Abstract
Rifampin resistance in Mycobacterium tuberculosis is largely determined by mutations in an 80-bp rifampin resistance determining region (RRDR) of the rpoB gene. We developed a rapid single-well PCR assay to identify RRDR mutations. The assay uses sloppy molecular beacons to probe an asymmetric PCR of the M. tuberculosis RRDR by melting temperature (Tm) analysis. A three-point Tm code is generated which distinguishes wild-type from mutant RRDR DNA sequences in approximately 2 h. The assay was validated on synthetic oligonucleotide targets containing the 44 most common RRDR mutations. It was then tested on a panel of DNA extracted from 589 geographically diverse clinical M. tuberculosis cultures, including isolates with wild-type RRDR sequences and 25 different RRDR mutations. The assay detected 236/236 RRDR mutant sequences as mutant (sensitivity, 100%; 95% confidence interval [CI], 98 to 100%) and 353/353 RRDR wild-type sequences as wild type (specificity, 100%; 95% CI, 98.7 to 100%). The assay identified 222/225 rifampin-resistant isolates as rifampin resistant (sensitivity, 98.7%; 95% CI, 95.8 to 99.6%) and 335/336 rifampin-susceptible isolates as rifampin susceptible (specificity, 99.7%; 95% CI, 95.8 to 99.6%). All mutations were either individually identified or clustered into small mutation groups using the triple Tm code. The assay accurately identified mixed (heteroresistant) samples and was shown analytically to detect RRDR mutations when present in at least 40% of the total M. tuberculosis DNA. This was at least as accurate as Sanger DNA sequencing. The assay was easy to use and well suited for high-throughput applications. This new sloppy molecular beacon assay should greatly simplify rifampin resistance testing in clinical laboratories.
INTRODUCTION
Multidrug-resistant (MDR) and extensively drug-resistant (XDR) Mycobacterium tuberculosis is increasing worldwide (9, 27, 37). Rapid methods to detect drug resistance are needed to quickly identify drug-resistant strains and to implement appropriate therapy (4, 16, 36). M. tuberculosis does not naturally contain plasmids, and almost all cases of clinical drug resistance are caused by single-nucleotide polymorphisms (SNPs) or small insertions/deletions in relevant genes (28). In the case of rifampin resistance, 95 to 98% of rifampin-resistant clinical strains have mutations in the 80-bp rifampin resistance determining region (RRDR) of the M. tuberculosis RNA polymerase beta (rpoB) gene (9, 11, 12, 15, 20, 28). PCR and probe-based molecular genotyping assays can be used to detect these resistance-inducing mutations. Such genotypic assays are potentially more rapid than labor-intensive culture-based drug susceptibility tests. Genotypic drug susceptibility testing has shown good overall concordance with the phenotypic antibiotic susceptibility tests of MDR and XDR clinical strains (6), and in a recent study, a genotypic test actually correlated better with clinical outcome than standard phenotypic susceptibility testing (36). When combined with automated sample processing systems, such as the Xpert MTB/RIF test (14), genotypic susceptibility tests can significantly reduce testing turnaround time, increasing patient notification rates and decreasing time to treatment (4, 32).
The Xpert MTB/RIF assay is one example of a genotypic test that is being increasingly used to screen for rifampin resistance (33). However, the single-use cartridge design of the Xpert assay limits its use for laboratory-based high-throughput testing. Several widely used reverse blot hybridization assays, such as the INNO-LIPA Rif.TB assay (Innogenetics, Belgium) and the MTBDRplus (Hain, Germany) assay (1, 5, 15, 19, 21, 23, 29, 34), are available for laboratory-based rifampin resistance screening; however, these assays are complicated by their open hybridization format. Open hybridization systems require a relatively cumbersome work process, including rigorous physical separation of different work areas (2, 23) due to the risk of handling open PCR amplicons in a molecular diagnostic laboratory. Open systems also require a relatively large number of probes to test for relevant resistance-associated mutations. This requirement complicates assay chemistry and hybridization parameters. In contrast to reverse blot hybridization assays, melting temperature (Tm)-based assays enable mutation detection in closed systems. These assays take advantage of the fact that fully complementary strands of DNA have higher melting temperatures than DNA heteroduplexes which contain one or more nucleotide mismatches. Tm assays are well suited for busy diagnostic laboratories, because they can be performed in homogeneous closed systems without the risk of carryover amplicon contamination, are amenable to multiplexing, and are easily adapted to a high-throughput format. Fluorescence resonance energy transfer (FRET) probes, dually labeled probes, TaqMan, and molecular beacon probes have all been used in Tm assays to detect drug resistance mutations in M. tuberculosis, as has the high-resolution Tm analysis (HRMA) of PCR products using DNA intercalating dyes (9, 10, 13, 18, 20, 24, 26, 27). However, M. tuberculosis drug resistance assays which use Tm analysis have been largely limited to tests of the most commonly encountered mutations (20, 24, 27). We have previously shown that a new generation of mismatch-tolerant probes, called sloppy molecular beacons (SMBs), can be efficiently used to identify mutations in the M. tuberculosis genome associated with fluoroquinolone resistance (8). The assay had high sensitivity and specificity, and the thermodynamic properties of SMBs made it possible to develop an assay with easily interpretable Tm curves. The assay was also able to detect fluoroquinolone resistance mutations in clinical samples that contained mixtures of drug-susceptible and drug-resistant DNA. We have now applied the same technology to detect rifampin resistance in M. tuberculosis. Here, we show that our approach enables the rapid, high-throughput, single-well detection of mutations in the rpoB RRDR of M. tuberculosis. We tested the ability of the assay to detect 44 different rpoB mutations that have been described previously to be associated with rifampin resistance, and we validated its performance on a panel of clinical samples representing a diverse collection in terms of geographical distribution and the mutation types. The ability of the assay to specifically identify rifampin-resistant clinical strains and to detect heteroresistance was also evaluated, and its performance was compared to conventional antibiotic susceptibility test results and verified with targeted sequencing.
MATERIALS AND METHODS
Clinical DNA samples.
Two clinical sample sets were tested to include a wide variety of mutations and geographic origins. The first sample set consisted of 440 sequential M. tuberculosis isolates cultured from patients enrolled in a natural history study of MDR tuberculosis (NCT00341601 at clinicaltrials.gov) in the National Masan Tuberculosis Hospital in Changwon, Republic of Korea, for which reliable conventional drug susceptibility tests and/or DNA sequencing of the M. tuberculosis RRDR were available. The second sample set consisted of 149 selected M. tuberculosis cultures obtained from the WHO TDR Tuberculosis Specimen Bank, maintained by the United Nations Children's Fund/UNDP/World Bank/WHO Special Program for Research and Training in Tropical Diseases, which includes cultures from Africa, Asia, Australia, Europe, and Latin America (8), for a total of 589 DNA samples tested. The TDR samples were chosen to represent a wide repertoire of RRDR mutants and a broad geographical distribution for both RRDR mutants and rifampin-susceptible controls. Conventional rifampin susceptibility was performed using the LJ proportions method with 40 μg/ml used as the critical rifampin concentration defining resistance (31). Rifampin resistance results were available for 561 out of the 589 samples. The RRDR sequence, and in most cases the entire rpoB gene sequence, was available for all 589 isolates. We resequenced a subset of RRDR mutants, representing all of the different mutations tested in our study, to confirm the original DNA sequencing results. The results of the DNA sequencing performed as part of this current study were used in the rare event of discordance with the prior sequencing result. Each DNA sample was quantified with a Nanodrop microvolume spectrophotometer (Thermo Scientific), and 2 to 5 ng of the DNA sample was used for PCR. All samples were independently coded and randomly redistributed to blind the sample sequence and origin before performing each test assay. The results of the assay used in this investigation were not reported to the treating physicians and were not used to guide treatment decisions.
