Abstract
Multidrug-resistant Mycobacterium tuberculosis strains are widespread and present a challenge to effective treatment of this infection. The need for a low-cost and rapid detection method for clinically relevant mutations in Mycobacterium tuberculosis that confer multidrug resistance is urgent, particularly for developing countries. We report here a novel test that detects the majority of clinically relevant mutations in the beta subunit of the RNA polymerase (rpoB) gene that confer resistance to rifampin (RIF), the treatment of choice for tuberculosis (TB). The test, termed TB ID/R, combines a novel target and temperature-dependent RNase H2-mediated cleavage of blocked DNA primers to initiate isothermal helicase-dependent amplification of a rpoB gene target sequence. Amplified products are detected by probes arrayed on a modified silicon chip that permits visible detection of both RIF-sensitive and RIF-resistant strains of M. tuberculosis. DNA templates of clinically relevant single-nucleotide mutations in the rpoB gene were created to validate the performance of the TB ID/R test. Except for one rare mutation, all mutations were unambiguously detected. Additionally, 11 RIF-sensitive and 25 RIF-resistant clinical isolates were tested by the TB ID/R test, and 35/36 samples were classified correctly (96.2%). This test is being configured in a low-cost test platform to provide rapid diagnosis and drug susceptibility information for TB in the point-of-care setting in the developing world, where the need is acute.
INTRODUCTION
The global incidence of drug-resistant tuberculosis (TB), particularly multidrug-resistant (MDR) and extremely drug resistant (XDR) strains, is a major worldwide issue. Rates of MDR TB have been estimated to be 4.8% of the 9.8 million TB infections (42), but rates as high as 55% have been observed for previously treated patients (4). TB can be treated effectively if properly identified (28, 29). However, delayed initiation of appropriate treatment in suspected MDR TB cases is associated with excess morbidity and nosocomial transmission (39). It has been determined that the main contributor to delay in treatment is poor sensitivity of diagnostic tests (26); the average sensitivity of sputum microscopy is <60% for immunocompetent patients and lower for HIV infected patients. The frequency of smear-negative disease increases the difficulty of detecting HIV-associated TB as well (33). While mycobacterium culture is much more sensitive, it has a very slow turnaround time of 2 to 8 weeks and is technically complex (33). Nucleic acid amplification-based tests have improved detection sensitivity and time to results but historically have been difficult to implement effectively (21, 30–32, 37–38). A recently described real-time PCR approach brings ease of use but at a high cost (6).
To address the need to bring sensitive and specific diagnostic testing closer to the patient in the developing world, we have designed a novel approach for the detection of Mycobacterium tuberculosis and mutations within the rpoB gene that confer resistance to the first-line drug rifampin (RIF). Described here is the performance of the benchtop version of the assay, termed TB ID/R. Target DNA sequences within the rpoB gene are amplified using a novel method, blocked-primer-mediated helicase-dependent amplification (bpHDA), which utilizes the isothermal amplification method HDA (2) coupled with a blocked-primer/RNase H2-mediated target-specific “hot start” (10) to exponentially amplify target DNA sequences. bpHDA utilizes modified blocked primers, which are constructed with a single ribonucleotide linkage inserted 4 bases upstream of a 3′-end block added to prevent primer extension. Once blocked primers hybridize to complementary target sequences, thermostable RNase H2 derived from Pyrococcus abyssi is activated, cleaving the ribonucleotide linkage in the primer present in duplex DNA. The short segment of the primer 3′ of the ribonucleotide dissociates, liberating the block and creating a free 3′-hydroxyl that is now capable of primer extension. The RNase H2 used here has very little activity at temperatures below 40°C and is highly active at 65°C, the temperature required for HDA to amplify target sequences optimally (10). Because primer/primer hybrids are unstable at elevated temperatures, no primer artifact is amplified. After bpHDA, the resultant amplicons are detected by hybridization to a probe set arrayed on a modified silicon chip surface that detects mutations in the amplified region of the rpoB gene. Intermolecular interactions on the chip trigger colorimetric intensity changes, permitting visual detection of attomole quantities of nucleic acids (19).
MATERIALS AND METHODS
Capture probe design.
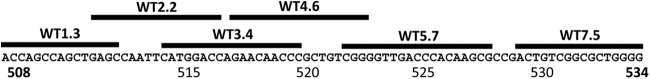
A set of overlapping probes was designed covering the core region of the rpoB gene, where the majority of mutations that confer rifampin resistance occur and which is highly conserved in the Mycobacterium tuberculosis complex (Fig. 1). We designed the probe set to be perfectly matched to the wild-type (WT) rpoB gene sequence of Mycobacterium tuberculosis (27) (GenBank accession no. L27989). DNA capture probes were designed using MeltCalc (36, 41), which uses nearest-neighbor calculations to optimize the discrimination of all potential mutations. Criteria were set for a melting temperature (Tm) of 58 to 60°C under our assay conditions of 825 mM monovalent cation. Higher Tms (68°C) were required for probes covering the 3′ end of the amplicon (probes 5.7 and 7.5), due to the presence of competing secondary structure in the amplicon. Each probe was screened to maximize the discrimination (ΔTm) of the major rifampin resistance mutations that it was designed to detect.
Fig 1.
Alignment of probes with the 81-bp core sequence of the rpoB gene. The rpoB gene of M. tuberculosis has an 81-bp core sequence (including codons 508 to 534) that harbors the majority of the mutations clinically relevant for reported drug-resistant TB cases. A 128-bp fragment containing this core region was amplified using bpHDA technology as described above. Probes hybridize to the indicated regions of the amplicon.
Chip production.
