14 endpoint, MRSA swabs held at room temperature had a mean of 2.02 × 104 CFU/ml for MRSA recovered and swabs held at 4°C had a mean of 1.04 × 105 CFU/ml for MRSA recovered. VRE swabs held at room temperature had a recovery of 6.73 × 104 CFU/ml on average for VRE, and swabs held at 4°C on average had a recovery of 8.96 × 104 CFU/ml for VRE. There was no relevant change in organism recovery in swabs stored from day 0 to day 14.
Fig 2.
Comparison of average colonies recovered per day for swabs inoculated with MRSA and VRE at different temperatures. A significantly different result was defined by CLSI guidelines as a difference of 3 log.
This was the first study to examine extended transport time past 7 days for MRSA and VRE specimens. Morosini et al. assessed transport times at room temperature for Staphylococcus aureus ATCC 44330 and Enterococcus faecium for up to 7 days using 4 different Amies transport swabs (6). Their results were consistent with our study. However, our study used multiple clinical strains and assessed recovery of strains up to day 14 at 4°C and at room temperature. Surveillance cultures for MRSA or VRE are standard, high-volume samples that are often grown for all patients in a unit or an entire hospital. For example, in Department of Veterans Affairs hospitals, all patients are swabbed at admission, transfer, and discharge at a significant cost (4). These cultures are often the outcome measure in large multicenter trials in which surveillance cultures have been processed at a reference laboratory (3). Our finding that culture results are stable under different conditions validates the use of a central laboratory and could potentially lead to decreased costs for hospital systems.
A limitation to this study was that it did not assess multiple organisms on one swab. The CLSI guidelines do not address the overgrowth of bacteria such as Gram-negative organisms and the loss of fastidious organisms in a polymicrobial environment (11). The organisms selected for this study were chosen because they are often cultured with swab transport systems for the purposes of surveillance. These organisms are also less demanding compared to the more fastidious bacteria such as Haemophilus influenzae and Streptococcus pneumoniae.
Our study showed that MRSA and VRE can survive in Amies transport media refrigerated or at room temperature for up to 14 days. We found that commonly collected infection control surveillance samples can be shipped using standard swabs without a significant decrease in yield of MRSA or VRE. Extended transit time of this swab transport system does not adversely affect the number of viable organisms or their morphology and may be a cost-saving mechanism for research and clinical laboratories.
ACKNOWLEDGMENTS
This study was funded by the Agency for Healthcare Research and Quality (AHRQ) under contract number HHSA290200600015I Task Order no. 5.
The content is solely the responsibility of the authors and does not necessarily represent the official views of AHRQ.
The BUGG Study Group consists of Carol Sulis, Connie Price, Dan Kett, Dave Warren, David Calfee, Deborah Yokoe, Deverick Anderson, Jason Bowling, Jesse Jacob, Joseph Gadbaw, Lisa Maragakis, Loreen Herwaldt, Marci Drees, Marcus Zervos, Matthew Lissauer, Nasia Safdar, Robin Carver, Stephanie Levine, and Syed Shahyrar.
Footnotes
Published ahead of print 25 April 2012
REFERENCES
- 1. CLSI 2010. Performance standards for antimicrobial susceptibility testing; 20th informational supplement. CLSI document M100-S20, vol 30 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 2. Donabedian S, Chow JW, Shlaes DM, Green M, Zervos MJ. 1995. DNA hybridization and contour-clamped homogeneous electric field electrophoresis for identification of enterococci to the species level. J. Clin. Microbiol. 33:141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huskins WC, et al. 2011. Intervention to reduce transmission of resistant bacteria in intensive care. N. Engl. J. Med. 364:1407–1418 doi:10.1056/NEJMoa1000373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jain R, et al. 2011. Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N. Engl. J. Med. 364:1419–1430 doi:10.1056/NEJMoa1007474 [DOI] [PubMed] [Google Scholar]
- 5. McDougal LK, et al. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morosini M, et al. 2006. Evaluation of 4 swab transport systems for the recovery of ATCC and clinical strains with characterized resistance mechanisms. Diagn. Microbiol. Infect. Dis. 56:19–24 doi:10.1016/j.diagmicrobio.2006.02.011 [DOI] [PubMed] [Google Scholar]
- 7. NCCLS 2003. Quality control of microbiological transport systems; approved standard. NCCLS document M40-A, 22 (11) ed, vol 23 NCCLS, Wayne, PA [Google Scholar]
- 8. Perry JL. 1997. Assessment of swab transport systems for aerobic and anaerobic organism recovery. J. Clin. Microbiol. 35:1269–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peterson LR, et al. 2001. Role of clinical microbiology laboratories in the management and control of infectious diseases and the delivery of health care. Clin. Infect. Dis. 32:605–611 [DOI] [PubMed] [Google Scholar]
- 10. Roelofsen E, Van Leeuwen M, Meijer-Severs GJ, Wilkinson MHF, Degener JE. 1999. Evaluation of the effects of storage in two different swab fabrics and under three different transport conditions on recovery of aerobic and anaerobic bacteria. J. Clin. Microbiol. 37:3041–3043 doi:0095-1137/99/$04.00+0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tano E, Melhus A. 2011. Evaluation of three swab transport systems for the maintenance of clinically important bacteria in simulated mono- and polymicrobial samples. APMIS 119:198–203 doi:10.1111/j.1600-0463.2010.02710.x [DOI] [PubMed] [Google Scholar]
- 12. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Van Horn KG, Audette CD, Tucker KA, Sebeck D. 2008. Comparison of 3 swab transport systems for direct release and recovery of aerobic and anaerobic bacteria. Diagn. Microbiol. Infect. Dis. 62:471–473 doi:10.1016/j.diagmicrobio.2008.08.004 [DOI] [PubMed] [Google Scholar]
- 14. Van Horn KG, Rankin I. 2007. Evaluation and comparison of two Stuart's liquid swab transport systems tested by the CLSI M40 method. Eur. J. Clin. Microbiol. Infect. Dis. 26:583–586 doi:10.1007/s10096-007-0328-y [DOI] [PubMed] [Google Scholar]