DNA samples from human origin, NTM, and Gram-positive and Gram-negative bacteria.
Pure human genomic DNA obtained from the ATCC repository (Manassas, VA); 121 clinical nontuberculous mycobacterial (NTM) cultures, representing 26 species, isolated from patients at the National Masan Tuberculosis Hospital; a laboratory strain of M. smegmatis; 18 NTM isolates obtained from the ATCC repository (Manassas, VA), consisting of M. abscessus, M. scrofulaceum, M. celatum, M. haemophilum, M. asiaticum, M. kansasii, M. avium, M. flavescens, M. szulgai, M. terrae, M. fortuitum, M. intracellulare, M. marinum, M. xenopi, M. thermoresistibile, M. simiae, M. trivial, and M. malmoense; and DNA extracted from 18 species of Gram-negative and Gram-positive bacteria representing the most common bloodstream infections and nosocomial pathogens obtained from various sources (7) were selected to test for analytical specificity. DNA was isolated from pure clinical strains by boiling a loopful of culture in Instagene matrix solution (Bio-Rad). The DNA was quantified as described above and tested using 100 to 400 ng for each PCR. To test for the capability of the assay to detect M. tuberculosis RRDR mutations in the presence of a 10-fold excess of background NTM DNA, 105 genome equivalents of M. tuberculosis DNA harboring common RRDR mutations found in our study (516GTC, 526TAC, 531TTG, and 533CCG) was mixed with 106 genome equivalents of NTM DNA. Human DNA was tested in 105-fold excess above the level of the target M. tuberculosis DNA.
SMBs and primers.
A 172-bp fragment (nucleotides 1226 to 1397, with numbering based on the gene start site according to the Escherichia coli DNA nomenclature) containing the rpoB RRDR was amplified using the target primer rpoB-F (5′-agacgttgatcaacatccg-3′) and the antisense primer rpoB-R (5′-acctccagcccggcacgctcacgt-3′). These primers were designed to be specific to the M. tuberculosis complex and were verified by an alignment of M. tuberculosis and all of the NTM rpoB sequences available in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/). Three SMB probes, rpo1 (5′-tetramethylrhodamine-cgaccgCccatgaattggctcagctggctggtgAcggtcg-BHQ2-3′), rpo2 (5′-cyanine 5-ggcgcgaaccAcgacagcgggttgttctggtccatgaacgcgcc-BHQ2-3′), and rpo3 (5′ −6 carboxyfluorescein-cgcgcgcaTcAccAacagtcggTgcttgtgggtcaacccgcgcg-BHQ1-3′) (where the underlined boldface sequences represent the stem portion of the SMB, the lowercase sequences represent the probe portion of the SMB, uppercase letters represent the mutations introduced into the probe region to obtain a stable stem-loop structure, and BHQ represents Black Hole Quencher), were targeted against the rpoB RRDR. The SMBs were designed using the in silico DNA folding program at http://mfold.rna.albany.edu/?q=mfold/dna-folding-form, and the probe-target hybrid folding program at http://mfold.rna.albany.edu/?q=DINAMelt/Two-state-melting was used to predict the possible probe-target hybrid structures and Tms. The probes were designed to generate a maximum Tm difference between wild-type and mutant sequences in their respective target regions of the RRDR so as to enable their unambiguous identification. Primers were obtained from Sigma Genosys, and the SMB probes were from Biosearch, CA.
Using artificial oligonucleotides containing rpoB RRDR mutations to test assay probes.
Not all of the RRDR mutations described in the literature were available to us in the form of clinical M. tuberculosis DNA. To enable us to test the assay against the broadest range of RRDR mutations, we created oligonucleotides containing either wild-type or mutant RRDR sequences, including 44 clinically relevant mutations identified as being present at a frequency of at least 0.1% in clinical settings (based on an extensive literature search), to test the ability of the assay probes to identify different RRDR sequence types. The 44 different mutations spanning the RRDR from codons 507 to 533 included SNPs, insertions, and double mutants (Table 1). The Tm of each of the three SMB probes in the presence of each RRDR oligonucleotide target was measured. Approximately 500 ng of individual oligonucleotide targets was added to a reaction mixture containing 0.8 ng/μl of each SMB probe, 4 mM MgCl2, 1× PCR buffer (10× Stoffel Buffer; Applied Biosystems), and 5% glycerol. Tm analysis was performed using a Light Cycler 480 II real-time PCR system (Roche Molecular Systems Inc.) using the following assay parameters; denaturation at 95°C for 5 min, followed by cooling down to 45°C and then gradual heating to 85°C, with continuous monitoring of fluorescence during the process at a rate of 10 data acquisitions per degree centigrade. Tm calls were performed at the end of the reaction using the automated Tm calling software (Light Cycler 480 software), and resulting Tm values for each probe were determined. The three Tm values (one for each SMB probe) that were generated in the presence of an RRDR target where then used to define a three-point Tm code for each RRDR mutant or wild-type sequence.
Table 1.
