Abstract
Two genetically unrelated OXA-163-carrying Klebsiella pneumoniae strains were identified from two infection cases in June 2009 and May 2010 in Cairo, Egypt. OXA-163-producing Enterobacteriaceae had been previously reported in Argentina only. Both patients had no history of travel abroad. The emergence of this newly recognized OXA-48-related β-lactamase able to hydrolyze cephalosporins and carbapenems is especially worrying in a geographic area where OXA-48 is endemic and effective surveillance for antibiotic resistance is largely unaffordable.
TEXT
Carbapenems are first-line drugs for severe infections caused by extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. However, the emergence and rapid diffusion of carbapenemase-producing Gram negatives in the recent years is compromising the therapeutic efficacy of this class of antimicrobial drugs (2, 11, 16).
Recently, two blaOXA-163-producing isolates of Klebsiella pneumoniae and Enterobacter cloacae have been identified in Argentina (15). OXA-163 differs from OXA-48 by a single amino acid substitution and a 4-amino-acid deletion only, but it works as an extended-spectrum oxacillinase showing a weaker ability to hydrolyze carbapenems than OXA-48 but a concurrent significant ability to hydrolyze extended-spectrum cephalosporins (15). The blaOXA-163 gene has been putatively proposed as the equivalent in South America of the blaOXA-48 gene that is increasingly identified in the southern and eastern Mediterranean countries (1, 6, 8, 14).
Here, we report two autochthonous cases of infection caused by two genetically unrelated OXA-163-carrying K. pneumoniae strains in Cairo, Egypt, in 2009 and 2010.
Case 1 was a 31-year-old woman hospitalized in Cairo (Egypt) on June 5, 2009, due to acute myelocytic leukemia. She was on therapy by doxorubicin and cytarabine. After 16 days since admission, she was transferred to an intensive care unit because of heart and respiratory failure. Initial antibiotic therapy was started with vancomycin, azithromycin, and imipenem. Blood culture was positive for a carbapenem-resistant strain of K. pneumoniae (isolate 18). The patient died on June 23.
Case 2 was a 25-year-old man with an 18-month history of acute lymphocytic leukemia. He was in outpatient follow-up until May 2010, when he was admitted to the hospital due to a lower respiratory tract infection. He was started on cefepime and amikacin on May 14, but the clinical course was complicated. A sputum culture grew carbapenem-resistant K. pneumoniae (isolate 78). No further data about the treatment outcome were available for this patient.
Both patients had never traveled outside Egypt.
The two carbapenem-resistant K. pneumoniae strains were sent to the molecular epidemiology laboratory of the Department of Sciences for Health Promotion “G. D'Alessandro,” University of Palermo, Italy, for confirmation and typing. The two isolates were initially tested for antimicrobial susceptibility by disk diffusion according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (3). Susceptibility testing for extended-spectrum cephalosporins and carbapenems was performed with Etest strips (bioMérieux, Marnes-La-Coquette, France). Results were interpreted according to the CLSI interpretative criteria (3).
PCR screening for carbapenemase-encoding genes of classes A (blaKPC, blaGES), B (blaVIM, blaIMP), and D (blaOXA-48) and for ESBL-encoding genes blaTEM, blaOXA, blaSHV, and blaCTX-M was performed as described previously (4). PCR products were purified and sequenced using the BigDye Terminator version 1.1 cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom) and the ABI Prism 310 Genetic Analyzer (Applied Biosystems, Foster City, CA). The nucleotide (nt) and deduced protein sequences were analyzed with the software available from the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov). Outer membrane protein (OMP) gene amplification was conducted using ompK35-F and -R and ompK36-F and -R, and PCR products were sequenced using OMP primers according to Kaczmarek et al. (8). The sequences were analyzed by comparison with reported nucleotide sequences in GenBank. Expression levels of the ompK35 and ompK36 genes were not investigated.
To evaluate their clonal relationship, the two K. pneumoniae isolates were submitted to pulsed-field gel electrophoresis (PFGE) after XbaI DNA digestion. Strain differentiation by PFGE was based on the criteria of Tenover et al. (17). The rep-PCR DiversiLab Microbial Typing System (bioMérieux, Marcy l'Étoile, France) was also used with the DiversiLab Klebsiella kit. DNA fragment separation and detection were done using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), and results were analyzed and interpreted using the Kullback-Leibler method, as previously reported (13). Moreover, multilocus sequence typing (MLST) was performed on both isolates according to the protocol described on the K. pneumoniae MLST website (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html). MLST results were compared with the international K. pneumoniae MLST database at the Pasteur Institute in Paris, France.
The two K. pneumoniae isolates exhibited resistance to extended-spectrum cephalosporins (cefotaxime, ceftazidime, cefepime), aztreonam, and carbapenems (meropenem and imipenem) (Table 1). Additionally, both isolates were resistant to ciprofloxacin and gentamicin and susceptible to colistin.
Table 1.
