Abstract
Candida rugosa is a poorly known fungal species occasionally involved in human infections. A molecular analysis of the sequences of the D1/D2 domains and the internal transcribed spacer (ITS) region of the ribosomal genes of 24 clinical isolates phenotypically identified as C. rugosa demonstrated that only 10 (41.6%) isolates belonged to that species. The other isolates were identified as Candida pararugosa (41.6%) and Candida pseudorugosa (8.3%). The remaining two isolates, from human and equine infections, respectively, were clearly different from the others and represent a new species proposed here as Candida neorugosa. The closest species by D1/D2 sequences was the type strain of C. rugosa, with only 92.3% similarity. C. neorugosa can also be differentiated from all other species of the C. rugosa complex by phenotypic features. The eight antifungal drugs tested showed high in vitro activity against the 24 isolates included in the study.
INTRODUCTION
The incidence of Candida infection has increased in recent years, representing an important cause of morbidity and mortality. Although candidiasis is caused mainly by Candida albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, and Candida krusei (15), infections caused by rarer species have increased in recent years (7, 18). Among these less common species, Candida rugosa has been recognized as an emerging fungal pathogen capable of causing invasive infection in immunocompromised patients (12), mostly related to the use of catheters but also by other modes of nosocomial acquisition (6, 8, 12). This species represents only 0.2% of the Candida isolates in the global ARTEMIS DISK Antifungal Surveillance Program (18) but shows a high prevalence in Latin America (18) and India (19). The fungus has decreased susceptibility to fluconazole (FLC) (17, 18), amphotericin B (AMB) (7), and the echinocandins (7). Considering its increasing pathogenic role and the potential development of resistance to antifungals, the reliable identification of C. rugosa is an important issue. However, the biochemical systems currently used for yeast identification in clinical laboratories commonly fail to identify the less frequent Candida spp. (23). Some genetic heterogeneity in C. rugosa has been reported (2, 13, 21), and the novel species Candida pseudorugosa, closely related to C. rugosa, has been recently proposed (11).
We have analyzed the D1/D2 domains and the intergenic transcribed spacer (ITS) sequences of the rRNA genes of a set of clinical isolates phenotypically identified as C. rugosa in order to assess the genetic heterogeneity of the species.
MATERIALS AND METHODS
Fungal isolates.
A total of 24 clinical isolates, received as C. rugosa by the Fungus Testing Laboratory in the Department of Pathology at the University of Texas Health Science Center (UTHSC) at San Antonio, TX, for identification and/or antifungal susceptibility determination, were included in the study. In addition, several ITS and D1/D2 sequences from type or reference strains, retrieved from GenBank, were also included in the phylogenetic analyses (Table 1).
Table 1.
Isolates and sequences of C. rugosa species complex included in the study
| Species | Isolatea | Origin | GenBank accession no. |
|
|---|---|---|---|---|
| ITS | D1/D2 | |||
| C. rugosa | CBS 613T | Feces of man | AY500374b | GU246244b |
| C. pseudorugosa | CBS 10433T | Sputum, Tianjin, China | DQ234792b | DQ234791b |
| C. catenulata | NRRL Y-1508T | Feces of man, Puerto Rico | AY493436b | CCU45714b |
| Clavispora lusitaniae | CBS 4413T | Cecum of pig, Portugal | EU568907b | AY190538b |
| C. pararugosa | NRRL Y-17089T | Feces of man | AF421856b | CPU62306b |