Three-probe Tm (°C) code of the assay in tests of artificial oligonucleotide targets with wild-type and mutant RRDR sequencesa
| Sample |
Mutation |
Tm (°C) of: |
dTm (°C) of: |
|||||
|---|---|---|---|---|---|---|---|---|
| No. | Name | rpo1 | rpo2 | rpo3 | rpo1 | rpo2 | rpo3 | |
| 1 | WT | 76.0 | 75.3 | 71.2 | ||||
| 2 | 507 | GGC-GAC | 71.3 | 75.2 | 71.2 | 4.7 | 0.0 | 0.0 |
| 3 | 510 | CAG-CAC | 69.7 | 74.4 | 71.9 | 6.3 | 0.8 | −0.7 |
| 4 | 511 A | CTG-CCG | 70.7 | 75.0 | 71.3 | 5.2 | 0.2 | −0.1 |
| 5 | 511 B | CTG-CGG | 71.3 | 75.0 | 71.4 | 4.7 | 0.3 | −0.2 |
| 6 | 512 | AGC-ACC | 70.7 | 75.2 | 71.4 | 5.3 | 0.1 | −0.3 |
| 7 | 513 A | CAA-AAA | 70.9 | 75.0 | 71.2 | 5.1 | 0.3 | 0.0 |
| 8 | 513 B | CAA-CCA | 71.1 | 74.9 | 71.6 | 4.9 | 0.4 | −0.4 |
| 9 | 513 C | CAA-CTA | 71.1 | 74.9 | 71.6 | 4.9 | 0.4 | −0.4 |
| 10 | 514 A | TTC insertion | 71.3 | 75.7 | 71.3 | 4.7 | −0.5 | −0.1 |
| 11 | 514 B | TTC-TTG | 70.2 | 74.5 | 71.0 | 5.8 | 0.8 | 0.1 |
| 12 | 514 C | TTC-TTT (silent) | 70.5 | 74.5 | 71.0 | 5.5 | 0.8 | 0.2 |
| 13 | 515 | ATG-ATT | 73.5 | 72.5 | 71.7 | 2.5 | 2.8 | −0.5 |
| 14 | 516 A | GAC-GTC | 75.6 | 71.3 | 71.4 | 0.4 | 7.0 | −0.2 |
| 15 | 516 B | GAC-TAC | 74.8 | 72.1 | 71.4 | 1.2 | 3.2 | −0.2 |
| 16 | 516 C | GAC-GGC | 77.0 | 73.0 | 71.3 | −01.0 | 2.3 | −0.1 |
| 17 | 516 D | GAC-GCC | 75.4 | 71.8 | 71.5 | 0.6 | 3.5 | −0.3 |
| 18 | 516 E | GAC-TTC | 75.1 | 71.4 | 71.5 | 0.9 | 3.9 | −0.4 |
| 19 | 516 F | GAC-AAC | 74.9 | 72.6 | 71.2 | 1.1 | 2.7 | −0.1 |
| 20 | 517 | CAG-CTA | 75.2 | 67.3 | 72.0 | 0.8 | 8.0 | −0.8 |
| 21 | 518 | AAC-TAC | 74.9 | 70.0 | 71.7 | 1.1 | 5.3 | −0.6 |
| 22 | 522 A | TCG-TTG | 76.2 | 73.2 | 70.2 | −0.2 | 2.1 | 1.0 |
| 23 | 522 B | TCG-CCG | 75.1 | 71.9 | 72.5 | 0.9 | 3.4 | −1.3 |
| 24 | 522 C | TCG-TGG | 76.0 | 72.4 | 71.2 | 0.0 | 2.9 | −0.1 |
| 25 | 526 A | CAC-TAC | 76.0 | 75.2 | 65.8 | 0.0 | 0.1 | 5.4 |
| 26 | 526 B | CAC-GAC | 75.7 | 75.0 | 64.1 | 0.3 | 0.3 | 7.1 |
| 27 | 526 C | CAC-CGC | 75.8 | 75.0 | 67.9 | 0.2 | 0.3 | 3.3 |
| 28 | 526 D | CAC-CTC | 76.4 | 75.4 | 65.0 | −0.5 | −0.1 | 6.2 |
| 29 | 526 E | CAC-CAG | 76.3 | 75.3 | 65.1 | −0.3 | −0.1 | 6.1 |
| 30 | 526 F | CAC-AAC | 75.9 | 75.1 | 65.7 | 0.0 | 0.2 | 5.5 |
| 31 | 526 G | CAC-TGC | 75.1 | 75.6 | 63.0 | 0.9 | −0.4 | 8.1 |
| 32 | 526 H | CAC-CCC | 76.3 | 75.1 | 64.0 | −0.3 | 0.2 | 7.2 |
| 33 | 526 I | CAC-ACC | 76.0 | 75.2 | 62.9 | −0.1 | 0.1 | 8.3 |
| 34 | 527 | AAG-AGG | 76.0 | 75.2 | 67.6 | 0.0 | 0.1 | 3.6 |
| 35 | 529 | CGA-CAA | 76.6 | 75.3 | 65.5 | −0.6 | 0.0 | 5.7 |
| 36 | 531 A | TCG-TTG | 75.2 | 75.4 | 74.7 | 0.8 | −0.1 | −3.5 |
| 37 | 531 B | TCG-TGG | 75.5 | 75.4 | 73.8 | 0.4 | −0.2 | −2.6 |
| 38 | 531 C | TCG-GCG | 76.2 | 75.5 | 71.9 | −0.2 | −0.2 | −0.7 |
| 39 | 531 D | TCG-TAC | 76.3 | 75.4 | 74.0 | −0.3 | −0.2 | −2.8 |
| 40 | 531 E | TCG-CAG | 75.5 | 75.7 | 74.9 | 0.5 | −0.4 | −3.8 |
| 41 | 531 F | TCG-TAG | 75.4 | 75.3 | 72.7 | 0.6 | −0.1 | −1.5 |
| 42 | 533 A | CTG-CCG | 75.6 | 75.6 | 72.1 | 0.4 | −0.3 | −1.0 |
| 43 | 533 B | CTG-ATG | 74.7 | 75.6 | 73.3 | 1.3 | −0.3 | −2.1 |
| 44 | 514, 531 | TTC-TTT, TCG-TTG | 71.1 | 75.7 | 74.9 | 4.9 | −0.4 | −3.7 |
| 45 | 516, 522 | GAC-GGC, TCG-TTG | 77.5 | 69.8 | 70.7 | −01.5 | 5.5 | 0.5 |
Each Tm value represents an average of 10 separate reactions. WT, wild-type RRDR sequence. The shaded cells represent the dTm values obtained for the probes detecting the corresponding mutations.
Sloppy molecular beacon Tm assay on clinical DNA.
All PCR-Tm analyses of DNA from clinical samples were performed and decoded by two experimenters who were blinded to the nature and identity of the samples. Each DNA sample was quantified using a Nanodrop microvolume spectrophotometer (Thermo Scientific), and 2 to 5 ng of the DNA sample was used for each PCR except where indicated otherwise. PCR was performed in 384-well plates using a Roche Light Cycler 480 II real-time PCR system (Roche Molecular Systems Inc.) in 20-μl reaction volumes containing 1 μM target primer and 50 nM antisense primer, 0.8 ng/μl of each SMB probe, 4 mM MgCl2, 250 mM deoxynucleoside triphosphates (dNTPs), 1× PCR buffer, 5% glycerol, 0.06 U/μl of AmpliTaq Gold Stoffel DNA polymerase (Applied Biosystems), and 2 to 5 ng of sample DNA or an equivalent volume of water. PCR was carried out with the following steps: activation of the enzyme for 2 min at 95°C, followed by 45 cycles of denaturation at 95°C for 15 s, annealing at 65°C for 30 s, and extension at 72°C for 10 s. Following PCR cycling, post-PCR-Tm analysis was performed by denaturation at 95°C for 5 min, followed by cooling down to 45°C and then gradual heating to 85°C, with continuous monitoring of fluorescence during the process at a rate of 10 data acquisitions per degree centigrade. Tm calls were performed at the end of the reaction using the automated Tm calling software (Light Cycler 480 software), and resulting Tm values for each probe were determined. A nontemplate control using sterile water instead of DNA as the template was used as the DNA-negative control, and a DNA-positive control using 1 ng of genomic DNA from M. tuberculosis H37Rv as the template was also included in each assay plate.
Assay LOD.
The analytical sensitivity of the assay was determined using serial dilutions of three different genomic DNA samples. One sample had a wild-type RRDR sequence, and one sample each had the RRDR mutations 516GTC and 531TTG, which were chosen because these two mutations occurred frequently in our clinical DNA panel and in the literature. Each DNA sample was serially diluted to represent a concentration range from 106 to 10 genome equivalents per reaction. The rpoB SMB assay was performed 10 times at each dilution, and the minimum DNA concentration which produced correct Tm codes 100% of the time was determined as the limit of detection (LOD) of the assay for that RRDR type.