Crystalline silicon wafers were coated with the polymer amino functional T-structure polydimethyl siloxane (TSPS; United Chemical Technologies, Bristol, PA) and were cured at 150°C for 24 h. The TSPS-coated wafer was further prepared as described previously (44) to create an aldehyde-functionalized surface and was stored at room temperature.
Probes were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA). The 5′ end of the probes was modified with a reactive hydrazide group designed to interact and attach to the aldehyde-functionalized surface of the silicon wafers (44). Probes in spotting buffer (0.1 M phosphate buffer [pH 7.8], 10% glycerol) were printed (75 nl) on the succinimidyl formyl benzoate (SFB)-coated silicon wafer using a BioDot dispenser (model AD5000). A biotin-labeled probe was spotted as a detection control (DC) to control for the activity of the horseradish peroxidase (HRP)-conjugated anti-biotin antibody and for tetramethylbenzidine (TMB) performance. A hybridization control (HC), which controls for the stringency of the hybridization step by reacting with a biotin-labeled complementary probe present in the hybridization buffer (αHC), was also spotted. After incubation for 2 h, the wafers were washed with 0.1% sodium dodecyl sulfate (SDS) solution, dried, scribed into 6.5-mm2 chips (DynaTek), and stored in nitrogen-purged bags prior to use.
DNA templates and genomic DNA samples.
Single-stranded 128-base synthetic DNA templates covering the region of the rpoB gene were amplified using the TB ID/R assay. The wild-type sequence and 28 mutant sequences were designed and synthesized by Integrated DNA Technologies (7, 8, 12, 13, 14, 16, 20, 23–25, 34, 35, 43). Within a DNA template, the mutated base is denoted by a capital letter, with wild-type bases in lowercase letters. The reference wild-type M. tuberculosis H37Ra genomic DNA (gDNA) template, strains of noncognate mycobacterium (NTM) species, and other bacterial genera for the specificity test were provided by the American Type Culture Collection (ATCC) (Table 1). All other genomic DNA templates (from the 10 wild-type and 26 mutant clinical isolates) were isolated from clinical samples by ZeptoMetrix (Buffalo, NY). Detailed genotypic information for those isolates is given in Table 2.
Table 1.
List of strains for specificity testing
| Species or straina | Source | Hybridization for TB |
|---|---|---|
| Mycobacterium bovis BCG | ATCC 19015 | Wild type |
| Mycobacterium microti | ATCC 11152 | Wild type |
| Mycobacterium africanum | ATCC 35711 | Wild type |
| ATCC 25420 | Wild type | |
| Mycobacterium abscessus | ATCC 19977 | Negative |
| Mycobacterium fortuitum | ATCC 35754 | Negative |
| Mycobacterium genavense | ATCC 51234 | Negative |
| Mycobacterium chelonae | ATCC 35749 | Negative |
| Mycobacterium celatum | ATCC 51131 | Negative |
| Clostridium difficile | ATCC BAA-1382D-5 | Negative |
| Bacillus subtilis | ATCC 23857D-5 | Negative |
| Staphylococcus aureus | ||
| MRSA | ATCC 1005-22-03 | Negative |
| MSSA | ATCC 3555D-5 | Negative |
| Homo sapiens | Roche (catalog no. 11691112001) | Negative |
| Saccharomyces cerevisiae | Novagen (catalog no. 69240-3) | Negative |
MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus.
Table 2.
Genotypic information for clinical isolates
| Sample IDa | rpoB gene sequencing result | TB ID/R assay result |
|
|---|---|---|---|
| WT/mutant ratio(s) | Determination | ||
| Single infections | |||
| H37Ra | WT | 1 | WT |
| 8545 | WT | 1 | WT |
| 24504 | WT | 1 | WT |
| 24507 | WT | 1 | WT |
| 24609 | WT | 1 | WT |
| 26933 | WT | 1 | WT |
| 26934 | WT | 1 | WT |
| 26938 | WT | 1 | WT |
| 27625 | WT | 1 | WT |
| 27628 | WT | 1 | WT |
| 27632 | WT | 1 | WT |
| 18918 | L511P | 143.3 | Mutant |
| 21479 | L511P | 186.3 | Mutant |
| 1868 | 514-515 insert | 171.7 | Mutant |
| 18460 | D516V | 22.8 | Mutant |
| 20626 | H526D | 24.6 | Mutant |
| 21072 | H526D | 87.4 | Mutant |
| 17718 | H526L | 38.6 | Mutant |
| 14191 | H526R | 28.7 | Mutant |
| 14269 | H526R | 57.5 | Mutant |
| 19752 | H526R | 125.6 | Mutant |
| 565 | H526Y | 87.4 | Mutant |
| 4557 | H526Y | 23.1 | Mutant |
| 13554 | H526Y | 53.63 | Mutant |
| 10460 | S531L | 13.8 | Mutant |
| 20324 | S531L | 12.9 | Mutant |
| 26848 | S531L | 29.0 | Mutant |
| 8600 | L533P | 5.3 | Mutant |
| Mixed infections | |||
| 8094 | L511P, D516G | 234.2, 233.2, 232.4 | Mutant |
| 18740 | S512R, S531W | 13.0, 13.0 | Mutant |
| 27950 | D515G, L533P | 8.1, 112.1 | Mutant |
| 27951 | D515G, L533P | 6.9, 154.8 | Mutant |
| 15606 | H526N, L533V | 15.7 | Mutant |
| 11230 | H526N, WT | 2.6 | Mutant |
| 19680 | H526R, WT | 85.6 | Mutant |
| 16866 | H526Y, WT | 1.6 | WT |
ID, identification number.
