Abstract
The rapid and accurate diagnosis of tuberculosis (TB) in children and extrapulmonary TB in adults continues to be a challenge. In this study, we determined the lower limit of detection (LOD) of the GeneXpert MTB/RIF assay with nonrespiratory specimens and investigated the utility of flotation procedures for concentrating the bacilli. Clinical specimens (9 cerebrospinal fluid [CSF], 13 gastric aspirate, 8 tissue, and 17 stool) were spiked with single-celled Mycobacterium tuberculosis, and the LOD of the GeneXpert assay was determined. Flotation studies were conducted with sucrose and NaCl, and the cycle thresholds of the MTB/RIF assay were compared between treated and untreated samples. There was no significant difference between the LODs of the GeneXpert assay with saline solution (median, 33 CFU/ml) and CSF (median, 25 CFU/ml) (P > 0.05) or gastric aspirate samples (median, 58 CFU/ml) (P > 0.05). The LOD with spiked tissue (median, 1,525 CFU/ml) and stool samples (median, 6,800 CFU/ml) was significantly elevated compared to that determined with saline solution (P ≤ 0.05 and ≤ 0.0005, respectively). Flotation studies with sucrose or NaCl did not consistently result in lowered cycle thresholds in stool or gastric aspirates, but a cycle reduction of >10 was achieved in two of the three pooled CSF samples. Unlike the results seen with tissue and stool samples, there was no significant PCR inhibition in the MTB/RIF assay with CSF and gastric aspirates. Although preconcentration of CSF samples with sucrose and NaCl may enhance detection of M. tuberculosis by PCR, further advances are needed to concentrate the bacilli and eliminate PCR inhibitors in paucibacillary nonrespiratory samples.
INTRODUCTION
The self-contained and automated GeneXpert MTB/RIF nucleic acid amplification test (NAAT) represents a significant advancement for laboratory diagnosis of tuberculosis (TB) and detection of rifampin resistance (3, 10). The GeneXpert technology allows rapid and relatively sensitive diagnosis of pulmonary TB in sputum (4, 16). Diagnosis of TB in children and others who cannot produce sputum remains challenging; sputum induction and gastric aspiration are invasive and uncomfortable procedures with relatively low yield (17). The ability to test stool as a surrogate specimen could be advantageous. Diagnosis of extrapulmonary TB by conventional methods such as microscopy and culture remains challenging in cases of paucibacillary TB (18). While the performance of the GeneXpert MTB/RIF assay with nonrespiratory samples such as tissue, body fluids, stool, pus, gastric aspirate, and cerebrospinal fluid (CSF) has revealed excellent sensitivity in smear-positive specimens, its sensitivity in smear-negative specimens has been variable (2, 5, 11, 12, 14, 15, 18, 20). In the largest study published to date, the sensitivity of the GeneXpert MTB/RIF assay was 96% (139/145) in smear-positive and 64% (89/138) in smear-negative extrapulmonary specimens (18). Thus, further studies are needed to define the impeding factors and to identify a simple and efficient way to enhance the sensitivity of the GeneXpert MTB/RIF assay for detection of Mycobacterium tuberculosis in nonrespiratory samples, particularly in resource-poor settings.
In this study, we first determined the lower limit of detection (LOD) of the GeneXpert MTB/RIF assay with nonrespiratory samples spiked with live single-celled bacilli. We then investigated the effectiveness of flotation procedures with sucrose and NaCl to determine if bacilli in extrapulmonary samples can be concentrated in the top layer prior to testing with the GeneXpert assay.
MATERIALS AND METHODS
Bacterial cultures.
A single-celled suspension of M. tuberculosis H37Ra was freshly prepared by concentrating mid-logarithmic-phase broth cultures in Middlebrook 7H9 broth (Hardy Diagnostics, Santa Maria, CA) and passage through a 5-μm-pore-size filter by gravity. The presence of single cells and the absence of clumps were confirmed by microscopic inspection. The bacterial concentration was targeted to be ∼1 × 107 CFU/ml based on the optical density and the volume of the starting culture. The actual counts were confirmed by serial dilution in normal saline solution and plating on Middlebrook 7H11 agar (Hardy Diagnostics).