DNA mixtures containing wild-type and 531TTG mutant DNA.
DNA mixtures containing wild-type and mutant DNA were prepared to evaluate the performance of the assay in mixed samples containing both wild-type and RRDR mutant DNA (heteroresistance). This situation might occur clinically in a dual M. tuberculosis infection or during the in vivo evolution of rifampin resistance caused by tuberculosis treatment (17). Various amounts of the 531TTG mutant genomic DNA (a representative highly prevalent RRDR mutation) was added to wild-type M. tuberculosis DNA to generate DNA mixtures containing 10 to 90% mutant DNA (in 10% increments) in a total DNA amount of 10 ng (106 genome equivalents). SMB Tm shift assays and Sanger sequencing of the RRDR were performed on each mixed DNA type to compare the ability of the SMB assay to detect the RRDR mutant in this mixture to that of Sanger DNA sequencing.
Sanger DNA sequencing.
DNA samples were amplified using rpoB gene-specific primers as described above, except for the fact that 0.5 μM both forward and reverse primers, 2.5 mM MgCl2, and 0.03 U/μl of AmpliTaq Gold DNA polymerase (Applied Biosystems) were used. The amplified products were checked by gel electrophoresis and then purified using a PCR purification kit (Qiagen) by following the manufacturer's instructions. The purified PCR products were subjected to bidirectional sequencing using the rpoB gene-specific forward and reverse primers in a 3130XL Genetic Analyzer (Applied Bio-systems) using a BigDye Terminator, version 3.1, cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions.
Human subject approval.
This study was approved by the National Masan Tuberculosis Hospital, NIAID, and UMDNJ institutional review boards, and all subjects gave written informed consent (UMDNJ IRB protocol number 0120090104).
RESULTS
SMB design parameters produce a gradient of Tm values which differentiate mutant from wild-type RRDR sequences.
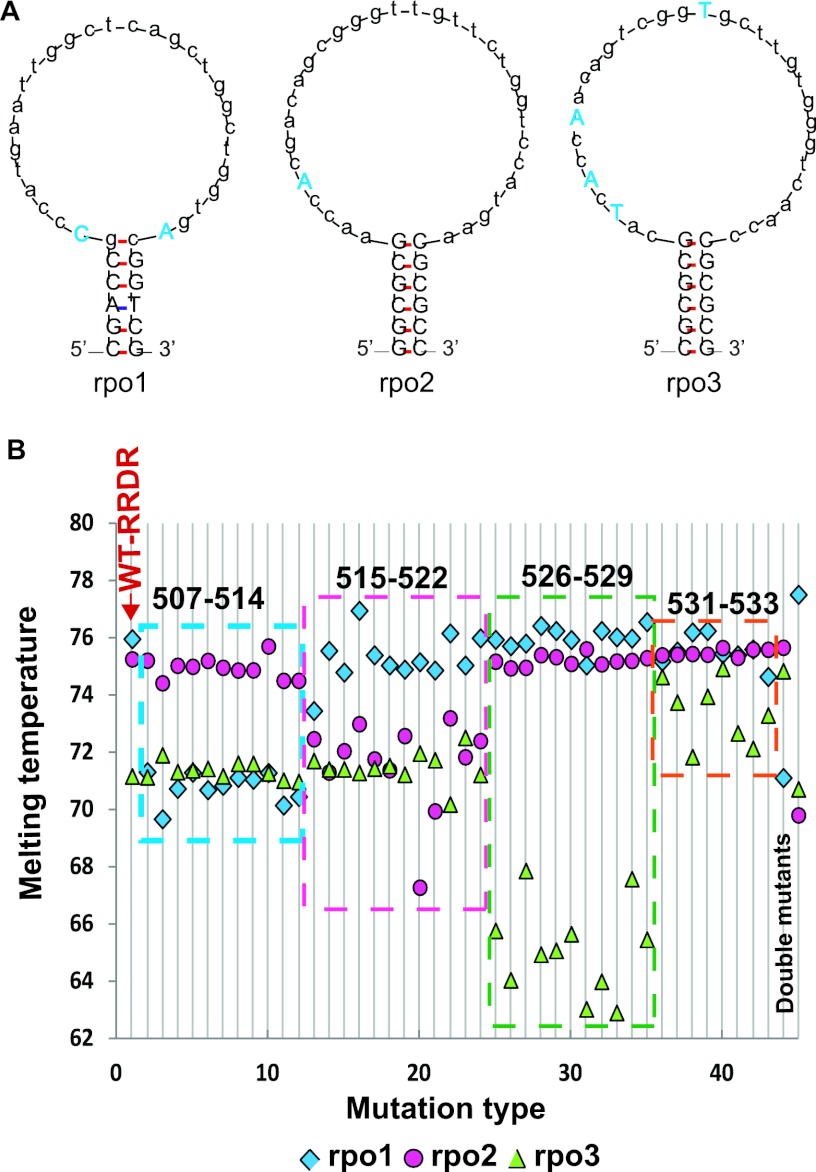
We have previously demonstrated the utility of SMB Tm-based assays to detect fluoroquinolone resistance-associated mutations in M. tuberculosis (8). To identify rifampin resistance-associated mutations in the M. tuberculosis RRDR, we designed three SMB probes, rpo1, rpo2, and rpo3, which contained 28- to 32-bp probe sequences and 5- to 6-bp stem sequences. The probes were designed to generate stable stem-loop structures while avoiding the formation of secondary structures in the loop region. The rpo1 probe targeted RRDR codons 507 to 516, rpo2 targeted codons 514 to 523, and rpo3 targeted codons 524 to 533. Each probe was designed to specifically detect all clinically relevant resistance-associated RRDR mutations spanning the codons targeted by the respective probes. A stable stem-loop structure is necessary for optimal binding kinetics between the SMB probes and the M. tuberculosis DNA target and for the production of distinct Tm values corresponding to each RRDR mutation. We stabilized the stem-loop structures for all three probes by introducing mutations into the probe loop to disrupt secondary structures and palindromes (Fig. 1A) as described previously (7, 8). The mutations introduced were chosen either to be complementary to a known RRDR mutation or to be unrelated to any known nucleotide at that position, depending on assay requirements and on the resulting probe structure. Among the rifampin resistance-associated mutations introduced, probe rpo1 included a 516GGC mutation instead of wild-type sequence and rpo3 included 531TTG and 533ATG mutations instead of wild-type sequence. The inclusion of RRDR resistance-associated mutations within the SMB probes produced a gradient of probe-target relatedness, with certain common RRDR mutations producing Tm values higher than wild-type RRDR values and other (less commonly occurring) RRDR mutations producing Tm values lower than that of wild-type RRDR (Table 1 and Fig. 1B).
Fig. 1.