Primers and blocked-primer helicase-dependent amplification (bpHDA).
Primers were designed against the wild-type rpoB gene sequence of Mycobacterium tuberculosis by using previously published parameters for HDA design (2), with Primer 3 software. Due to the high GC content of the M. tuberculosis genome, constraints were relaxed for primer GC content, product Tm, and product length. The blocked primers are rpoB1502F63 (5′-CGA TCA AGG AGT TCT TCG GCrA CCA G/iSpC3-3′) and rpoB1629F52 (5′-/5BioTEG/GGC ACG CTC ACG TGA CAG ArCC GCC/iSpC3-3′), where iSpC3 indicates a C3 (3-carbon spacer arm) block at the 3′ end of the primer sequence. The unblocked primers used were rpoB1502F1 (5′-CGA TCA AGG AGT TCT TCG GC-3′) and mtb-9R1 (5′-GGC ACG CTC ACG TGA CAG A-3′). All primers were synthesized by Integrated DNA Technologies, Inc.
Amplification reactions were performed at 65°C using 1× ABII buffer (3.85 mM MgSO4, 40 mM NaCl, 0.4 mM IsoAmp deoxynucleoside triphosphates [dNTPs]), 1× IsoAmp enzyme mixture (BioHelix), 10 μl RN2 master mix (Great Basin) containing the enzyme RNase H2, 0.01% Tween 20, 0.01% Triton X-100, 200 nM rpoB1502F63, and 400 nM rpoB1629F52. For real-time amplification, EvaGreen dye (Biotium, Inc.) was added to a final amount of 0.2× for each reaction, and fluorescence was monitored using the LC 480 instrument (Roche). To determine amplification efficiency, the amount of input genomic DNA for the amplification reaction was plotted against the crossing time (the amplification time required to generate a detectable fluorescence signal) and fitted to a linear curve fit. The slope of the curve was used to calculate efficiency 10(−1/slope) − 1.
The natural log of the amount of genomic DNA was plotted against the natural log of the crossing time. The doubling rate (in minutes) is determined from the slope of a linear curve fit.
Chip assay and imaging.
The chip assay was performed using chips immobilized in 96-well plates with flat, square-bottom wells (Whatman). Twenty microliters of the amplicon and 80 μl of the hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5 mg/ml alkaline-treated casein, 0.05% Tween 20, 0.03% ProClin 300 preservative, and 250 pM biotin-labeled reverse complementary sequence for the hybridization control probe [αΗC]) were added and briefly mixed in the well for each chip, and then the plate was incubated for 6 min in an oven (Torrey Pines Scientific) set at 95°C to denature the amplicon. After denaturation, the plate was immediately transferred to a second hybridization oven set at 53°C for 10 min. After hybridization, the wells were briefly washed 3 times (200 μl each time) with wash buffer A (0.1× SSC, 0.1% SDS), followed by 3 washes (200 μl each time) with wash buffer B (0.1× SSC, 0.01% Tween 20). After the wash, 100 μl of a peroxidase-conjugated mouse monoclonal antibody against biotin (Jackson ImmunoResearch Laboratory, Inc.) in 75 mM sodium citrate, 500 mM sodium chloride, 10% fetal bovine serum, 5 mg/ml alkaline-treated casein, and 0.5% ProClin 300 preservative was first added to the well and then incubated at room temperature for 10 min. The wells were further washed briefly 3 times (200 μl each time) with wash buffer B at room temperature. Then 100 μl of TMB substrate (BioFX/SurModics) was added, and the mixture was incubated at room temperature for 5 min. Finally, the wells were rinsed twice with distilled water and methanol, respectively. The chips were dried using compressed air, and images were taken using a charge-coupled device (CCD) camera controlled by the μEye software package (IDS Imaging Development Systems).
Quantitative analysis of chip data.
ImageJ (National Institutes of Health) (http://imagej.nih.gov/ij/) was used to quantify the chip spot signal intensity. A circle was drawn around the spot, and the average signal pixel intensity was measured. The same circle was then dragged out to the neighboring nonspot area, and the average pixel intensity was measured to generate a background pixel value. The adjusted average spot signal intensity was obtained by subtracting the background value from the reacted spot signal intensity. For each probe with a signal intensity of >100 pixels, the result was determined to be wild type. Those values were then averaged to determine a mean wild-type signal. Any probe signal with an intensity of <50 pixels was determined to cover a mutant allele. The mean wild-type signal was divided by the mutant signal to determine a signal-to-noise ratio for each mutation. For a result to be considered a valid test for the presence of the M. tuberculosis complex, at least three probes had to have signals of >100 pixels within 2-fold of each other. A ratio of >2 indicated the presence of a mutation.
rpoB gene sequencing.
A DNA sequencing template (440 bp) containing the rpoB amplicon region was PCR amplified from 100 ng of genomic DNA using 500 nM primers (rpoB1375F [5′-CTGATCCAAAACCAGATCCG-3′] and rpoB1814R [5′-TACACGATCTCGTCGCTAAC-3′]) in the Roche LightCycler 480 SYBR green I master kit and was then gel purified. The DNA templates were bidirectionally sequenced (SeqWright, Inc.) using the rpoB gene-specific primers rpoB1402F (5′-ATGTCGCGGATGGAGCGGGTG-3′) and rpoB1721R (5′-GAGCCGATCAGACCGATGTTG-3′).
RESULTS
bpHDA principle and characterization.