| C. rugosa | LYSM3 | Soil from forest, Thailand | AB498988b | |
| C. rugosa | EB2 | Water in mangrove forest, Thailand | AB436404b | |
| C. rugosa | EF1 | Water in mangrove forest, Thailand | AB436406b | |
| C. rugosa | STC4 | Blood, Kuala Lumpur, Malaysia | HQ412590b | |
| C. rugosa | STC1 | Blood, Kuala Lumpur, Malaysia | HQ412589b | |
| C. rugosa | L154 | Feces of man, Brazil | FJ768915b | |
| C. rugosa | L2683B | Blood, Brazil | FJ768918b | |
| C. rugosa | L387A | Rectal swab, Brazil | FJ768920b | |
| C. rugosa | L412D | Catheter, Brazil | FJ768919b | |
| C. rugosa | L69D | Blood, Brazil | FJ768917b | |
| C. pseudorugosa | MZKI K-259 | Coastal Arctic, Norway | EU056285b | |
| C. pseudorugosa | MZKI K-269 | Coastal Arctic, Norway | EU056286b | |
| C. pararugosa | CBS 9121 | Saliva, sarcoma patient, Japan | AB112430b | |
| C. pararugosa | CBS 9122 | Saliva, sarcoma patient, Japan | AB112432b | |
| C. rugosa | UTHSC 01-2568 | Toe, USA | HE716177 | |
| C. rugosa | UTHSC 05-205 | Blood, USA | HE716180 | |
| C. rugosa | UTHSC 05-646 | Jackson-Pratt drain, USA | HE716182 | |
| C. rugosa | UTHSC 05-1919 | Ankle, USA | HE716179 | |
| C. rugosa | UTHSC 06-3729 | Ear, USA | HE716760 | HE716180 |
| C. rugosa | UTHSC 06-3931 | Stool, USA | HE716181 | |
| C. rugosa | UTHSC 06-3976 | Sputum, USA | HE716178 | |
| C. rugosa | UTHSC 09-1289 | Dolphin, USA | HE716183 | |
| C. rugosa | UTHSC 09-1402 | Blood, USA | HE716176 | |
| C. rugosa | UTHSC R-3412 | Unknown | HE716759 | HE716175 |
| C. pararugosa | UTHSC 03-344 | Bronchial wash, USA | HE716167 | |
| C. pararugosa | UTHSC 03-1143 | Blood, USA | HE716170 | |
| C. pararugosa | UTHSC 04-1051 | Blood, USA | HE716173 | |
| C. pararugosa | UTHSC 05-1693 | Bronchial wash, USA | HE716172 | |
| C. pararugosa | UTHSC 06-538 | Blood, USA | HE716174 | |
| C. pararugosa | UTHSC 07-2797 | Blood, USA | HE716171 | |
| C. pararugosa | UTHSC 07-3133 | Blood, USA | HE716169 | |
| C. pararugosa | UTHSC 08-442 | Urine, USA | HE716757 | HE716166 |
| C. pararugosa | UTHSC 09-2953 | Vaginal, USA | HE716758 | HE716165 |
| C. pararugosa | UTHSC 10-2648 | Blood, USA | HE716168 | |
| C. pseudorugosa | UTHSC 06-3641 | Catheter urine, USA | HE716755 | HE716163 |
| C. pseudorugosa | UTHSC 08-707 | Knee, USA | HE716756 | HE716164 |
| C. neorugosa | SK75 | Ulcerated lesion, Brazil | GQ176145b | GQ176145b |
| C. neorugosa | UTHSC 10-2054T | Leg wound, USA | HE716762 | HE716185 |
| C. neorugosa | UTHSC 10-121 | Wound on left forelimb of horse, USA | HE716761 | HE716186 |
CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; UTHSC, Fungus Testing Laboratory, University of Texas Health Science Center at San Antonio, San Antonio, TX; NRRL, National ARS Culture Collection, Peoria, IL.
Sequences retrieved from GenBank database.
DNA extraction, amplification, and sequencing.
The fungal isolates were grown on potato dextrose agar (PDA) (Pronadisa, Madrid, Spain) at 28°C for 24 h, and DNA was extracted using a PrepMan Ultra sample preparation reagent (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol. The DNA was quantified using GeneQuant Pro (Amersham Pharmacia Biotech, Cambridge, England). The D1/D2 domains of the 24 isolates and the ITS regions of two isolates from the different clades obtained in the D1/D2 phylogenetic analysis were amplified with the primer pairs NL1/NL4 and ITS5/ITS4, respectively, following the protocols described by Cano et al. (4) and Gilgado et al. (10). The PCR products were purified and sequenced with the same primers used for amplification at Macrogen Europe Inc. (Amsterdam, The Netherlands) with a 3730XL DNA analyzer (Applied Biosystems). The program SeqMan (Lasergene, Madison, WI) was used to obtain consensus sequences.