Probe structures and Tm profiles against artificial targets. (A) The stable stem-loop structures of the three SMB probes used in the assay are shown. Related and unrelated mutations in the loop region introduced to obtain a stable stem-loop structure are shown in blue uppercase letters. Uppercase letters in black show the stem regions. (B) Three-probe Tm code of the assay tested against artificial targets with the wild-type RRDR or known RRDR mutations. Each horizontal line containing a square, circle, and triangle represents a unique 3-point Tm code corresponding to a single RRDR sequence. Line 1 shows results for a wild-type RRDR target, and lines 2 to 45 show results for mutant RRDR targets. The sequences of the mutations tested correspond to those shown in rows 1 to 45 of Table 1. Mutants are detectible by the presence of a substantial shift in the Tm value of either one or more of the three Tm points. Lines 2 to 12 show RRDR sequences harboring mutations in codons 507 to 514. Lines 13 to 24, RRDR sequences harboring mutations in codons 515 to 522; lines 25 to 35, RRDR sequences harboring mutations in codons 526 to 529; lines 36 to 43, RRDR sequences harboring mutations in codons 531 to 533; lines 44 and 45 show two RRDR double mutants.
We measured the Tm of all three SMBs in the presence of wild-type RRDR and each of the RRDR mutant targets using artificial oligonucleotides. A three-point Tm code for each oligonucleotide target was generated by this procedure (Table 1). We found that the Tm code for the wild-type target could be distinguished from RRDR mutant targets in all cases. For 41 of the 44 RRDR mutants, at least one of the three SMBs had a Tm value difference (dTm) of at least 2.1°C from the wild-type RRDR value (Table 1 and Fig. 1B). Many dTms were even higher. Targets containing common double mutants also had at least one probe with a dTm of at least 2.1°C (Table 1 and Fig. 1B). Three less commonly occurring RRDR mutations, 531GCG, 531TAG, and 533CCG, were detected with dTms of −0.7, −1.5, and −1.0°C, respectively. However, these mutants produced larger dTms when chromosomal DNA from clinical M. tuberculosis isolates was tested (described below). The RRDR mutants could also be conveniently grouped into Tm code clusters corresponding to the codons tested by the individual probes (Fig. 1B and Fig. 2).
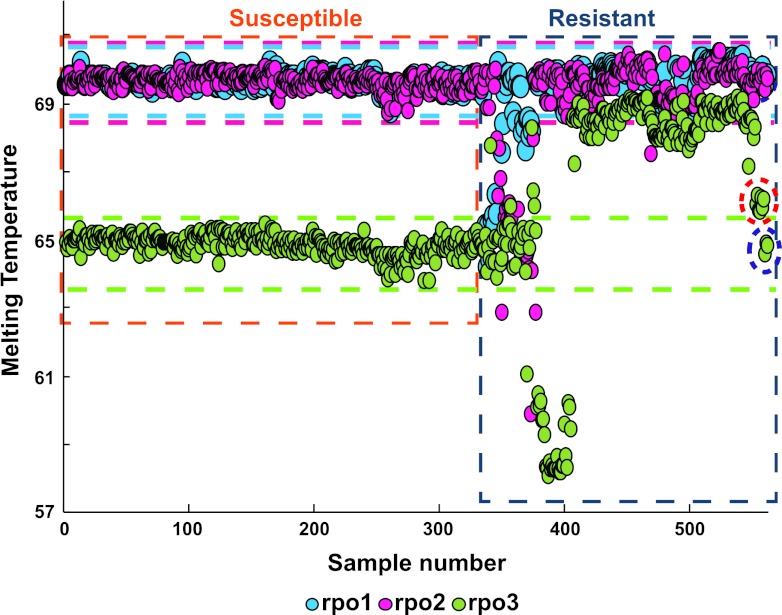
Fig. 2.
Three-point Tm profile of 589 clinical DNA study samples. The results of the assay tested against the clinical M. tuberculosis study isolates are shown. The horizontal green, pink, and light-blue lines represent the Tm zones corresponding to the wild-type RRDR sequence for each of the three individual probes. Samples with one or more of the three Tm values that fall outside these Tm zones are easily identified as rifampin resistant. Rifampin-susceptible and rifampin-resistant isolates can be distinctly clustered based on their individual Tm profiles, as shown enclosed within the orange and blue rectangles. The red circle indicates the samples with the 533CCG mutation, which was distinctly and independently clustered from the wild-type samples in spite of having a relatively low dTm value. The purple circle indicates the three rifampin-resistant samples with mutations in the rpoB gene outside the RRDR.
Assay Tm profiles of clinical samples compared to DNA sequencing.
We evaluated the ability of the SMB assay to detect RRDR mutations against a panel of 589 clinical samples of diverse geographical origins (Table 2). The test panel included 236 RRDR mutant samples and 25 different RRDR mutation genotypes distributed over 11 codons, including the common clinical mutations, double mutants, codon insertions, and deletions (Table 2). The predominant mutation type was 531TTG (62%), followed by 516GTC (8.8%), 526TAC (7%), and 533CCG (4%), with the other genotypes being contained in the remaining 18% of the RRDR mutants. The assay identified 236/236 (sensitivity, 100%; 95% confidence intervals [CIs], 98.0 to 100.0%) RRDR mutants correctly, including 234/234 samples that contained only mutant DNA and 2/2 samples that were identified as heteroresistant mixed infections, by DNA sequencing. For 24 out of the 25 mutations, the average dTm values were between 2.4 and 9.7°C, as either a positive or a negative shift from the Tm of the wild-type sequence, for at least one of the three probes (Table 2). The 10 samples with a 533CCG mutation had dTms ranging from −1.0 to −1.5°C. Thus, our tests of clinical samples identified all RRDR mutants, in contrast to our tests of oligonucleotides where 3/44 RRDR mutations showed smaller dTms, making them more difficult to detect. Assay specificity was also high, with 353/353 (100%; 95% CI, 98.7 to 100.0%) samples with wild-type RRDR sequences being correctly identified as having wild-type RRDR by the SMB assay. The 353 wild-type samples had mean dTm values for rpo1, rpo2, and rpo3 of 69.7 ± 0.18, 69.7 ± 0.21, and 64.9 ± 0.29°C, respectively (Table 2).
Table 2.