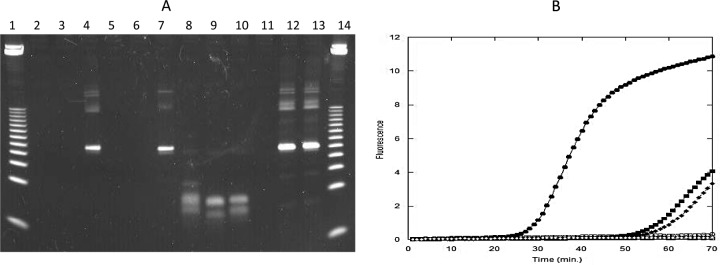
Because the M. tuberculosis genome has a high GC content (65%) (data available at http://tuberculist.epfl.ch/), a primer artifact is a major potential competing side reaction in target amplification approaches lacking “hot-start” capabilities, impacting assay sensitivity and the ability to multiplex. To demonstrate the impact on target amplification performance of the utilization of RNase H2 in bpHDA reactions to unblock primers at elevated temperatures, we amplified sequences in wild-type M. tuberculosis genomic DNA both by bpHDA and by the standard HDA approach. bpHDA and HDA reactions using 50 nM primer concentrations displayed similar amplification kinetics. Under these conditions, amplification reactions were slow, requiring ∼60 min to obtain detectable signal for 30,000 copies of genomic DNA in real-time amplification experiments (Fig. 2B). Additionally, sensitivity was poor, with the inability to amplify genomic DNA amounts below 100 copies even after a 90-min reaction time. In an effort to speed the amplification reactions and improve the limits of detection, higher primer concentrations were tested. In standard HDA reactions, only a primer artifact was amplified, even with larger amounts of the genomic DNA template (Fig. 2A). In bpHDA reactions, a specific product was produced with little competing artifact; moreover, the bpHDA reactions were more rapid, with a crossing time of 29.7 min versus 57.1 min at lower primer concentrations (Fig. 2B). Additionally, it was possible to amplify small amounts of genomic DNA, with amplification of 1 copy detectable by real-time analysis within 50 min. The speed and efficiency of the blocked-primer approach were measured by amplifying various known quantities of wild-type genomic DNA in a real-time detection instrument. Plotting of genomic DNA amounts versus crossing time reveals a highly efficient exponential amplification system (95 to 100%), with a rapid doubling time of 78 s. Even after the amplification reaction mixture was allowed to sit at room temperature for 30 min, no primer artifact was generated by use of blocked primers, further confirming that the RNase H2 must be at elevated temperatures to become active.
Fig 2.
Amplification reactions were performed as described in Materials and Methods. (A) Gel analysis of amplicons. Lanes 1 and 14, 25-bp DNA ladder; lanes 2 to 4, 50 nM/50 nM unblocked HDA primers; lanes 5 to 7, 50 nM/50 nM bpHDA primers; lanes 8 to 10, 200 nM/400 nM unblocked HDA primers; lanes 11 to 13, 200 nM/400 nM bpHDA primers. Lanes 2, 5, 8, and 11, no-template control; lanes 3, 6, 9, and 12, 30 copies of input genomic DNA from M. tuberculosis H37a; lanes 4, 7, 10, and 13, 30,000 copies of H37a genomic DNA. (B) Real-time amplification analysis. Circles, 200 nM/400 nM bpHDA primers; squares, 50 nM/50 nM bpHDA primers; diamonds, 50 nM/50 nM unblocked HDA primers. All open symbols indicate that the reaction mixtures contained no-template controls, and all filled symbols indicate that they contained 30,000 copies H37a genomic DNA.
Analytical performance of the TB ID/R assay.
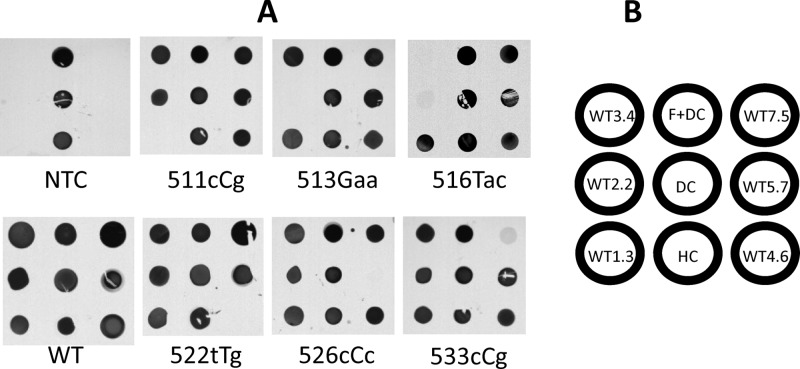
To detect mutations present within the region of the rpoB gene amplified here, we created a set of overlapping probes with perfect complementarity to wild-type Mycobacterium tuberculosis, arrayed on a modified silicon chip. If a mutation is present in the amplicon, the hybridization signal for the probe that is complementary to the mutated region will be dramatically reduced or eliminated. For those mutations that are covered by two overlapping probes, both probes could be affected (Fig. 1). To validate the sensitivity and specificity of the final array, a set of full-length, single-stranded templates was designed representing >95% of all known mutations in the rpoB core sequence of M. tuberculosis (7, 8, 12, 13, 16, 20, 23–25, 34, 35, 43). Each was tested by the TB ID/R assay, and each chip image was captured using a CCD camera. As seen in Fig. 3, probe signals were visually unambiguous; the wild-type amplicon displayed clear and balanced signals for all probes. For the target sequences with mutations in codons 509 to 531, a complete loss of signal was observed in the probes that covered the mutant allele. Significant, but not complete, reductions in signals were observed for the various mutations in codon 533. For all of the point mutations tested, a single probe lost signal except in the case of mutations within codons 516 and 518, which affected the two probes that overlap those alleles. One mutation, 508Gcc, could not be distinguished with this probe set.
