Abstract
Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a potentially useful tool for the detection of antimicrobial resistance, especially that conferred by β-lactamases. Here we describe a modification of a previously reported MALDI-TOF MS meropenem hydrolysis assay. The modified method was validated on 108 carbapenemase-producing members of the Enterobacteriaceae, two NDM-1-producing Acinetobacter baumannii isolates, and 35 carbapenem-resistant enterobacteria producing no carbapenemase. The detection of carbapenemases by MALDI-TOF MS seems to be a powerful, quick, and cost-effective method for microbiological laboratories.
TEXT
Detection of carbapenemases has been one of the challenges in clinical microbiology diagnostics (9). Recently, new matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) assays for β-lactamase activity have been developed independently by at least three groups (2, 7, 12). These techniques are based on the detection of β-lactams and their degradation products. A similar assay was validated to detect carbapenemases in Acinetobacter baumannii (8).
However, for some β-lactams, like meropenem, visualization of degradation products by MALDI-TOF MS seemed to be problematic (7, 12). This might be due to binding of the molecules to cell lysate components. Here we describe a modification of one of these protocols (7) that allows the detection of degradation products and shortening of the turnaround time to ca. 2.5 h. The modified assay was validated with NDM-1-, VIM-1-, KPC-2-, KPC-3-, and OXA-48/-162-producing members of the Enterobacteriaceae and NDM-1-producing A. baumannii isolates.
(The data included in this article were presented in part as a poster at the 22nd European Congress of Clinical Microbiology and Infectious Diseases [ECCMID], London, United Kingdom, 31 March to 3 April 2012.)
One hundred ten carbapenemase-producing isolates were collected by the National Reference Laboratory for Antibiotics in Prague, Czech Republic, the National Reference Centre for Susceptibility Testing in Warsaw, Poland, the Robert Koch Institute in Wernigerode, Germany, and two military hospitals at Rawalpindi, Pakistan (Table 1). Almost all of the isolates had been well characterized previously on the molecular level (1, 5, 6, 10, 11). For the newly included isolates, carbapenemase production was verified by the imipenem hydrolysis spectrophotometric assay (13), followed by PCR and sequencing of the carbapenemase genes (1, 5, 6, 11). Species identification was performed using a MALDI Biotyper system, version 3.0 (Bruker Daltonics, Bremen, Germany). Thirty-five non-carbapenemase-producing isolates resistant to carbapenems (4), identified in the Czech Republic, were also included (Table 1). Most of these isolates had been characterized in detail previously (3); in several new ones, carbapenemase production was excluded by the spectrophotometric assay (13).
Table 1.
Carbapenemase-producing isolates used in the study
| Description | Carbapenemase | Species (na) | Peaks detected in the spectra (m/z) |
|---|---|---|---|
| Carbapenemase producing | NDM-1 | Escherichia coli (30) | 353.8, 358.6, 375.4, 380.5, 381.6, 382.7, 399.6, 402.6, 404.6, 424.5, 426.6, 446.6, 448.5, 468.5 |
| Enterobacter cloacae (22) | |||
| Citrobacter spp. (6) | |||
| Klebsiella pneumoniae (5) | |||
| Providencia rettgeri (2) | |||
| Acinetobacter baumannii (2) | 353.8, 358.6, 370.6, 375.4, 380.5, 381.6, 382.7,392.5, 402.6, 404.6, 424.5, 426.6, 446.6, 448.5 | ||
| KPC-2, KPC-3 | Klebsiella pneumoniae (10) | 353.8, 358.6, 375.4, 380.5, 381.6, 382.7, 399.6, 402.6, 404.6, 424.5, 426.6, 446.6, 448.5, 468.5 | |
| VIM-1 | Klebsiella pneumoniae (16) | 353.8, 358.6, 375.4, 380.5, 381.6, 382.7, 399.6, 402.6, 404.6, 424.5, 426.6, 446.6, 448.5, 468.5 | |
| Enterobacter cloacae (4) | |||
| Serratia marcescens (3) | |||
| OXA-48 | Escherichia coli (3) | 353.8, 358.6, 362.6, 365.6, 378.6, 380.5, 381.6, 382.7,392.5, 402.6, 404.6, 424.5, 426.6, 444.5, 446.6 | |
| Enterobacter cloacae (1) | |||
| Klebsiella pneumoniae (2) | |||
| OXA-162 | Escherichia coli (2) | 353.8, 358.6, 362.6, 365.6, 378.6, 380.5, 381.6, 382.7,392.5, 402.6, 404.6, 424.5, 426.6, 444.5, 446.6 | |
| Raoultella ornithinolytica (1) | |||
| Citrobacter freundii (1) | |||
| Total isolates | 110 | ||
| Non-carbapenemase-producing, resistant to carbapenems | Klebsiella pneumoniae (28) | 353.8, 381.6, 384.5, 397.6, 406.5, 422.6, 428.5, 444.5 | |
| Enterobacter cloacae (4) | |||
| Citrobacter spp. (3) | |||
| Total isolates | 35 |
n, no. of isolates.