Clinical specimens.
Deidentified specimens of TB-negative (by culture) gastric aspirates (n = 13), CSF (n = 9), tissue (n = 8), and stool (n = 17) were used. Use of samples for this purpose was approved by the institutional review board (IRB). CSF specimens included 7 clear colorless samples, 1 clear yellow sample, and 1 bloody sample. Tissue samples included 1 lung sample, 1 abscess sample, 5 bone/cartilage samples, and 1 soft tissue sample. Normally discarded gastric aspirates were retrieved after routine endoscopy procedures performed on children undergoing endoscopy for noninfectious workups. All specimens were stored at 4°C.
LOD studies.
Specimens were tested individually. CSF and gastric aspirates were used directly. Tissue samples 3 to 4 g in weight were homogenized in 3 ml of normal saline solution in a closed tissue grinder system (Dupont). A 0.25-g aliquot of fresh stool was emulsified in 4.5 ml of normal saline solution. The debris in homogenized tissue and stool samples was allowed to settle for 20 min, and the top 2 ml was removed for spiking studies. Serial 10-fold dilutions (1:10 to 1:1,000,000) of the single-celled bacterial suspensions were prepared in saline solution. A 100-μl volume of each bacterial dilution was added to 500 μl of each specimen to produce final M. tuberculosis concentrations of from ∼1.6 ×106 CFU/ml to ∼1.6 CFU/ml. A 100-μl volume of each bacterial dilution was also added to 500 μl of saline solution and used as a control (n = 13). The spiked samples were mixed thoroughly, and the MTB/RIF assay was performed immediately. Sample buffer (provided in the kit) (1.7 ml) was added to each sample and inverted 10 times. Tubes were incubated at room temperature for 15 min, with a second mixing performed halfway through. Using the pipette supplied with the kit, the entire sample volume (∼2.3 ml) was transferred to an MTB/RIF cartridge and placed in the Gene Xpert instrument and run per manufacturer's instructions. The lower limit of detection (LOD) determined with each spiked specimen was defined as the lowest concentration of M. tuberculosis in that specimen that was detected by the GeneXpert assay.
Flotation studies.
The flotation solutions were 56% sucrose (Sheather's solution), 36% NaCl, and normal saline solution. CSF and gastric samples from multiple patients were pooled to obtain sufficient volume for the flotation studies. For sucrose studies, 5 ml of each pool was combined with 5 ml of saline solution or 56% sucrose to obtain 0% and 28% sucrose concentrations. For NaCl studies, 0 or 1.8 g of NaCl was dissolved in 5 ml of pooled specimen to obtain 0% and 36% NaCl concentrations. CSF and gastric samples were spiked with 10 μl of a single-celled M. tuberculosis suspension containing ∼1 × 107 CFU/ml and subjected to a vortex procedure for 30 s. A 0.5-g volume of fresh stool sample was emulsified in 10 ml of each flotation solution to obtain 0%, 14%, and 28% sucrose and 0%, 24%, and 36% NaCl. Stool samples were spiked with 10 μl of bacterial suspension, subjected to a vortex procedure for 30 s, and poured over a sieve funnel (Medical Chemical Corporation, Torrance, CA) to remove fibrous debris. A 10-μl volume of bacterial suspension was also added to 10 ml of saline solution and used as a control. The debris in all samples was allowed to settle by gravity for 1 h or by centrifugation at 500 ×g for 1 min. Six successive 1-ml aliquots were collected from the volume at the top of the mixture. A total of 0.5 ml of the first volume (referred to as the “top”) and 0.5 ml of the sixth volume (referred to as the “middle”) were combined with 1.8 ml of sample buffer and tested with the Xpert MTB/RIF assay as described above.
Analysis.
The LOD of the GeneXpert MTB/RIF assay with spiked clinical samples was compared to the LOD of M. tuberculosis with spiked saline solution. Wilcoxon's test was used to compare differences in medians. The cycle threshold (CT) of probe B of the top and middle volumes of each spiked sample treated with sucrose and NaCl was compared to that of untreated samples.
RESULTS
Lower limit of detection of the GeneXpert MTB/RIF assay.