Fig 3.
rpoB gene SNP discrimination by the TB ID/R test. Synthetic 128-bp templates were amplified using bpHDA as described above and were hybridized to the chip, and CCD camera images were captured. (A) Representative images of mutations that affect each probe. NTC, no-template control; WT, wild-type M. tuberculosis. Capitalized bases indicate mutant positions. (B) Array map. HC, hybridization control; DC, detect control; F+DC, fiducial marker mixed with the detect control.
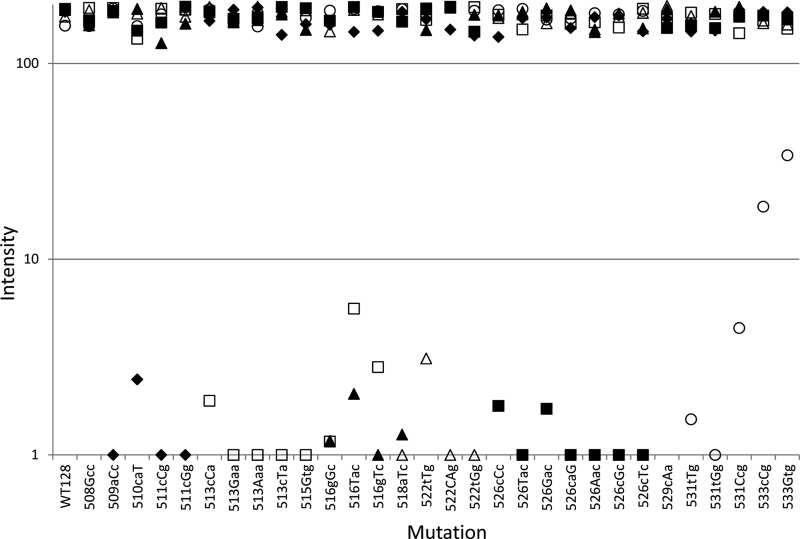
Due to the virtually binary responses of the probes to mutations, visual discrimination is straightforward: if a spot is missing or very faintly present, then a mutation is present. However, the array is still visually complex, so image analysis was also investigated to generate an automated call. Probe pixel intensities were determined from the CCD image, corrected for background, and plotted for each sample tested (Fig. 4). CCD image analysis verified the visual interpretation of the assay performance; all mutations tested except for those in codon 533 have signal dropout (<10 pixels). Analysis of each mutant sample reveals signal-to-noise ratios of >20 for the dropouts, 5.2 for 533Gtg, and 9.4 for 533cCg, allowing for unambiguous discrimination (Fig. 4).
Fig 4.
Performance of the TB ID/R assay for detecting synthetic mutants with mutations in the rpoB locus. Synthetic templates with mutations (indicated by capital letters) were amplified, tested by the TB ID/R assay, and subjected to CCD imaging, and images were analyzed as described above. Signals for each probe on each chip were plotted. Filled diamonds, probe WT1.3; open squares, probe WT2.2; filled triangles, probe WT3.4; open triangles, probe WT4.6; filled squares, probe WT5.7; open circles, probe WT7.5. Signals below 10 pixels are difficult to detect by the unaided eye.
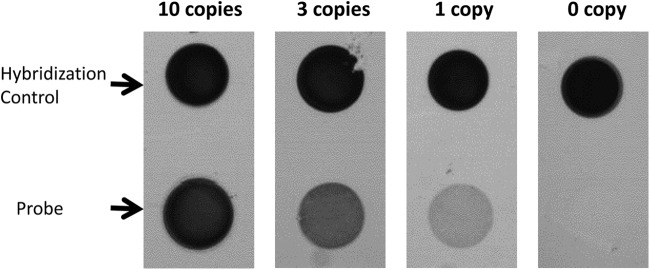
The limit of detection was determined by titrating known quantities of purified, wild-type M. tuberculosis genomic DNA using the TB ID/R assay. To detect a single copy of genomic DNA, 40 min of amplification time was required (Fig. 5).
Fig 5.
Limit of detection of the assay. The limit of detection was determined by using a wild-type (H37Ra) genomic template with an amplification time of 40 min. (Top) Hybridization control for assay performance. (Bottom) Signal from one probe (WT1.3) as a function of the input copy number of genomic DNA to the TB ID/R assay.
Additionally, we determined reactivity with other Mycobacterium species, including those within the M. tuberculosis complex (Table 1). All members of the M. tuberculosis complex tested, including Mycobacterium bovis, Mycobacterium microti, and two strains of Mycobacterium africanum, generated signals on all probes. By the criteria that at least three probes must have signals of >100 pixels and within 2-fold of each other, none of the 5 Mycobacterium species that do not belong to the M. tuberculosis complex were detected. However, Mycobacterium genavense yielded strong signals on two probes and a very weak signal on another. Testing of several non-Mycobacterium genomic DNA samples, including human DNA, displayed no signal (Table 1).
Clinical-isolate testing.
To further verify this assay, we tested genomic DNA from clinical isolates for 11 wild-type and 25 RIF-resistant specimens (Table 2). The TB ID/R assay correctly classified 11/11 wild-type samples (100% specific; 95% confidence interval [95% CI], 67.9 to 100%) and 24/25 RIF-resistant samples (96.0%; 95% CI, 77.7 to 98.7%) relative to the reference method of rpoB gene sequencing. The wild-type amplicons displayed strong signals on each probe, with less than 2-fold variability across 11 unique isolates (data not shown). For each of the clinical samples containing a single mutation, levels of discrimination similar to those obtained using synthetic templates were observed. Signal dropout was observed for mutations in codons 511 to 526. Discrimination for the amplicon from genomic DNA of the S531L mutant was not as good as that for the synthetic template, but the mutant was easily discriminated, with a signal-to-noise ratio of 19 as an average for three unique strains. The amplicon with the L533P mutation performed similarly to the synthetic template, with a signal-to-noise ratio of 5.3.