Three-probe Tm (°C) code of the assay in tests of clinical M. tuberculosis DNAa
| Sample |
Tm (°C) of: |
SD for probe no.: |
dTm (°C) for probe no.: |
No. of isolates | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Mutation type | rpo1 | rpo2 | rpo3 | 1 | 2 | 3 | 1 | 2 | 3 | |
| #1 | Wild type | 69.7 | 69.7 | 64.9 | 0.18 | 0.21 | 0.29 | 353 | |||
| 2 | 511CTG-CCG | 65.0 | 69.6 | 64.8 | 0.70 | 0.60 | 0.55 | 4.6 | 0.1 | 0.0 | 3 |
| 3 | 513CAA-CTA | 65.9 | 69.8 | 64.7 | 0.64 | 0.35 | 0.51 | 3.8 | −0.1 | 0.1 | 2 |
| 4 | 513CAA-AAA | 65.7 | 69.8 | 64.7 | 0.11 | 0.16 | 0.43 | 4.0 | −0.1 | 0.2 | 2 |
| 5 | 513-514CAA insertion | 64.4 | 68.9 | 64.5 | 0.00 | 0.00 | 0.00 | 5.3 | 0.8 | 0.4 | 1 |
| 6 | 516GAC-AAC | 69.8 | 67.7 | 65.1 | 0.00 | 0.00 | 0.00 | −0.1 | 2.0 | −0.2 | 1 |
| 7 | 516GAC-GTC | 69.3 | 65.4 | 64.9 | 0.44 | 0.50 | 0.49 | 0.4 | 4.3 | 0.0 | 21 |
| 8 | 516GAC-CTC | 67.6 | 62.9 | 64.5 | 0.00 | 0.00 | 0.00 | 2.1 | 6.8 | 0.4 | 1 |
| 9 | 516GAC-TAC | 68.3 | 64.7 | 64.5 | 0.36 | 0.33 | 0.87 | 1.4 | 5.0 | 0.4 | 7 |
| 10 | 518AAC deletion | 69.8 | 62.9 | 65.3 | 0.00 | 0.00 | 0.00 | −0.1 | 6.8 | −0.4 | 1 |
| 11 | 522TCG-TTG | 69.7 | 66.8 | 64.3 | 0.00 | 0.00 | 0.00 | 0.0 | 2.8 | 0.5 | 1 |
| 12 | 526CAC-CGC | 69.8 | 69.9 | 60.0 | 0.00 | 0.20 | 0.01 | −0.2 | −0.2 | 4.9 | 4 |
| 13 | 526CAC-TGC | 69.8 | 70.1 | 59.5 | 0.00 | 0.00 | 0.00 | −0.1 | −0.4 | 5.4 | 1 |
| 14 | 526CAC-CTC | 69.8 | 69.4 | 58.9 | 0.18 | 0.50 | 0.56 | −0.2 | 0.3 | 5.9 | 3 |
| 15 | 526CAC-TAC | 69.6 | 69.6 | 59.0 | 0.34 | 0.30 | 0.14 | 0.1 | 0.1 | 5.9 | 17 |
| 16 | 526CAC-GAC | 69.8 | 69.7 | 58.3 | 0.08 | 0.44 | 0.17 | −0.1 | 0.0 | 6.5 | 4 |
| 17 | 531TCG-CAG | 69.9 | 69.9 | 68.6 | 0.01 | 0.02 | 0.06 | −0.2 | −0.2 | −3.7 | 2 |
| 18 | 531TCG-TTG | 69.8 | 69.6 | 68.6 | 0.55 | 0.53 | 0.42 | −0.1 | 0.0 | −3.7 | 147 |
| 19 | 531TCG-TGG | 70.0 | 69.9 | 67.3 | 0.00 | 0.00 | 0.00 | −0.3 | −0.2 | −2.4 | 1 |
| 20 | 533CTG-CCG | 69.8 | 69.8 | 66.1 | 0.18 | 0.14 | 0.23 | −0.1 | −0.1 | −1.2 | 10 |
| 21 | 513CAA-CTA, 523GGG-GAG | 65.8 | 68.0 | 64.4 | 0.00 | 0.00 | 0.00 | 3.9 | 1.7 | 0.5 | 1 |
| 22 | 515ATG-ATT, 516GAC-TAC | 66.7 | 64.8 | 64.4 | 0.00 | 0.00 | 0.00 | 3.0 | 4.9 | 0.5 | 1 |
| 23 | 515ATG-GTG, 516GAC-GGC | 70.2 | 66.3 | 65.1 | 0.00 | 0.00 | 0.00 | −0.5 | 3.4 | −0.2 | 1 |
| 24 | 516GAC-GGC, 518AAC-CAC | 68.5 | 59.9 | 65.4 | 0.00 | 0.00 | 0.00 | 1.2 | 9.7 | −0.5 | 1 |
| 25 | 516GAC-TAC, 531TCG-TTG | 68.2 | 64.1 | 68.3 | 0.00 | 0.00 | 0.00 | 1.5 | 5.5 | −3.4 | 1 |
| 26 | 516GAC-GGC, 533CTG-CCG | 72.2 | 67.9 | 66.2 | 0.23 | 0.19 | 0.31 | −2.5 | 1.8 | −1.4 | 2 |
SD represents the standard deviations of the Tm values of each of the three probes for the different clinical samples harboring the same RRDR mutations. The shaded cells represent the dTm values obtained for the probes detecting the corresponding mutations.
All of the wild-type and mutant DNA samples generated an identifiable three-Tm code, which enabled the grouping of the RRDR mutants into smaller clusters that contained one or more possible mutations as observed with the artificial targets (Fig. 2 and Table 2). Specifically, the assay clustered single mutants, insertions, and deletions into the following groups: 511/513 mutant, 513-514CAA insertion, 516AAC/522TTG, 516GTC, 516CTC, 516TAC, 516GGC, 518AAC deletion, 526CGC, 526TGC/CTC/TAC, 526GAC, 531CAG/TTG, 531TGG, and 533CCG (Table 2 and Fig. 1B). Double mutants were identified by the presence of characteristic dTms from more than one probe (Table 2).
Comparison to phenotypic drug susceptibility testing.
The performance of the rpoB SMB assay was compared to phenotypic drug susceptibility testing in the 561 samples for which rifampin drug susceptibility testing results were available (Fig. 2). Sensitivity for resistance was 222/225 (98.7%; 95% CI, 95.8 to 99.6%), and specificity was 335/336 (99.7%; 95% CI, 98.1 to 99.9%). Among the rifampin-resistant false negatives, three contained rpoB mutations outside the RRDR (251TTC, 331CCC, and 572TTC). The single rifampin-resistant false positive occurred in a sample that was found to have an rpoB 511CCG mutation upon DNA sequencing. Clinical isolates with 511CCG mutations are prone to be misreported as rifampin susceptible by phenotypic rifampin susceptibility assays (31). Thus, this particular clinical strain may have been truly rifampin resistant. In fact, two other isolates in our study with the 511CCG mutation were reported to be phenotypically rifampin resistant. Unfortunately, cultures of this isolate were not available for repeat phenotypic susceptibility testing. The SMB assay and phenotypic drug susceptibility results were concordant for all of the other 557 samples tested.
Analytical specificity of the assay.
The analytical specificity of the assay was further tested using DNA extracted from 121 clinical and 18 reference strains and 1 laboratory NTM strain, as well as 18 species of Gram-positive and Gram-negative bacteria. None of the NTMs, except for M. malmoense, generated a perceptible signal from any of the 3 probes. Very high concentrations of M. malmoense DNA (100 to 200 ng) caused the rpo2 probe to generate a Tm of 70°C, but no measurable Tm values were generated for the rpo1 and rpo3 probes. M. smegmatis also generated a profile similar to that of M. malmoense which is an rpo2 Tm of 70°C, even at lower DNA concentrations of up to 1 ng. This triple-Tm code (0, 70, and 0) was quite distinct from all of the other Tm codes that we had obtained on tests of both wild-type and mutant RRDR M. tuberculosis DNA. None of the Gram-positive or Gram-negative bacteria tested produced a measurable signal. To test for possible interference from NTM DNA, we mixed 106 genome equivalents of M. malmoense, M. abscessus, M. avium, M. chelonae, M. gordonae, M. intracellulare, M. kansasii, and M. smegmatis DNA with 105 genome equivalents of M. tuberculosis DNA containing RRDR mutations at 516GTC, 526TAC, 531TTG, or 533CCG. Each RRDR mutant was correctly identified despite the presence of excess NTM DNA (see Table S1 in the supplemental material). Further increasing the M. malmoense DNA concentration to a 20- to 50-fold excess compared to the concentration of M. tuberculosis DNA caused the rpo2 probe to generate a double Tm peak in the presence of the M. tuberculosis 516GTC and 516AAC RRDR mutants (see Table S1). When a double peak was visualized, one of the double peaks corresponded to the Tm expected for the M. tuberculosis RRDR mutant, and the other peak corresponded to the Tm expected for M. malmoense DNA (data not shown). M. malmoense DNA alone, when used at a 20- to 50-ng concentration, generated a single peak with a Tm of 70°C corresponding to the rpo2 probe, and this did not mimic the wild-type profile or any of the RRDR mutant three-probe Tm profiles (see Fig. S1A in the supplemental material). Results identical to those for M. malmoense were obtained when 10 ng of the laboratory strain of M. smegmatis DNA was spiked into M. tuberculosis DNA containing the 516GTC and 516AAC mutations (see Table S1). These results strongly suggest that NTM coinfections will not affect assay results regarding M. tuberculosis rifampin susceptibility. The assay was also found to be unaffected by the presence of as much as a 105-fold excess of human genomic DNA compared to the concentration of M. tuberculosis DNA. The assay's capacity for mutation detection also remained unaltered, as tested with a mixture containing a large excess of human DNA mixed with clinical sample DNA containing the RRDR mutation 516TAC, suggesting that human genomic DNA from sputum should not interfere with the assay's performance (see Fig. S1B).