To determine if the relative insensitivity of the GeneXpert assay in smear-negative extrapulmonary specimens (18) is due to the presence of PCR inhibitors or is due to paucibacillary disease, we compared the lower limit of detection (LOD) of the GeneXpert MTB/RIF assay with spiked samples to the LOD with saline solution. We used single-celled suspensions of live bacilli to ensure an accurate measure of LOD. As shown in Fig. 1, the median LOD of GeneXpert MTB/RIF assay was 33 CFU/ml (range, 8 to 325) with saline, 25 CFU/ml (range, 8 to 33) with CSF, 58 CFU/ml (range, 25 to 775) with gastric aspirate, 1,525 CFU/ml (range, 150 to 2,450) with homogenized tissue, and 6,800 CFU/ml (range, 267 to 193,333) with emulsified stool. There was no significant difference between the LODs of the GeneXpert MTB/RIF assay with saline solution and CSF or gastric aspirate samples (P > 0.05). However, the LOD of the GenXpert assay determined with spiked tissue (P ≤ 0.05) and stool (P ≤ 0.0005) was significantly higher than that determined with saline solution.
Fig 1.
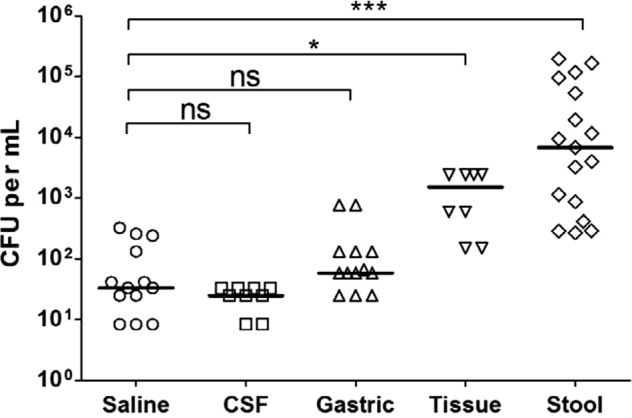
Comparison of lower limits of detection of the GeneXpert MTB/RIF assay in seeded samples. Clinical specimens were spiked with dilutions of single-celled suspensions of M. tuberculosis. The lower limit of detection of the GeneXpert MTB/RIF assay in CSF (n = 9), gastric aspirate (n = 13), tissue (n = 8), and stool (n = 12) samples was compared to that in saline solution (n = 13). Wilcoxon's test was used to compare differences in medians. The asterisks indicate statistically significant difference. *, P ≤ 0.05; ***, P ≤ 0.0005; ns, not significant.
Flotation studies.
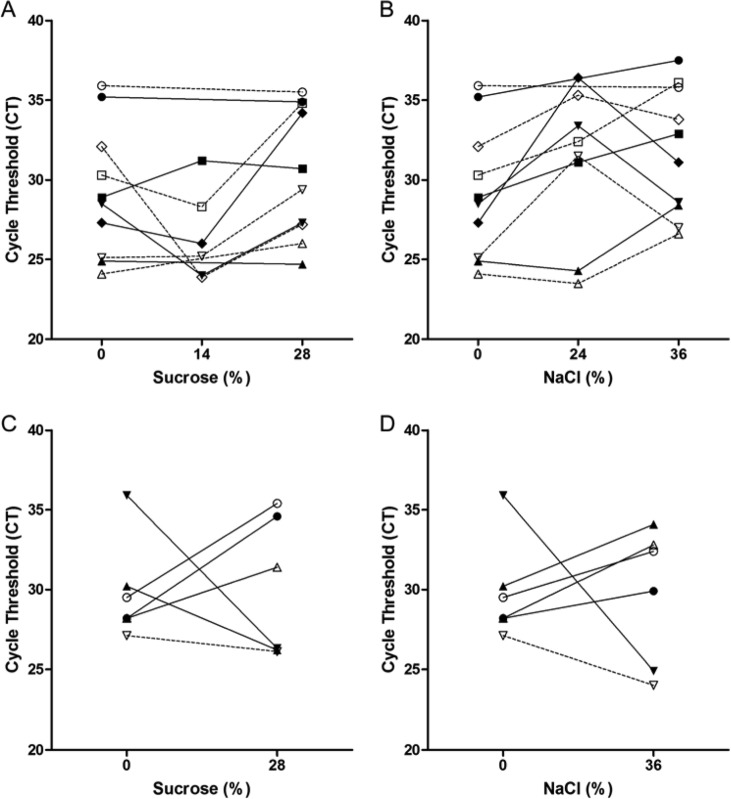
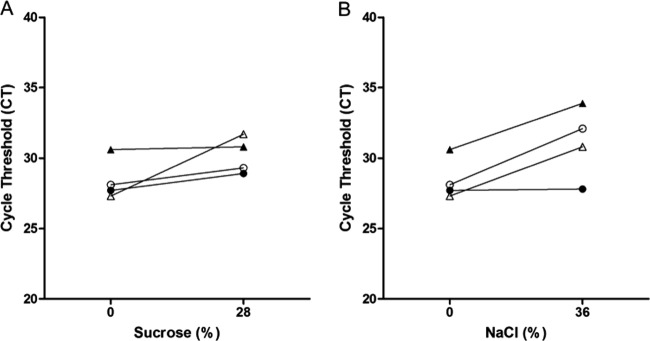
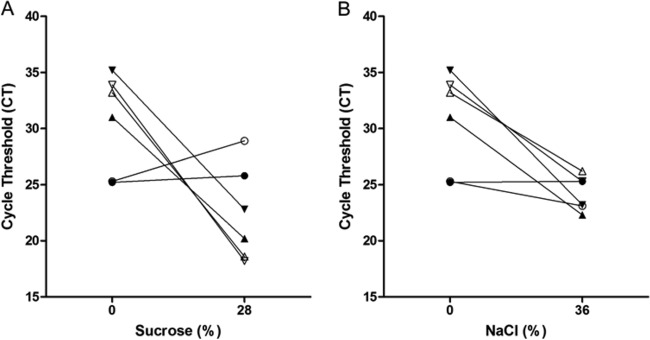