The TB ID/R assay was further challenged with 8 clinical samples that contained multiple mutations in the rpoB core region as determined by rpoB gene sequencing. The TB ID/R assay correctly classified 7/8 samples as mutant. An unexpected probe response in a sample with H526N and L533V (sample 15606) was observed, with signal loss observed only with probe 7.5, when both probes 5.7 and 7.5 should have been affected. Sample 11230 was correctly reported as mutant by the TB ID/R test, but with a very low ratio, 2.6. Sequencing of this sample revealed a mixed base at the mutant position, indicating the presence of two unique alleles, explaining the low ratio for this mutation. The mixed sample (sample 16866) that was discordant between the TB ID/R assay and the initial rpoB gene sequencing was subjected to repeat sequencing of the rpoB gene. Sample 16866 was misidentified as wild type by the TB ID/R assay. Resequencing confirmed the initial sequencing determination for the sample to be a mixed WT/H526Y result.
DISCUSSION
Here we have described a novel amplification method that adds a hot-start component to the isothermal amplification method bpHDA, permitting more-rapid amplification and a potentially greater level of multiplexing. By use of this blocked-primer approach, initiation of DNA amplification is DNA target dependent, improving sensitivity and specificity by mitigation of competing primer artifact amplification. Analysis revealed a highly efficient exponential amplification system with short doubling times, suggesting that the initiation of HDA by RNase H2-mediated cleavage of blocked primers is rapid and not rate-limiting. Amplified products were detected using a silicon chip modified with a polymer coating to create a surface with optical properties such that the surface-bound DNA hybrids effect a permanent change in color intensity on the chip surface visible to the unaided eye. Additionally, the surface is molecularly flat and chemically inert, reducing nonspecific interactions and thus allowing for highly sensitive and specific detection. This allows for discrimination of single-nucleotide polymorphisms (SNPs) as demonstrated here, and with picomolar limits of detection, only short hybridization reactions are required with target sequences amplified by bpHDA to generate detectable surface-bound hybrids. Because the signal response to mutations is effectively binary, results can be determined using either a noninstrumented visual approach or an inexpensive CCD or complementary metal oxide semiconductor (CMOS) camera-based imaging system.
The analytical performance of the TB ID/R assay is similar to those of other recently described molecular methods for the detection of M. tuberculosis complex strains, including those with mutations in the rpoB gene associated with RIF resistance. A comprehensive set of mutant templates was constructed based on a survey of the literature (7, 8, 12, 13, 16, 20, 23–25, 34, 35, 43), and all mutations tested here were distinguished except for one, 508Gcc. Nearest-neighbor analysis revealed that this probe set was unable to distinguish this mutation (ΔTm, 0°C). Fortunately, this is a very rare mutation, accounting for less than 0.1% of the reported rifampin resistance cases in TB. Alternative designs, including the addition of secondary mutations in the probe or primer sequences, are currently being tested to improve the discrimination of this mutation as well as that of codon 533, where the discrimination is also not binary. Testing of genomic DNA isolated from clinical specimens displayed excellent sensitivity for the detection of RIF-resistant strains and excellent specificity for RIF-sensitive strains. In contrast to other recently described molecular diagnostic methods, such as real-time PCR (13) and loop-mediated isothermal amplification (LAMP) (17), with limited multiplex detection capabilities, more information can be added to the TB ID/R assay to detect additional M. tuberculosis resistance mechanisms, potentially aiding in appropriate management and treatment of greater numbers of infected patients, and further reducing transmission risks. Diagnosis of MDR TB requires information on isoniazid (INH) resistance in addition to RIF resistance, and mutations in the katG and inhA genes have been described for the detection of INH resistance (15), while XDR TB may be diagnosed by detecting mutations in the gyrA and gyrB genes for fluoroquinolone resistance and the rrs gene for resistance to aminoglycosides, in addition to the RIF and INH mutations. These markers have been shown to detect XDR TB with high sensitivity using another scalable test platform, the reverse line blot hybridization (RLBH) assay (1, 3, 15). However, the RLBH test is time-consuming and technically complex, limiting its usefulness in the point-of-care setting.
Multiple mutations within a sample, presumably from infections with more than one TB strain or from individual strains with multiple mutations, present potential challenges for genotypic detection approaches. If two or more mutations exist within a single TB isolate, detection is straightforward, but if there is a mixture of multiple unique RIF-resistant strains or mixtures of RIF-resistant and RIF-sensitive populations, resistance detection could be complicated. To illustrate this point, we tested two isolates that had a mixture of wild-type and mutant genomic templates (H526R/WT and H526Y/WT) and obtained very different results. Both the H526R and H526Y mutations create a signal dropout for probe 5.7 in an isolate with a single mutation; thus, they are strongly discriminated. We detected the H526R/WT sample unambiguously, indicating that the assay is tolerant of the presence of some amount of competing wild-type allele. However, the identification of the H526Y/WT sample as wild type by the TB ID/R assay suggests that if the ratio of the wild-type allele to the mutant allele is higher, the potential for missing a mutation exists. Similarly, sample 15606 was correctly identified as a mutant sample, but not all of the mutations were detected, suggesting that more than one infecting organism was the source of the mutations. In the two samples with D516G and L533P mutations, we were able to detect each mutation with the same level of discrimination as if they were singly mutated isolates, suggesting that they both occurred in the same organism. It has been observed previously, in an assay with a similar probe design, that the strength of discrimination of a given mutation has a strong effect on the ability to detect mutations in a mixed sample (5). This factor, combined with the fraction of each allele present, is critical for correct detection in RIF-resistant samples containing more than one infecting organism.