Alignment and phylogenetic analysis.
The sequences were aligned using the ClustalX (version 1.8) computer program (22) with default parameters, followed by manual adjustments with a text editor. The phylogenetic analysis was performed with the software program MEGA 5.0 (20), using Maximum Likelihood (ML) with General-Time-Reversible (GTR) as a substitution model. Gaps were treated as pairwise deletion. Support for internal branches was assessed by a search of 1,000 bootstrapped sets of data.
Phenotypic studies.
Morphological, biochemical, and physiological characterization of a representative number of isolates of the different clades obtained in the molecular study was performed using methods and protocols previously described (24). The tests included growth on Sabouraud chloramphenicol agar (Bio-Rad, Marnes-LaCoquette, France) at 30°C, 37°C, and 45°C; growth in liquid culture medium; germ tube tests; assessment of chlamydospore and ascospore production; hydrolysis of esculin; and the urease test. The ability of the isolates to assimilate carbohydrate source compounds was determined for glucose, d-xylose, melibiose, l-arabinose, d-ribose, l-sorbose, galactose, salicin, raffinose, sucrose, d-mannitol, trehalose, glycerol, 2-ceto-d-gluconate, ribitol, xylitol, inositol, sorbitol, α-metil-d-glucoside, N-acetyl-d-glucosamine, cellobiose, lactose, maltose, melezitose, and citric acid. Chromogenic testing of the colonies was performed on Chromagar Candida (Chromagar Company, Paris, France). The API ID 20C yeast identification kit (bioMérieux SA, Lyon, France) was also used for identification according to the manufacturer's instructions.
In vitro studies.
We evaluated the antifungal susceptibility of the fungal isolates to AMB, FLC, itraconazole (ITC), posaconazole (PSC), voriconazole (VRC), caspofungin (CAS), micafungin (MCF), and anidulafungin (AND). The tests were performed in duplicate using a broth microdilution method according to the M27-A3 guidelines for yeasts (5). MIC results for echinocandins and FLC were read after 24 h and the others after 48 h of incubation. Two reference strains, C. parapsilosis ATCC 22019 and C. krusei ATCC 6258, were included as quality controls for all testing.
Nucleotide sequence accession numbers.
The new DNA sequences generated in this study were deposited in the GenBank database (Table 1).
RESULTS
Molecular analysis.
The phylogenetic tree inferred from the ML analysis of the D1/D2 sequences revealed that the 24 isolates tested were distributed in four statistically well-supported groups, each representing a phylogenetic species (Fig. 1). The first group (bootstrap support [bs], 81%) was formed by 10 clinical isolates, the sequence of the type strain of C. rugosa, and several sequences of clinical and environmental strains retrieved from GenBank, some of which were previously reported as atypical isolates of C. rugosa (21). The isolates' sequence similarities ranged from 99.3 to 100%. The second group (bs, 100%) was comprised of three identical sequences; two belonged to our clinical isolates (UTHSC 10-2054 and UTHSC 10-121) and the third (SK75; GQ176145), retrieved from GenBank, was from an ulcerated lesion in Brazil. The members of that group showed low similarity to the type strains of the species C. rugosa (92.3%), C. pseudorugosa (92%), and C. pararugosa (67.3%). The third group (bs, 99%) consisted of two clinical isolates, the type strain of C. pseudorugosa, and sequences of two environmental isolates retrieved from GenBank. The sequence similarity of the members of this group ranged from 99.2 to 100%. Finally, 10 clinical isolates were nested with the type strain of C. pararugosa, along with two sequences from clinical isolates deposited in GenBank (bs, 100%).
Fig 1.
ML tree obtained from the D1/D2 domains of the 28S rRNA gene sequences of the strains of C. rugosa and related taxa. Bootstrap support values above 70% are indicated at the nodes. Clinical isolates from UTHSC are indicated in boldface. The bar indicates genetic distance. Sequences retrieved from GenBank are marked with a superscript “a.”