The meropenem hydrolysis assay (7) was performed as previously described, with modifications. An overnight bacterial culture on Mueller-Hinton agar (Bio-Rad Laboratories, Prague, Czech Republic) was suspended in 20 mM Tris-HCl–20 mM NaCl, pH 7.0 (Sigma-Aldrich, Prague, Czech Republic), to a density equivalent to a 3.0 McFarland standard. In our experience, other common cultivation media (e.g., Columbia agar) can be used as well. The 1-ml aliquot of the suspension was centrifuged; the pellet was resuspended in 50 μl of a reaction buffer (20 mM Tris-HCl, 0.01% sodium dodecyl sulfate [SDS], pH 7.0; Sigma-Aldrich), supplemented with 0.1 mM meropenem (Astra Zeneca, Macclesfield, United Kingdom). After incubation at 35°C for 2 h, the reaction mixture was centrifuged; 1 μl of the supernatant was immediately mixed with 1 μl of dihydroxybenzoic acid solution (DHB) (Sigma-Aldrich) and allowed to dry on a target. Spectra were measured after drying between m/z 160 and 600, using a Microflex LT mass spectrometer (Bruker Daltonics). The measurements must be performed immediately after drying of the samples because a delay of more than 1 h may cause degradation of meropenem and its variants. The laser range was set up individually to obtain spectra with intensities between 0.2 × 104 and 2.0 × 104 (ca. 60 to 90%). Calibration was performed using meropenem and its two sodium salt variants (m/z 384.5, 406.5, and 428.5).
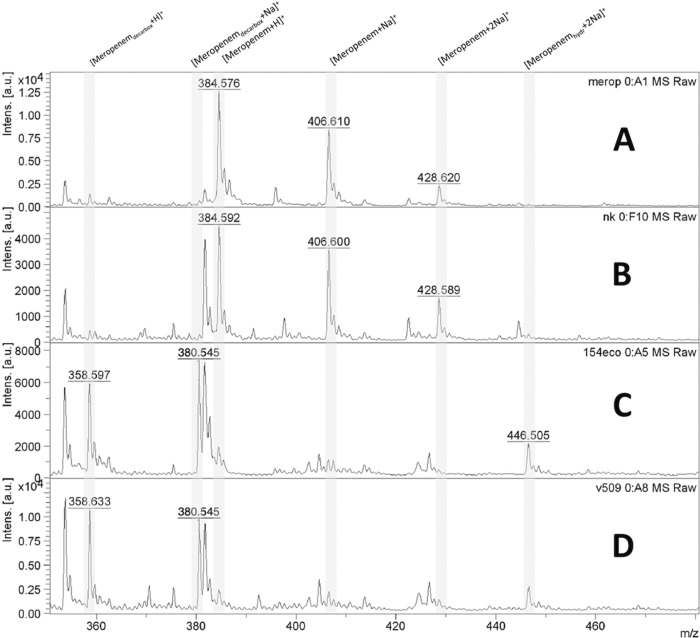
Spectra were analyzed by using the software program flexAnalysis, version 3.3 (Bruker Daltonics), in the range between m/z 350 and 480 (Fig. 1). For the carbapenemase-producing isolates, two products of meropenem degradation were identified, with m/z values of 358.5 (the decarboxylated product) and 380.5 (sodium salt of the decarboxylated product). For some carbapenemase-positive strains, other degradation products were observed (e.g., three sodium salt variants with m/z 424.5, 446.5, and 468.5) (Table 1). Meropenem (m/z 384.5) and its monosodium salt (m/z 406.5) were not detected. For the non-carbapenemase-producing isolates, no peaks with m/z 358.5 and 380.5 were recorded, but meropenem and its monosodium salt were present.
Fig 1.