Main characteristics of two OXA-163-producing K. pneumoniae isolates detected in Egypt, June 2009 and May 2010
| K. pneumoniae strain (patient) | MIC (μg/ml)a |
bla genes | Sequence type | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cefotaxime | Ceftriaxone | Ceftazidime | Aztreonam | Imipenem | Meropenem | Colistin | |||
| 18 (case 1) | >256 | >256 | >256 | >256 | 6 | 8 | 0.25 | TEM-1, SHV-1, CTX-M15, OXA-163 | 37 |
| 78 (case 2) | >256 | >256 | >256 | >256 | 6 | 12 | 0.25 | TEM-1, SHV-1, CTX-M15, OXA-163 | 20 |
MICs assessed by Etest.
For both isolates, positive PCR results were obtained for the blaTEM, blaSHV, and blaCTX-M genes, and sequencing identified the narrow-spectrum β-lactamase TEM-1 and SHV-1 genes and the ESBL CTX-M-15. A blaOXA-48-like gene was detected by PCR in both isolates that, by sequencing, was identified as blaOXA-163 (www.lahey.org).
The ompK35 and ompK36 genes of isolates 18 and 78 were amplified, sequenced, and aligned with those of OMP gene sequences of K. pneumoniae MGH 78578. The ompK35 gene of both isolates showed a frameshift mutation due to a nucleotide deletion at nt 873. Moreover, in the ompK35 gene of isolate 18, sequencing revealed one substitution, T→A, at nucleotide position 108, generating a TAA nonsense mutation which resulted in a premature stop codon at amino acid position 36. Silent point mutations were also found at nt 303, 420, 537, 648, and 786 on the nucleotide sequence of both isolates. Comparison of the ompK36 gene sequence with the wild-type sequence revealed that both had an insertion of six nucleotides (5′-GGCGAC-3′) at positions 405 to 410, due to a duplication of the adjacent region at nucleotide positions 399 to 404. Sequencing of the ompK36 sequence of isolate 78 detected also a 9-nucleotide deletion at positions 556 to 564.
PFGE and rep-PCR showed that the two isolates were genetically unrelated to each other (Fig. 1). Additionally, MLST attributed two different sequence types, ST37 and ST20, respectively, to the strains 18 and 78.
Fig 1.
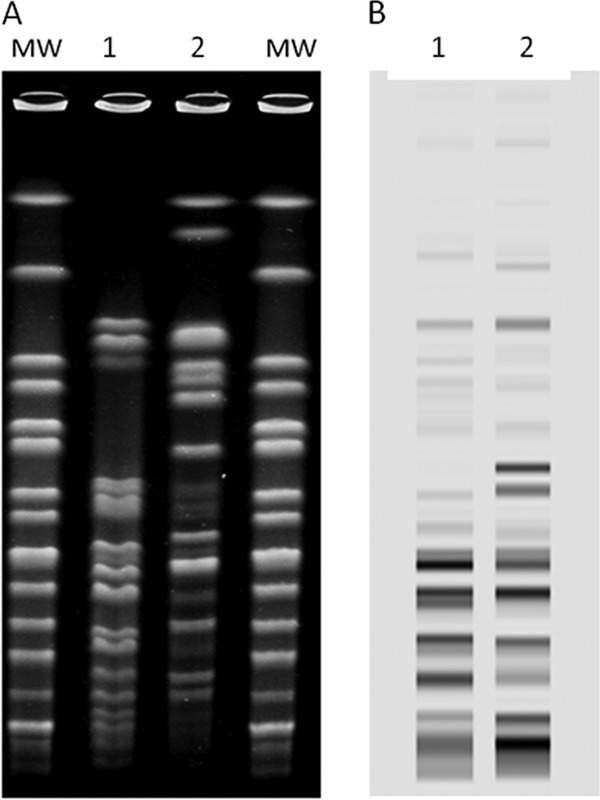
XbaI pulsed-field gel electrophoresis patterns and rep-PCR patterns of two OXA-163-carrying Klebsiella pneumoniae strains, Cairo, Egypt, 2009 and 2010. (A) Lanes: MW, XbaI-digested DNA of Salmonella Braenderup H9812; 1, K. pneumoniae strain 18; 2, K. pneumoniae strain 78. (B) Lanes: 1, K. pneumoniae strain 18; 2, K. pneumoniae strain 78.
The main characteristics of the two isolates of K. pneumoniae are summarized in Table 1.