The phylogenetic tree inferred from the ML analysis of the ITS sequences (Fig. 2) showed topology and genetic relationships similar to those seen in the D1/D2 tree.
Fig 2.
ML tree obtained from the ribosomal ITS sequences of several isolates of C. rugosa and related taxa. Bootstrap support values above 70% are indicated at the nodes. Clinical isolates from UTHSC are indicated in boldface. The bar indicates genetic distance. Sequences retrieved from GenBank are marked with a superscript “a.”
Physiology.
Four of the physiological tests used were useful to distinguish the three phylogenetically most closely related species, i.e., C. rugosa, C. pseudorugosa, and the unidentified species represented by the isolates UTHSC 10-2054 and UTHSC 10-121. The two isolates of each species tested showed identical results (Table 2). C. rugosa was differentiated from C. pseudorugosa by the utilization of d-xylose, glycerol, and sorbitol. Clinical isolates UTHSC 10-2054 and UTHSC 10-121 differ from C. rugosa by their ability to assimilate ribitol and from C. pseudorugosa by their ability to assimilate d-xylose, glycerol, ribitol, and sorbitol. API ID 20C results confirmed those findings. On Chromagar, C. pseudorugosa and C. rugosa yielded dark blue-green and from white to light-blue colonies, respectively, while the isolates UTHSC 10-2054 and UTHSC 10-121 showed white to dark-blue colonies.
Table 2.
Key physiological features useful to distinguish the species of the C. rugosa complex
| Species | Isolatea | Assimilation testb |
|||
|---|---|---|---|---|---|
| d-Xylose | Glycerol | Ribitol | Sorbitol | ||
| C. pseudorugosa | UTHSC 06-3641 | − | − | − | − |
| UTHSC 08-707 | − | − | − | − | |
| C. rugosa | UTHSC 06-3729 | + | + | − | + |
| UTHSC R-3412 | + | + | − | + | |
| C. neorugosa sp. nov. | UTHSC 10-121 | + | + | + | + |
| UTHSC 10-2054 | + | + | + | + | |
UTHSC, Fungus Testing Laboratory, University of Texas Health Science Center at San Antonio, Texas, USA.
+, positive; −, negative.
Antifungal susceptibility.
The eight drugs tested showed high in vitro activity against all the isolates, with no significant differences noted among the different species. MIC ranges were <0.03 to 2 μg/ml for the azoles, 0.25 to 1 μg/ml for AMB, and 0.03 to 0.5 μg/ml for the echinocandins (Table 3).
Table 3.
Results of in vitro antifungal susceptibility testing
| Species (no. of isolates) | Antifungal agents | MIC (μg/ml)a |
||
|---|---|---|---|---|
| Range | GM | 90 | ||
| C. pseudorugosa (2) | Amphotericin B | 0.5–1 | 0.71 | − |
| Fluconazole | 1 | 1 | − | |
| Posaconazole | 0.06 | 0.06 | − | |
| Voriconazole | <0.03–0.03 | 0.04 | − | |
| Itraconazole | 0.12 | 0.12 | − | |
| Caspofungin | 0.06–0.5 | 0.17 | − | |
| Micafungin | 0.12–0.25 | 0.17 | − | |
| Anidulafungin | 0.03–0.06 | 0.04 | − | |
| C. rugosa (10) | Amphotericin B | 0.25–1 | 0.66 | 1 |
| Fluconazole | 0.25–2 | 0.81 | 2 | |
| Posaconazole | 0.03–0.12 | 0.07 | 0.12 | |
| Voriconazole | <0.03–0.03 | 0.02 | 0.03 | |
| Itraconazole | 0.03–0.5 | 0.06 | 0.12 | |
| Caspofungin | 0.06–1 | 0.38 | 1 | |
| Micafungin | 0.06–0.5 | 0.17 | 0.5 | |
| Anidulafungin | 0.03–0.5 | 0.12 | 0.25 | |
| C. pararugosa (10) | Amphotericin B | 0.25–1 | 0.47 | 1 |
| Fluconazole | 0.25–2 | 0.76 | 2 | |
| Posaconazole | 0.03–0.12 | 0.08 | 0.12 | |
| Voriconazole | <0.03–0.06 | 0.03 | 0.06 | |
| Itraconazole | 0.03–0.25 | 0.07 | 0.12 | |
| Caspofungin | 0.03–0.25 | 0.13 | 0.25 | |
| Micafungin | 0.06–0.12 | 0.1 | 0.12 | |
| Anidulafungin | 0.03–0.12 | 0.06 | 0.12 | |
| C. neorugosa sp. nov. (2) | Amphotericin B | 0.25–1 | 0.5 | − |
| Fluconazole | 1–2 | 1.41 | − | |
| Posaconazole | 0.12 | 0.12 | − | |
| Voriconazole | 0.06 | 0.06 | − | |
| Itraconazole | 0.25 | 0.25 | − | |
| Caspofungin | 0.12–0.25 | 0.17 | − | |
| Micafungin | 0.06 | 0.06 | − | |
| Anidulafungin | 0.03–0.06 | 0.04 | − | |
90, MIC90, the minimal concentration of drug capable of inhibiting the growth of 90% of assayed isolates; GM, geometric mean; −, value not calculated.