Next, we investigated whether simple flotation procedures used in clinical parasitology to concentrate helminthic eggs in stool preparations (6, 13) can be used to concentrate M. tuberculosis in paucibacillary specimens. The flotation experiments were based on the premise that, because the buoyant density of M. tuberculosis cells (1.02 to 1.13 g/cm3) is less than that of sucrose and salt solutions, it should be possible to float the bacilli to the surface in clinical samples treated with sucrose and salt (7). If the attempt were successful, one could sample the top layer of each flotation trial and thus lower the cycle threshold (CT) value. Treatment of M. tuberculosis with sucrose at final concentrations of 14% and 28% and with NaCl at 24% and 36% by either gravity or centrifugation did not consistently yield lower CT values in the top layer of stool preparations compared to untreated sample values (Fig. 2). Similar findings were obtained with gastric aspirates (Fig. 3 and data not shown). For CSF samples, however, sucrose and NaCl reduced the CT values in the top layer by an average of 11.6 and 10.4 cycles, respectively, in 2 out of 3 three pooled samples and resulted in a slight positive change in the third sample (Fig. 4).
Fig 2.
Comparison of cycle threshold values of the GeneXpert MTB/RIF assay on stool preparations floated with sucrose and NaCl. Stool preparations were treated with the indicated concentrations of sucrose (A and C) or NaCl (B and D). Bacilli were allowed to float by gravity (A and B) or subjected to centrifugation (C and D). Symbols represent unique patient samples. Closed symbols represent the top volume of stool preparations, while open symbols represent the middle volume.
Fig 3.
Comparison of cycle threshold values of the GeneXpert MTB/RIF assay on gastric aspirates floated with sucrose and NaCl. Pooled gastric aspirates were treated with the indicated concentrations of sucrose (A) or NaCl (B). Bacilli were floated by centrifugation. Symbols represent unique pooled samples. Closed symbols represent the top volume, while open symbols represent the middle volume.
Fig 4.