Assay LOD.
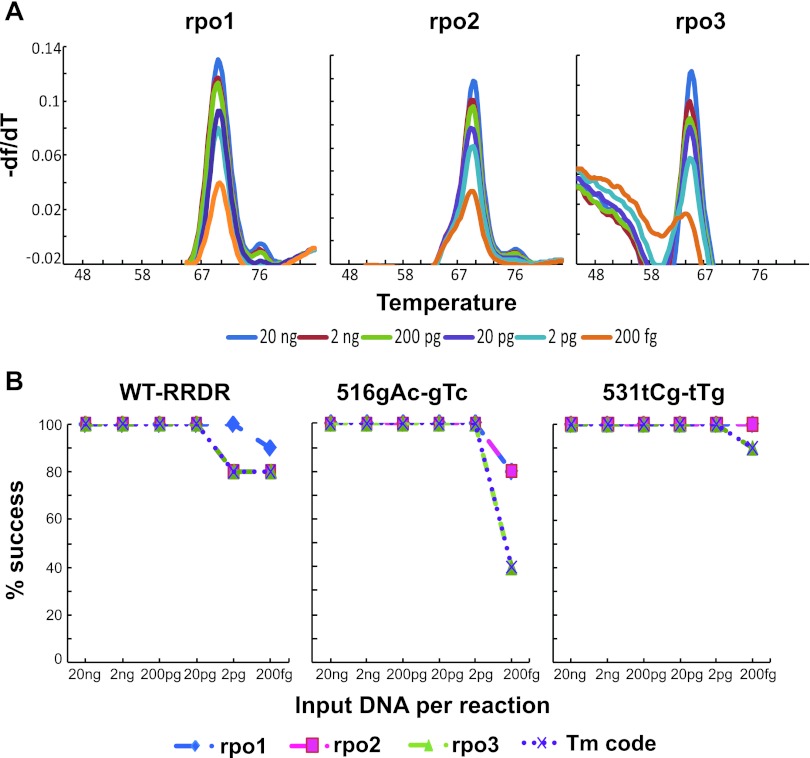
Genomic DNA from M. tuberculosis containing wild-type rpoB RRDR and the two most common RRDR mutations, 516GTC and 531TTG, were chosen to determine the limit of detection of the rpoB SMB assay. DNA samples were serially diluted from 10 ng to 100 fg per microliter in 10-fold decrements, and the SMB assay was performed on 2 μl of each individual dilution 10 times. We observed that the Tm values obtained at different concentrations were independent of the initial amount of target DNA put into the reaction for M. tuberculosis with wild-type (Fig. 3A) and mutant RRDR (data not shown), when Tm peaks could be measured. Each probe had a characteristic LOD below which no Tm value could be observed. We defined the assay LOD as the DNA concentration below which at least one probe could not consistently measure a Tm (probe rpo3 in this case). For the wild-type and mutant DNA samples, the assay could detect 20 and 2 pg of DNA correctly 100% of the time (Fig. 3B), respectively, which is approximately 2,000 and 200 M. tuberculosis genome equivalents, respectively. These results suggest that the assay is able to detect rifampin resistance in DNA extracted from all smear-positive and many smear-negative patient samples.
Fig. 3.
Analytical sensitivity of the assay. (A) Assay curves with limiting amounts of wild-type DNA target. Melting temperature profiles of the three SMB probes on serial dilutions of M. tuberculosis H37Rv genomic DNA with wild-type rpoB RRDR. (B) Limit of detection (LOD) of the assay against wild-type and RRDR mutant DNA. Tm values for each probe are shown for assays performed against the wild-type RRDR and two different RRDR mutant DNA samples. Assays were tested with a range of DNA concentrations as indicated. The LOD of the assay for each RRDR sequence type is defined by the LOD of the probe with the lowest analytical sensitivity. As shown, the LOD for wild-type RRDR was 20 pg, and the LOD for the 516GTC and 531TTG mutant RRDR samples was 2 pg.
Detection of 531TTG mutation in DNA mixtures.
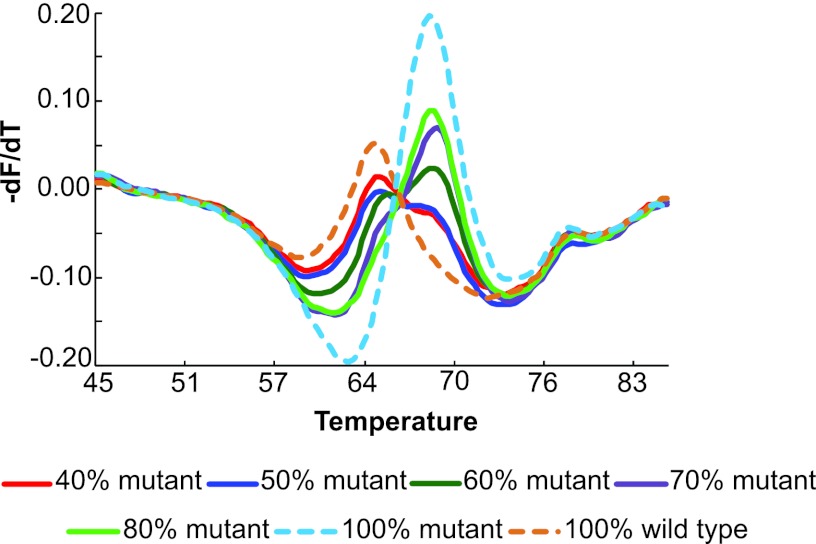
The ability of the rpoB SMB assay to detect the RRDR mutant in this mixture was compared to that of Sanger DNA sequencing. The SMB assay detected the presence of the RRDR mutation down to a concentration of 40% mutant DNA, as indicated by the double peak of Tm values (Fig. 4). The mutant Tm peak became increasingly prominent as more mutant DNA was present in the mixture. DNA mixtures with 10, 20, and 30% mutant DNA showed a single wild-type Tm peak and were indistinguishable from a sample containing 100% wild-type DNA (Fig. 4). Using the same DNA mixtures to perform the bidirectional sequencing of the rpoB amplicon, the unequivocal presence of the mutant DNA was apparent only on sequencing chromatograms when at least 50% mutant DNA was present. Thus, our SMB assay detected mixed infections at least as well as Sanger DNA sequencing as far as the most common 531TTG mutation was concerned.