MALDI-TOF MS spectrum showing meropenem, sodium salts of meropenem, and degradation products. (A) Spectrum of meropenem solution. (B) Negative control (non-carbapenemase-producing isolate of Klebsiella pneumoniae). (C) NDM-1-producing Escherichia coli. (D) NDM-1-producing Acinetobacter baumannii. [Meropenemdecarbox+H]+, decarboxylated degradation product of meropenem after carbapenemase hydrolysis (m/z 358.5); [Meropenemdecarbox+Na]+, decarboxylated sodium salt of degradation product of meropenem after carbapenemase hydrolysis (m/z 380.5); [Meropenem+H]+, meropenem molecule (m/z 384.5); [Meropenem+Na]+, meropenem sodium salt (m/z 406.5); [Meropenem+2Na]+, meropenem disodium salt (m/z 428.5); [Meropenemhydr+2Na]+, disodium salt of degradation product of meropenem after carbapenemase hydrolysis (m/z 446.5).
According to the results, the detection interpretation criteria were established (see Table 2). Using these criteria, it was possible to identify carbapenemase activity in all carbapenemase-positive isolates used in the study. There were neither false-positive nor false-negative results.
Table 2.
Interpretation criteria based on presence/absence of peaks
| Criterion | Peak [m/z] (product) meeting criterion for: |
|
|---|---|---|
| Carbapenemase-producing isolate | Non-carbapenemase-producing isolate | |
| Presence of at least one peak | 358.5 (decarboxylated product) | 384.5 (meropenem) |
| 380.5 (sodium salt of decarboxylated product) | 406.5 (meropenem sodium salt) | |
| Absence of all peaks | 384.5 (meropenem) | 358.5 (decarboxylated product) |
| 406.5 (meropenem sodium salt) | 380.5 (sodium salt of decarboxylated product) | |
The main modification of the method published previously (7) was the supplementation of the reaction buffer with SDS, resulting in a decrease of the amount of bacterial cells and reduction of the incubation time to 2 h. The MALDI-TOF MS meropenem hydrolysis assay was validated mainly with carbapenemase-producing members of the Enterobacteriaceae, and in our opinion it can be used in microbiological laboratories for these organisms routinely.
ACKNOWLEDGMENTS
This work was supported by research project grants NT11032-6/2010 from the Ministry of Health of the Czech Republic and MSM0021620819 from the Ministry of Education of the Czech Republic.
We have no competing financial interest to declare.
Footnotes
Published ahead of print 2 May 2012
REFERENCES
- 1. Baraniak A, et al. 2011. Molecular characteristics of KPC-producing Enterobacteriaceae at the early stage of their dissemination in Poland, 2008-2009. Antimicrob. Agents Chemother. 55:5493–5499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burckhardt I, Zimmermann S. 2011. Using matrix-assisted laser desorption ionization–time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J. Clin. Microbiol. 49:3321–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chudáčková E, et al. 2010. Carbapenem-non-susceptible strains of Klebsiella pneumoniae producing SHV-5 and/or DHA-1 β-lactamases in a Czech hospital. FEMS Microbiol. Lett. 309:62–70 [DOI] [PubMed] [Google Scholar]
- 4. European Committee on Antimicrobial Susceptibility Testing 2012. Breakpoint tables for interpretation of MICs and zone diameters. Version 2.0, February. http://www.eucast.org
- 5. Hrabák J, et al. 2011. KPC-2-producing Klebsiella pneumoniae isolated from a Czech patient previously hospitalized in Greece and in vivo selection of colistin resistance. Folia Microbiol. 56:361–365 [DOI] [PubMed] [Google Scholar]
- 6. Hrabák J, et al. 2012. NDM-1 producing Acinetobacter baumannii isolated from a patient repatriated to the Czech Republic from Egypt, July 2011. Euro Surveill. 17:pii=20085 [PubMed] [Google Scholar]
- 7. Hrabák J, et al. 2011. Carbapenemase activity detection by matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry. J. Clin. Microbiol. 49:3222–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kempf M, et al. 2012. Rapid detection of carbapenem resistance in Acinetobacter baumannii using matrix-assisted laser desorption ionization-time of flight mass spectrometry. PLoS One 7:e31676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nordmann P, et al. 2012. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2012.03815.x [DOI] [PubMed] [Google Scholar]
- 10. Perry JD, et al. 2011. Prevalence of faecal carriage of Enterobacteriaceae with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J. Antimicrob. Chemother. 66:2288–2294 [DOI] [PubMed] [Google Scholar]
- 11. Pfeifer Y., et al. 2012. Emergence of OXA-48 type carbapenemase-producing enterobacteriaceae in German hospitals. Antimicrob. Agents Chemother. 56:2125–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sparbier K, et al. 2012. Matrix-assisted laser desorption ionization–time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. J. Clin. Microbiol. 50:927–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Woodford N, et al. 2004. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York medical center. Antimicrob. Agents Chemother. 48:4793–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]