Origins and clinical data of the isolates UTHSC 10-2054 and UTHSC 10-121.
UTHSC 10-2054 was originally recovered from a female patient who sustained leg injuries while in Africa. A culture from the initial wound debridement there yielded yeast that was identified as C. rugosa. The patient was subsequently transferred to Johns Hopkins Hospital, Baltimore, MD, where an isolate identified as C. rugosa using the API ID 20C kit (profile 2442104, 97.1%) was also recovered from a surgical wound. The wound infection was finally resolved without antifungal therapy.
UTHSC 10-121 was isolated from a 22-month-old male Arabian horse that was presented to the Veterinary Medical Teaching Hospital at Texas A&M University, College Station, TX, for evaluation of a laceration and puncture wound on the palmar medial aspect of the left forelimb. The lesion was surgically debrided, and cultures collected at 24 days of hospitalization grew Pseudomonas aeruginosa and a yeast identified as C. tropicalis using a commercial identification system (Vitek II YBC; bioMérieux, Durham, NC). The organism was also cultured from the wound 1 week later, along with Enterococcus spp. and Stenotrophomonas maltophilia. At 56 days of hospitalization, yeast was again isolated from the site and identified as C. rugosa (Vitek II YBC; bioMérieux, Durham, NC). The patient was taken to surgery for ankylosis of the joint and started with a daily oral dose of FLC (4 mg/kg of body weight) and broad-spectrum antibiotic therapy. After 79 days of hospitalization, the horse was discharged. C. tropicalis was not recovered again.
TAXONOMY
Based on molecular and phenotypic data, it is concluded that the isolates UTHSC 10-2054 and UTHSC 10-121 represent a novel species of the genus Candida, for which the name Candida neorugosa sp. nov. is proposed.
Candida neorugosa Paredes, D. A. Sutton, Cano, Guarro, sp. nov. (Fig. 3).
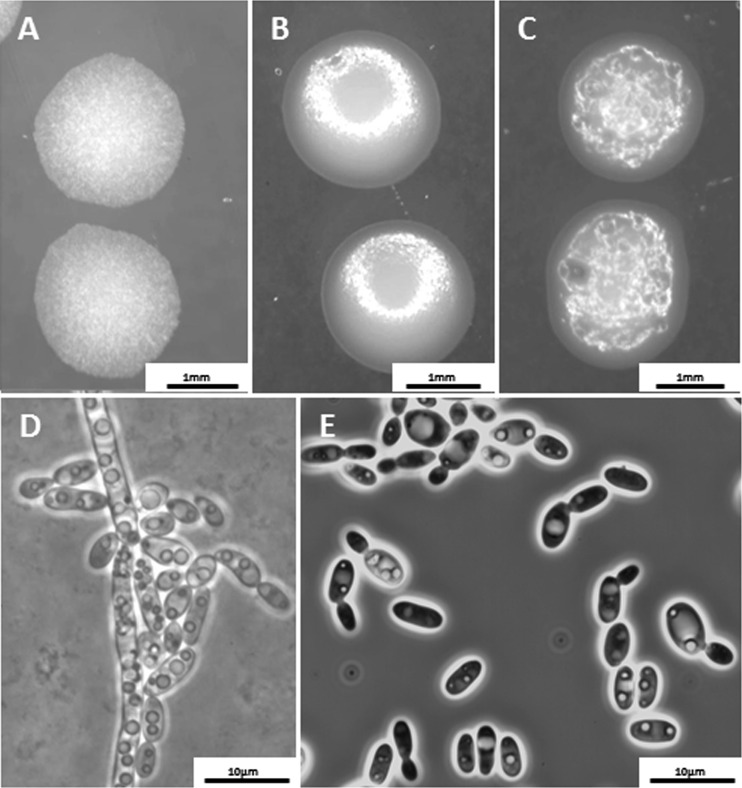
Fig 3.