Comparison of cycle threshold values of the GeneXpert MTB/RIF assay on CSF samples floated with sucrose and NaCl. Pooled CSF samples were treated with the indicated concentrations of sucrose (A) or NaCl (B). Bacilli were floated by centrifugation. Symbols represent unique pooled samples. Closed symbols represent the top volume, while open symbols represent the middle volume.
DISCUSSION
Our results showed that gastric aspirate and CSF samples had M. tuberculosis LODs similar to those of saline solution in the GeneXpert assay. The LOD with CSF samples (median, 25 CFU/ml) was lower than that reported for spiked sputum samples (131 CFU/ml; 95% confidence interval [CI], 106.2 to 176.4) (10). These findings suggest that the reduced sensitivity of the MTB/RIF assay in smear-negative CSF and gastric aspirate specimens is due to a paucity of organisms and not to inhibitory factors encountered in the PCR (2, 5, 11, 15, 18, 20). In contrast, the LOD of the GeneXpert assay was significantly elevated with spiked tissue and stool samples. The presence of soluble PCR inhibitors such as heme, bilirubin, and bile salts in tissue and stool is well known (1, 19). These PCR inhibitors interfere with the DNA polymerase and decrease the sensitivity of PCR-based NAATs. Although the Xpert system removes many inhibitors during the wash and filtration steps, it is possible that some substances pass into the reaction vial. Alternatively, the tubercle bacilli may have bound to and cosettled with debris or cells in tissue and stool samples rather than having floated to the top. The stool samples showed the broadest range of LOD (267 to 193,333 CFU/ml) compared to other sample types, suggesting that heterogeneity in stool makeup may variably affect the sensitivity of the MTB/RIF assay. Although the ability to use stool as an alternative to sputum samples would be greatly beneficial to pulmonary TB patients who cannot expectorate, there are limited clinical data on the sensitivity of the GeneXpert system with stool samples. The two prior studies that evaluated the sensitivity of the GeneXpert system with actual patient stool samples showed a sensitivity of 100% compared to culture, although only 3 samples were tested (11, 15). In contrast to our study, both studies used conventional digestion-decontamination with N-acetyl-l-cysteine-NaOH followed by concentration of the sediment by centrifugation. Our aim, however, was to develop a simple, centrifuge-free sample preparation protocol for use in minimally equipped laboratories and smear-microscopy centers in resource-poor settings where GeneXpert technology could have the greatest impact.
The low LOD observed with CSF and gastric aspirates indicates that the GeneXpert system is a viable diagnostic tool for detection of M. tuberculosis and rifampin resistance in these sample types. However, there is still a need for a simple and practical approach to concentrating the organisms in paucibacillary or complex samples prior to performing the MTB/RIF assay. The flotation of M. tuberculosis in clinical specimens based on its buoyancy relative to sucrose and NaCl solutions is an attractive methodology, because it requires simple reagents which can be easily obtained and prepared anywhere at very low cost and can be implemented with minimal equipment. Our efforts to concentrate M. tuberculosis in spiked gastric aspirate and stool samples by the use of flotation techniques were not successful. However, we were able to concentrate M. tuberculosis more than 30-fold by centrifugation in two of the three pooled CSF samples with either 28% sucrose or 36% NaCl. Although these results are encouraging, evaluation of individual samples from TB suspects is needed to validate these preliminary findings. In addition, further advances are needed to concentrate the bacilli and eliminate PCR inhibitors in other nonrespiratory samples such as tissue and stool. Recent technological advances for purification of microorganisms directly from patient specimens such as cellular imprinting (8, 9), an immunomagnetic capture system (NanoMR Inc., Albuquerque, NM), or ligand-coated paramagnetic beads (Microsens Biotechnologie, United Kingdom) are potential technologies that could allow rapid, simple, and affordable concentration of intact M. tuberculosis. Coupled with the simplicity of the GeneXpert platform, a successful concentration method could greatly extend the application of this platform for diagnosis of TB in extrapulmonary samples in resource-limited areas.
ACKNOWLEDGMENTS
We thank William Berquist and his staff for providing gastric aspirates and Cepheid for providing cartridges.