The findings of this study are of concern for a number of reasons. First, the detection of OXA-163 outside the continent where it has been firstly identified was unexpected. This finding seems to stand against the hypothesis of OXA-163 as the equivalent in South America of OXA-48 in the Mediterranean area (15). The two patients had no history of travel abroad. Moreover, possible contacts with source patients from Argentina, though it was not possible to investigate them in detail, appear to be unlikely due also to the 1-year-long interval of time between the two cases. An autochthonous emergence in Egypt of the OXA-163 resistance determinant appears to be the more plausible explanation. Second, the two K. pneumoniae isolates were genetically unrelated based upon PFGE and MLST. This rules out the clonal expansion of an OXA-163-producing strain in Egypt and, alternatively, supports the hypothesis of horizontal transmission of this genetic determinant via plasmids, according with the previous report (14). Notably, strains belonging to ST20 and ST37 have been previously identified in different countries as ESBL or carbapenemase producers (11, 12). Moreover, it is of special interest that OXA-163 has been identified in Egypt, a country of North Africa, a geographical area where OXA-48-producing Gram negatives are considered endemic (6, 9, 10). Third, both strains had much higher MICs for carbapenems than those reported by Poirel et al. (15). Studies suggest that outer membrane permeability defects coupled with ESBL production can confer resistance to carbapenems (5, 7, 9). This is likely to account for the higher level of carbapenem resistance detected in our strains. It is also noteworthy that the presence of similar mutations altering the properties of the OmpK35 and OmpK36 porins have been previously described in an ST37 K. pneumoniae clone carrying blaCTX-M-15 and exhibiting reduced susceptibility to carbapenems (5). However, our study did not definitively attribute the carbapenem resistance phenotype to OXA-163, because transfer of antimicrobial agent resistance experiments and plasmid analysis were not performed. This is a substantial limitation of our report.
As previously reported about OXA-163-producing strains in Argentina, because of its unique hydrolysis ability mimicking a class A ESBL, OXA-163 could be able to spread unrecognized as a carbapenemase (15). Surveillance of emergence and dissemination of enterobacteria with complex carbapenem resistance mechanisms is a serious public health problem where the costs can be unaffordable, such as low-resource countries. Further studies are urgently needed to assess prevalence of the blaOXA-163 genetic determinant worldwide.
Nucleotide sequence accession numbers.
The ompK35 of isolate 78 was assigned accession number JQ660370, and the ompK36 sequences of isolates 18 and 78 were assigned accession numbers JQ660371 and JQ660372, respectively, in the GenBank database.
Footnotes
Published ahead of print 18 April 2012
REFERENCES
- 1. Benouda A, Touzani O, Khairallah MT, Araj GF, Matar GM. 2010. First detection of oxacillinase-mediated resistance to carbapenems in Klebsiella pneumoniae from Morocco. Ann. Trop. Med. Parasitol. 104:327–330 [DOI] [PubMed] [Google Scholar]
- 2. Carmeli Y, et al. 2010. Controlling the spread of carbapenemase-producing Gram-negatives: therapeutic approach and infection control. Clin. Microbiol. Infect. 16:102–111 [DOI] [PubMed] [Google Scholar]
- 3. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 4. Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 65:490–495 [DOI] [PubMed] [Google Scholar]
- 5. García-Fernández A, et al. 2010. An ertapenem-resistant ESBL-producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob. Agents Chemother. 54:4178–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goren MG, Chmelnitsky I, Carmeli Y, Navon-Venezia S. 2011. Plasmid-encoded OXA-48 carbapenemase in Escherichia coli from Israel. J. Antimicrob. Chemother. 66:672–673 [DOI] [PubMed] [Google Scholar]
- 7. Jacoby GA, Mills DM, Chow N. 2004. Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:3203–3206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kaczmarek FM, Dib-Hajj F, Shang W, Gootz TD. 2006. High-level carbapenem resistance in a Klebsiella pneumoniae clinical isolate is due to the combination of blaACT-1 β-lactamase production, porin OmpK35/36 insertional inactivation, and downregulation of the phosphate transport porin phoe. Antimicrob. Agents Chemother. 50:3396–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ktari S, et al. 2011. Spread of Klebsiella pneumoniae isolates producing OXA-48 β-lactamase in a Tunisian university hospital. J. Antimicrob. Chemother. 66:1644–1666 [DOI] [PubMed] [Google Scholar]
- 10. Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17:1791–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Orsi GB, et al. 2011. Risk factors and clinical significance of ertapenem-resistant Klebsiella pneumoniae in hospitalised patients. J. Hosp. Infect. 78:54–58 [DOI] [PubMed] [Google Scholar]
- 12. Oteo J, et al. 2009. Emergence of CTX-M-15-producing Klebsiella pneumoniae of multilocus sequence types 1, 11, 14, 17, 20, 35 and 36 as pathogens and colonizers in newborns and adults. J. Antimicrob. Chemother. 64:524–528 [DOI] [PubMed] [Google Scholar]
- 13. Perez F, et al. 2010. Carbapenem-resistant Acinetobacter baumannii and Klebsiella pneumoniae across a hospital system: impact of post-acute care facilities on dissemination. J. Antimicrob. Chemother. 65:1807–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poirel L, Bonnin RA, Nordmann P. 2012. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56:559–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Poirel L, et al. 2011. OXA-163, an OXA-48-related class D β-lactamase with extended activity toward expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 55:2546–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poirel L, Naas T, Nordmann P. 2010. Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob. Agents Chemother. 54:24–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tenover FC, et al. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]