(A) C. rugosa UTHSC 06-3729. (B) C. pseudorugosa UTHSC 06-3641. (C to E) C. neorugosa sp. nov. UTHSC 10-2054. (A, B, and C) Colonies on PDA after 2 days of incubation at 25°C. (D) Pseudohyphae on rice Tween agar after 3 days of incubation at 28°C. (E) Yeast cells on YPD broth after 3 days of incubation at 37°C.
MycoBank MB 564807.
In medio liquido YPD post dies 3 ad 25°C sedimentum et annulus formatur, cellulae elipsoideae, ovoidae, 5 to 7 by 3 to 4 μm, singulae, binae et adhaerents aut in racemis brevibus. Per gemmationem multipolarem reproducents. In agaro PDA post dies 2 ad 25°C, coloniae convexa, involvit, album ad cremea. Germina tubulata non formantur. Chlamydosporas et pseudohyphas non fert. Ascosporae non fiunt. Assimilat glucosum, trehalosum, galactosum, d-xylosum, ribitolum, sorbitolum et glycerolum. Fermentatio nulla. Ureum et aesculinum non hydrolysatur. Holotypus ex humana crus vulnus in collectione CBS deposita est, CBS H-20946 (cultura viva UTHSC 10-2054, CBS 12627).
Etymology: Refers to its similarity to C. rugosa.
Candida neorugosa Paredes, D. A. Sutton, Cano, Guarro, sp. nov.
On YPD broth, after 3 days at 25°C, sediment is formed and turbidity of the medium is visible; the cells are ellipsoidal or ovoid, 5 to 7 μm by 3 to 4 μm, single, in pairs, chains or small groups, with multilateral budding. On PDA, the colonies are convex, folded, and white to cream colored. Germ tubes and chlamydospores are not formed. Abundant pseudohyphae are formed on rice agar. Ascospores are not detected in acetate agar or Gorodkowa medium incubated at 25°C for up to 4 weeks. Assimilation of trehalose, glucose, d-xylose, galactose, ribitol, sorbitol, and glycerol is produced. No sugars are fermented. Susceptible to cycloheximide. Unable to hydrolyze esculin and urea.
The type strain CBS 12627 (= UTHSC 10-2054) was isolated in 2010 in the Johns Hopkins University School of Medicine, Baltimore, MD, from a human leg wound. The strain CBS 12628 (= UTHSC 10-121) was isolated in 2010 in the Veterinary Medical Teaching Hospital at Texas A&M University, College Station, TX, from the left forelimb of an Arabian horse.
DISCUSSION
Our molecular analysis revealed that the 24 isolates initially identified as C. rugosa belonged to at least four species, three of which were phylogenetically close to each other, i.e., C. rugosa, C. pseudorugosa, and the undescribed species, while a fourth species, C. pararugosa, was very distant from the other three. The lack of agreement between phenotypic and molecular identification of the isolates included in this study can be explained, as indicated above, by the limitations of the commercial identification systems used in clinical laboratories, which do not allow the identification of uncommon yeasts (1, 23). This can lead to an overestimation of the incidence of some species to the detriment of others that are less common.