The need for improved point-of-care testing for drug resistance in M. tuberculosis is acute. Increasing the initial test sensitivity from 35% (microscopy sensitivity for an HIV-positive patient) to 95% (molecular diagnostic approaches) would decrease the mean delay in diagnosis by approximately 25 days and reduce the dropout rate (the proportion of infected individuals who cease seeking medical treatment) by approximately 30% (26). By providing drug resistance information during the initial diagnostic test, the delay to appropriate treatment, a critical factor in reducing mortality attributable to infectious diseases, would be reduced further (22). To address the need for an easy-to-use, highly sensitive diagnostic test for M. tuberculosis identification and drug susceptibility information, we are developing the TB ID/R test in a disposable cartridge on a small, inexpensive, electromechanically simple, and potentially battery-powered device. This test is being automated in a manner that is consistent with the ASSURED goals for developing world point-of-care testing for drug-resistant TB (40). The combination of isothermal amplification with chip-based, eye-visible detection allows for the use of low-cost heaters and imaging equipment. Additionally, we are utilizing low-cost, highly stable, and robust reagents, with measured stability at 37°C for 9 months so far, with no loss in activity (data not shown). These reagents have been used in commercially successful assays performed in the point-of-care setting for test-and-treat indications such as viral and respiratory pathogens that require rapid, accurate results (11). Furthermore, the chips can be produced inexpensively on a very large scale by utilizing well-established semiconductor processes (18). Early work with an appreciably similar device we are developing has shown excellent performance for identifying staphylococcal species and the presence of the mecA gene in Gram-positive cocci in cluster-positive blood cultures from hospitalized patients (9). A low-cost, easy-to-use device that can detect RIF resistance could have a tremendous impact on reducing the community spread of the disease, as well as improving outcomes for infected patients by providing appropriate treatment sooner.
ACKNOWLEDGMENTS
We thank Laura Geyer for technical assistance, ZeptoMetrix for clinical isolates and DNA isolation, and the ATCC for isolation of genomic DNA as well. We also thank the members of the Research and Development Group at Great Basin for thoughtful and productive discussions.
Footnotes
Published ahead of print 18 April 2012
REFERENCES
- 1. Ajbani K, Shetty A, Mehta A, Rodrigues C. 2011. Rapid diagnosis of extensively drug-resistant tuberculosis by use of reverse line blot hybridization assay. J. Clin. Microbiol. 49:2546–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. An L, et al. 2005. Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J. Biol. Chem. 280:28952–28958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bártfai Z, et al. 2001. Molecular characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Hungary by DNA sequencing and the line probe assay. J. Clin. Microbiol. 39:3736–3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Becerra MC, et al. 2010. Recurrence after treatment for pulmonary multidrug-resistant tuberculosis. Clin. Infect. Dis. 51:709–711 [DOI] [PubMed] [Google Scholar]
- 5. Blakemore R, et al. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J. Clin. Microbiol. 48:2495–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boehme CC, et al. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caws M, et al. 2006. Mutations prevalent among rifampin- and isoniazid-resistant Mycobacterium tuberculosis isolates from a hospital in Vietnam. J. Clin. Microbiol. 44:2333–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan RC, et al. 2007. Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. J. Antimicrob. Chemother. 59:866–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Denys GA, Renzi PB, Wissel CM, Koch KM. 2011. Portrait Staph ID/R: a novel molecular diagnostic test for simultaneous identification of Staphylococcus species and detection of the mecA gene directly from positive blood cultures, abstr 40-D. Abstr. 2nd ASMET Conf American Society for Microbiology, Washington, DC [Google Scholar]
- 10. Dobosy JR, et al. 2011. RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnol. 11:80 doi:10.1186/1472-6750-11-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerber MA, et al. 1997. Optical immunoassay for group A β-hemolytic streptococcal pharyngitis: an office-based, multicenter investigation. JAMA 277:899–903 [PubMed] [Google Scholar]
- 12. Heep M, et al. 2001. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:107–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Helb D, et al. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Herrera L, Jiménez S, Valverde A, GarcíA-Aranda MA, Sáez-Nieto JA. 2003. Molecular analysis of rifampin-resistant Mycobacterium tuberculosis isolated in Spain (1996–2001). Description of new mutations in the rpoB gene and review of the literature. Int. J. Antimicrob. Agents 21:403–408 [DOI] [PubMed] [Google Scholar]
- 15. Hillemann D, Weizenegger M, Kubica T, Richter E, Niemann S. 2005. Use of the genotype MTBDR assay for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis complex isolates. J. Clin. Microbiol. 43:3699–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang H, Jin Q, Ma Y, Chen X, Zhuang Y. 2002. Characterization of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis isolated in China. Tuberculosis (Edinb.) 82:79–83 [DOI] [PubMed] [Google Scholar]
- 17. Iwamoto T, Sonobe T, Hayashi K. 2003. Loop-mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J. Clin. Microbiol. 41:2616–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jenison R, Yang S, Haeberli A, Polisky B. 2001. Interference-based detection of nucleic acid targets on optically coated silicon. Nat. Biotechnol. 19:62–65 [DOI] [PubMed] [Google Scholar]