Footnotes
Published ahead of print 2 May 2012
REFERENCES
- 1. Akane A, Matsubara K, Nakamura H, Takahashi S, Kimura K. 1994. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J. Forensic Sci. 39:362–372 [PubMed] [Google Scholar]
- 2. Armand S, Vanhuls P, Delcroix G, Courcol R, Lemaitre N. 2011. Comparison of the Xpert MTB/RIF test with an IS6110-TaqMan real-time PCR assay for direct detection of Mycobacterium tuberculosis in respiratory and nonrespiratory specimens. J. Clin. Microbiol. 49:1772–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Blakemore R, et al. 2010. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J. Clin. Microbiol. 48:2495–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehme CC, et al. 2010. Rapid molecular detection of tuberculosis and rifampin resistance. N. Engl. J. Med. 363:1005–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Causse M, Ruiz P, Gutierrez-Aroca JB, Casal M. 2011. Comparison of two molecular methods for rapid diagnosis of extrapulmonary tuberculosis. J. Clin. Microbiol. 49:3065–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cox DD, Todd AC. 1962. Survey of gastrointestinal parasitism in Wisconsin dairy cattle. J. Am. Vet. Med. Assoc. 141:706–709 [PubMed] [Google Scholar]
- 7. den Hertog AL, Klatser PR, Anthony RM. 2009. Buoyant density of Mycobacterium tuberculosis: implications for sputum processing. Int. J. Tuberc. Lung Dis. 13:466–471 [PubMed] [Google Scholar]
- 8. Dickert FL, Hayden G. 2002. Bioimprinting of polymers and sol-gel phases. Selective detection of yeasts with imprinted polymers. Anal. Chem. 74:1302–1306 [DOI] [PubMed] [Google Scholar]
- 9. Guo P, et al. 2012. Microfluidic capture and release of bacteria in a conical nanopore array. Lab Chip 12:558–561 [DOI] [PubMed] [Google Scholar]
- 10. Helb D, et al. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hillemann D, Rusch-Gerdes S, Boehme C, Richter E. 2011. Rapid molecular detection of extrapulmonary tuberculosis by the automated GeneXpert MTB/RIF system. J. Clin. Microbiol. 49:1202–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ioannidis P, et al. 2011. Cepheid GeneXpert MTB/RIF assay for Mycobacterium tuberculosis detection and rifampin resistance identification in patients with substantial clinical indications of tuberculosis and smear-negative microscopy results. J. Clin. Microbiol. 49:3068–3070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuczynska E, Shelton DR. 1999. Method for detection and enumeration of Cryptosporidium parvum oocysts in feces, manures, and soils. Appl. Environ. Microbiol. 65:2820–2826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Malbruny B, Le Marrec G, Courageux K, Leclercq R, Cattoir V. 2011. Rapid and efficient detection of Mycobacterium tuberculosis in respiratory and non-respiratory samples. Int. J. Tuberc. Lung Dis. 15:553–555 [DOI] [PubMed] [Google Scholar]
- 15. Moure R, et al. 2011. Rapid detection of Mycobacterium tuberculosis complex and rifampin resistance in smear-negative clinical samples by use of an integrated real-time PCR method. J. Clin. Microbiol. 49:1137–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nicol MP, et al. 2011. Accuracy of the Xpert MTB/RIF test for the diagnosis of pulmonary tuberculosis in children admitted to hospital in Cape Town, South Africa: a descriptive study. Lancet Infect. Dis. 11:819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oberhelman RA, et al. 2010. Diagnostic approaches for paediatric tuberculosis by use of different specimen types, culture methods, and PCR: a prospective case-control study. Lancet Infect. Dis. 10:612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vadwai V, et al. 2011. Xpert MTB/RIF: a new pillar in diagnosis of extrapulmonary tuberculosis? J. Clin. Microbiol. 49:2540–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Widjojoatmodjo MN, Fluit AC, Torensma R, Verdonk GP, Verhoef J. 1992. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J. Clin. Microbiol. 30:3195–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zeka AN, Tasbakan S, Cavusoglu C. 2011. Evaluation of the GeneXpert MTB/RIF assay for rapid diagnosis of tuberculosis and detection of rifampin resistance in pulmonary and extrapulmonary specimens. J. Clin. Microbiol. 49:4138–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]