Of the 24 isolates investigated, only 10 that nested with the type strain of C. rugosa were identified as that species. An identical number of isolates were identified as C. pararugosa. The latter species was first isolated from human feces and, after the initial description, has been recovered from the oral cavity of a denture wearer (9) and the saliva of a sarcoma patient (14), suggesting its contribution to the oral microbiota. In both cases, the isolates were misidentified by using phenotypic methods, and their true pathogenic role was not established. Our study has expanded the host range of C. pararugosa, with several additional isolates recovered from various anatomical sites. The present study also identified two isolates, one recovered from a urine catheter and the other from a wound infection of the knee, as C. pseudorugosa. This is the first report of this species from clinical specimens, as the only two other isolates are from subglacial ice from arctic coastal environments (3). It is also remarkable that two of the isolates tested, UTHSC 10-2054 and UTHSC 10-121, were clearly genetically different from known species of Candida whose sequences have been deposited in GenBank, Centraalbureau voor Schimmelcultures (CBS), or National Institute of Technology and Evaluation Biological Resource Center (NITE) databases. The D1/D2 sequences of these two isolates clustered in the D1/D2 phylogenetic tree with a third sequence of a Candida sp. from a Brazilian patient with an ulcerated lesion. The three sequences were identical and formed a clearly separated branch. Unfortunately, the last isolate is not available for comparative studies. The ITS sequence analysis confirmed those results. Previously, divergent ITS2 sequences (30 to 35 bp), obtained by pyrosequencing, were reported in some clinical isolates of C. rugosa (2, 13). Our studies confirmed such variability and showed that C. pseudorugosa also displays divergence in that fragment, i.e., a different sequence for each isolate (CBS 613T, GTCAAAAGTGGTTAGTCGGCGACTTACTTGA; UTHSC 06-3729, GTCGATATTGGTTAGTCTGCGACTTACTTGA; and UTHSC R-3412, GTCAACATCTAAAAGTCGGCGACTTACTTGA), and all were different from those of C. rugosa and C. neorugosa. The three strains of C. neorugosa showed the same sequence (GTCGACGTTCAAAAGCCGGCGACTACACTAA) for the referred fragment, which suggests that this new species could be identified by pyrosequencing.
While there are no data on the in vitro antifungal susceptibility of C. pseudorugosa and C. pararugosa, several in vitro susceptibility studies have reported reduced susceptibility of C. rugosa to AMB (6, 7), FLC and VRC (17, 18), and the echinocandins (7). However, in our case, all the antifungal drugs tested, with the exception of CAS against C. rugosa, were active against the 24 isolates. Although C. rugosa is not included in the proposed CLSI breakpoints for antifungal susceptibility testing, its GM MIC (0.38 mg/liter) and MIC90 (1 mg/liter) would be interpreted as inferring resistance to CAS (16), as opposed to the other species of the C. rugosa complex, which exhibit lower CAS MICs.
In conclusion, the present study confirmed the value of molecular tools for the identification of cryptic species within the C. rugosa species complex. Prior to this evaluation, only C. rugosa had been documented as a human etiologic agent. This study confirms that a significant number of isolates of other species have also been recovered from clinical samples. Now that reliable methods for the identification of these species are available, an important objective for future studies would be to elucidate the true prevalences, susceptibilities, and clinical roles of these species.
ACKNOWLEDGMENT
This study was supported by the Spanish Ministerio de Educación y Ciencia, grant CGL 2009-08698/BOS.