- 19. Jenison R, La H, Haeberli A, Ostroff R, Polisky B. 2001. Silicon-based biosensors for rapid detection of protein or nucleic acid targets. Clin. Chem. 47:1894–1900 [PubMed] [Google Scholar]
- 20. Kapur V, et al. 1995. Rapid Mycobacterium species assignment and unambiguous identification of mutations associated with antimicrobial resistance in Mycobacterium tuberculosis by automated DNA sequencing. Arch. Pathol. Lab. Med. 119:131–138 [PubMed] [Google Scholar]
- 21. Kapur V, et al. 1994. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase beta subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J. Clin. Microbiol. 32:1095–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kollef MH, Sherman G, Ward S, Fraser VJ. 1999. Inadequate antimicrobial treatment of infections: a risk factor for hospital mortality among critically ill patients. Chest 115:462–474 [DOI] [PubMed] [Google Scholar]
- 23. Lipin MY, Stepanshina VN, Shemyakin IG, Shinnick TM. 2007. Association of specific mutations in katG, rpoB, rpsL and rrs genes with spoligotypes of multidrug-resistant Mycobacterium tuberculosis isolates in Russia. Clin. Microbiol. Infect. 13:620–626 [DOI] [PubMed] [Google Scholar]
- 24. Mani C, Selvakumar N, Narayanan S, Narayanan PR. 2001. Mutations in the rpoB gene of multidrug-resistant Mycobacterium tuberculosis clinical isolates from India. J. Clin. Microbiol. 39:2987–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mani C, Selvakumar N, Kumar V, Narayanan S, Narayanan PR. 2003. Comparison of DNA sequencing, PCR-SSCP and PhaB assays with indirect sensitivity testing for detection of rifampin resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 7:652–659 [PubMed] [Google Scholar]
- 26. Millen SJ, Uys PW, Hargrove J, van Helden PD, Williams BG. 2008. The effect of diagnostic delays on the drop-out rate and the total delay to diagnosis of tuberculosis. PLoS One 3:e1933 doi:10.1371/journal.pone.0001933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miller LP, Crawford JT, Shinnick TM. 1994. The rpoB gene of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 38:805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mitnick CD, Appeton SC, Shin SS. 2008. Epidemiology and treatment of multidrug resistant tuberculosis. Semin. Respir. Crit. Care Med. 29:499–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mitnick CD, et al. 2008. Comprehensive treatment of extensively drug-resistant tuberculosis. N. Engl. J. Med. 359:563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nahid P, Pai M, Hopewell PC. 2006. Advances in the diagnostics and treatment of tuberculosis. Proc. Am. Thorac. Soc. 3:103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Noordhoek GT, van Embden JD, Kolk AH. 1996. Reliability of nucleic acid amplification for detection of Mycobacterium tuberculosis: an international collaborative quality control study among 30 laboratories. J. Clin. Microbiol. 34:2522–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pai M, Kalantri S, Dheda K. 2006. New tools and emerging technologies for the diagnosis of tuberculosis. Part II. Active tuberculosis and drug resistance. Expert Rev. Mol. Diagn. 6:423–432 [DOI] [PubMed] [Google Scholar]
- 33. Perkins MD, Cunningham J. 2007. Facing the crisis: improving the diagnosis of tuberculosis in the HIV era. J. Infect. Dis. 196:S15–S27 [DOI] [PubMed] [Google Scholar]
- 34. Rossau R, et al. 1997. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob. Agents Chemother. 41:2093–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sajduda A, et al. 2004. Molecular characterization of rifampin- and isoniazid-resistant Mycobacterium tuberculosis strains isolated in Poland. J. Clin. Microbiol. 42:2425–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schutz E, von Ahsen N. 1999. Spreadsheet software for the thermodynamic melting point prediction of oligonucleotide hybridization with and without mismatches. Biotechniques 27:1218–1224 [DOI] [PubMed] [Google Scholar]
- 37. Suffys P, et al. 2000. Evaluation of the polymerase chain reaction for the detection of Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 4:179–183 [PubMed] [Google Scholar]
- 38. Telenti A, et al. 1993. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647–650 [DOI] [PubMed] [Google Scholar]
- 39. Telzak EE, et al. 1995. Multidrug-resistant tuberculosis in patients without HIV infection. N. Engl. J. Med. 333:907–911 [DOI] [PubMed] [Google Scholar]
- 40. Urdea M, et al. 2006. Requirements for high impact diagnostics in the developing world. Nature 444:73–79 [DOI] [PubMed] [Google Scholar]
- 41. von Ahsen N, Oellerich VW, Armstrong VW, Schutz E. 1999. Application of thermodynamic nearest neighbor mode to estimate nucleic acid stability and optimize probe design: prediction of melting points of different mutations of apolipoprotein B 3500 and factor V Leiden with a hy-bridization probe genotyping assay in the LightCycler. Clin. Chem. 45:2094–2101 [PubMed] [Google Scholar]
- 42. World Health Organization 2008. Anti-tuberculosis drug resistance in the world: fourth global report. The WHO/IUATLD Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Publication no. WHO/HTM/TB/2008.394. World Health Organization, Geneva, Switzerland [Google Scholar]
- 43. Yang B, et al. 1998. Relationship between antimycobacterial activities of rifampin, rifabutin and KRM-1648 and rpoB mutations of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 42:621–628 [DOI] [PubMed] [Google Scholar]
- 44. Zhong XB, et al. 2003. Single-nucleotide polymorphism genotyping on optical thin-film biosensor chips. Proc. Natl. Acad. Sci. U. S. A. 100:11559–11564 [DOI] [PMC free article] [PubMed] [Google Scholar]