Footnotes
Published ahead of print 2 May 2012
REFERENCES
- 1. Alcoba-Flórez J, Méndez-Álvarez S, Cano J, Guarro J, Pérez-Roth E, Arévalo MP. 2005. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J. Clin. Microbiol. 43:4107–4111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Borman AM, et al. 2010. Rapid molecular identification of pathogenic yeasts by pyrosequencing analysis of 35 nucleotides of internal transcribed spacer 2. J. Clin. Microbiol. 48:3648–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Butinar L, Strmole T, Gunde-Cimerman N. 2011. Relative incidence of ascomycetous yeasts in arctic coastal environments. Microb. Ecol. 61:832–843 [DOI] [PubMed] [Google Scholar]
- 4. Cano J, Guarro J, Gené J. 2004. Molecular and morphological identification of Colletotrichum species of clinical interest. J. Clin. Microbiol. 42:2450–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard, 3rd ed, document M27-A3. CLSI, Wayne, PA [Google Scholar]
- 6. Colombo AL, et al. 2003. Outbreak of Candida rugosa candidemia: an emerging pathogen that may be refractory to amphotericin B therapy. Diagn. Microbiol. Infect. Dis. 46:253–257 [DOI] [PubMed] [Google Scholar]
- 7. Diekema DJ, et al. 2009. In vitro activity of seven systemically active antifungal agents against a large global collection of rare Candida species as determined by CLSI broth microdilution methods. J. Clin. Microbiol. 47:3170–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dubé MP, Heseltine PN, Rinaldi MG, Evans S, Zawacki B. 1994. Fungemia and colonization with nystatin-resistant Candida rugosa in a burn unit. Clin. Infect. Dis. 18:77–82 [DOI] [PubMed] [Google Scholar]
- 9. Giammanco GM, Melilli D, Pizzo G. 2004. Candida pararugosa isolation from the oral cavity of an Italian denture wearer. Res. Microbiol. 155:571–574 [DOI] [PubMed] [Google Scholar]
- 10. Gilgado F, Cano J, Gené J, Guarro J. 2005. Molecular phylogeny of the Pseudallescheria boydii species complex: proposal of two new species. J. Clin. Microbiol. 43:4930–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li J, Xu J, Bai F. 2006. Candida pseudorugosa sp. nov., a novel yeast species from sputum. J. Clin. Microbiol. 44:4486–4490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Minces LR, Ho KS, Veldkamp PJ, Clancy CJ. 2009. Candida rugosa: a distinctive emerging cause of candidaemia. A case report and review of the literature. Scand. J. Infect. Dis. 41:892–897 [DOI] [PubMed] [Google Scholar]
- 13. Montero CI, et al. 2008. Evaluation of pyrosequencing technology for the identification of clinically relevant non-dematiaceous yeasts and related species. Eur. J. Clin. Microbiol. Infect. Dis. 27:821–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakagawa Y, et al. 2004. Recurrent isolation of an uncommon yeast, Candida pararugosa, from a sarcoma patient. Med. Mycol. 42:267–271 [DOI] [PubMed] [Google Scholar]
- 15. Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 20:133–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pfaller MA, et al. 2011. Clinical breakpoints for the echinocandins and Candida revisited: integration of molecular, clinical, and microbiological data to arrive at species-specific interpretive criteria. Drug Resist. Updat. 14:164–176 [DOI] [PubMed] [Google Scholar]
- 17. Pfaller MA, et al. 2006. Candida rugosa, an emerging fungal pathogen with resistance to azoles: geographic and temporal trends from the ARTEMIS DISK Antifungal Surveillance Program. J. Clin. Microbiol. 44:3578–3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pfaller MA, et al. 2010. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48:1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singh RI, et al. 2011. Epidemiology of candidemia in critically ill trauma patients: experiences of a level I trauma centre in North India. J. Med. Microbiol. 60:342–348 [DOI] [PubMed] [Google Scholar]
- 20. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tay ST, Tan HW, Na SL, Lim SL. 2011. Phenotypic and genotypic characterization of two closely related subgroups of Candida rugosa in clinical specimens. J. Med. Microbiol. 60:1591–1597 [DOI] [PubMed] [Google Scholar]
- 22. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vijgen S, et al. 2011. Comparison of Vitek identification and antifungal susceptibility testing methods to DNA sequencing and Sensititre YeastOne antifungal testing. Med. Mycol. 49:107–110 [DOI] [PubMed] [Google Scholar]
- 24. Yarrow D. 1998. Methods for isolation, maintenance and classification of yeasts. p 77–100 In Kurtzman C. P., Fell J. W. (ed), The yeasts, a taxonomic study, 4th ed Elsevier Science BV, Amsterdam, Netherlands [Google Scholar]