Fig. 4.
Detection of mixed-sequence heteroresistance. The assay was performed with various mixtures of wild-type and mutant DNA, as indicated. Double Tm peaks, indicating mixtures of wild-type and mutant target, were seen when as little as 40% mutant sequence was present.
DISCUSSION
The assay used in this study detected the reported RRDR mutations with high sensitivity and specificity. The assay was also analytically very specific for M. tuberculosis when tested against a large panel of NTM and other bacteria. Heteroresistance was detected at least as well as that detected by Sanger DNA sequencing. Finally, the assay LOD appeared to be sufficiently sensitive for use in all smear-positive and many smear-negative sputum samples from tuberculosis patients. To our knowledge, our study describes the first Tm shifting assay for rifampin-resistant M. tuberculosis with this high a level of performance. The mismatch-tolerant SMB probes used in this study offer several advantages over the current generation of probe-based drug susceptibility tests. Unlike open hybridization assays, our assay is performed in a closed system, which greatly simplifies sample testing and minimizes cross-contamination. Unlike HRMA detection methods, which detect mutations by recognizing subtle melting curve variants (9, 27), our assay produces clear and consistent Tm peaks, which enables highly reproducible Tm value identification.
Several assays have been described previously which utilize FRET probes or dually labeled probes and post-PCR Tm shifts to identify resistance-associated mutations (9, 10, 13, 18, 20, 24, 26, 27). A recent study by Luo et al. used dually labeled probes and a common fluorophore to label and identify mutations associated with isoniazid and rifampin resistance using Tm curve patterns, which is similar to our approach. However, the dually labeled probes used by those authors produced overlapping Tm peaks that are potentially difficult to analyze (24). This system is also unlikely to be able to identify most mixtures of wild-type and mutant DNA sequences. In contrast, the peaks generated by SMBs were sharp and consistent even when our assay was performed starting from very small numbers of genomic DNA copies. Assays which use FRET probes are constrained by the requirement to design probes in pairs (one anchor and one sensor probe). This requirement can cause some mutations to be masked by the anchor probes (18) and in some cases results in Tm differences that are less than 0.5°C between wild-type and mutant sequences (25). FRET probes also require the monitoring of fluorescence ratios, which generates complicated Tm patterns that can be easily confounded by DNA mixtures (20). A recent study used shared-stem molecular beacons and TaqMan probes to identify rpoB RRDR mutations using post-PCR Tm calculations (18). However, the assay required at least two independent reactions and four different probes to cover the entire RRDR. Furthermore, the Tm differences and the resolution provided by the shared-stem molecular beacons was much smaller than that observed with other dually labeled probes and by our assay, decreasing mutant discriminatory capacity (18).
Mismatch-tolerant SMB probes created according to our design principles allow versatility in sequence recognition while maintaining a robust capacity to discriminate mutations. The three-point Tm code produced by our assay either specifically identified RRDR mutations or grouped them into easily identifiable mutation clusters. This ability to subtype different RRDR mutations may prove useful in epidemiological investigations or where different rifamycins confer different levels of resistance (20, 28, 30, 35). The assay did not depend on high-resolution Tm capabilities. Unlike other assays which focus mainly on identifying the most common clinically prevalent mutations (20, 24, 27), we designed our assay to detect both the common and uncommon mutations over the entire RRDR sequence. The large probe regions permitted by SMB design made it possible for us to use just three probes to target the entire 80-bp rpoB RRDR sequence. The assay identified every type of mutation tested, including nucleotide transitions, transversions, deletions, and insertions, along with several double mutations.
There are some limitations to the current form of our assay. First, our assay cannot specifically differentiate an infrequently occurring synonymous mutation at RRDR codon 514 from nonsynonymous mutations at codon 514, which are associated with rifampin resistance (13). Should synonymous mutations appear more frequently than are currently reported in clinical settings, our assay could be redesigned to generate a unique three-point Tm code for that particular mutation. Second, the average dTm of the rpo3 probe for the 533CCG mutant was 1.16°C. While sufficient to reliably detect 533CCG mutations, this dTm was the smallest of all the dTms generated by mutations in our assay. It is possible that the dTm for 533CCG mutations could be improved by additional rounds of probe redesign. Third, we have not yet tested our assay on DNA directly extracted from human sputum samples. It is possible that PCR inhibitors present in sputum degrade the assay LOD. However, many methods exist to extract M. tuberculosis DNA from sputum for sensitive real-time PCR assays (3, 22). Therefore, we do not expect this to be a major developmental hurdle.
In summary, we have developed a sensitive, specific, and rapid assay that is strongly amenable to a high-throughput format which can detect all of the clinically significant mutations in the rpoB RRDR of M. tuberculosis. This assay is specific for M. tuberculosis, simple, and robust and does not require any high-resolution melting software, thus it is compatible with various real-time PCR platforms. The SMB Tm coding approach we describe is likely to be generally useful for rapid mutation detection in clinical microbiology.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health grants AI082174 and AI080653 (to D.A., S.C., H.K., and S.S.R.); Northeast Bio-Defense Career training grant 3185-07 (to B.A.); the Intramural Research Program of the NIAID, NIH (to C.E.B.); and the South Korean Ministry of Health and Welfare (to S.-N.C. and C.E.B.).
Clinical DNA samples obtained from the WHO TDR Tuberculosis Specimen Bank are gratefully acknowledged. We thank Regina Wilson for kindly providing us with frozen stock of a laboratory strain of M. smegmatis.
D. Alland is among a group of inventors who earn royalties for molecular beacon usage.
Footnotes
Published ahead of print 25 April 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Ahmad S, Al-Mutairi NM, Mokaddas E. 2009. Comparison of performance of two DNA line probe assays for rapid detection of multidrug-resistant isolates of Mycobacterium tuberculosis. Indian J. Exp. Biol. 47:454–462 [PubMed] [Google Scholar]
- 2. Albert H, et al. 2010. Rapid screening of MDR-TB using molecular Line Probe Assay is feasible in Uganda. BMC Infect. Dis. 10:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aldous WK, Pounder JI, Cloud JL, Woods GL. 2005. Comparison of six methods of extracting Mycobacterium tuberculosis DNA from processed sputum for testing by quantitative real-time PCR. J. Clin. Microbiol. 43:2471–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boehme CC, et al. 2011. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet 377:1495–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brossier F, Veziris N, Truffot-Pernot C, Jarlier V, Sougakoff W. 2006. Performance of the genotype MTBDR line probe assay for detection of resistance to rifampin and isoniazid in strains of Mycobacterium tuberculosis with low- and high-level resistance. J. Clin. Microbiol. 44:3659–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bwanga F, Hoffner S, Haile M, Joloba ML. 2009. Direct susceptibility testing for multi drug resistant tuberculosis: a meta-analysis. BMC Infect. Dis. 9:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chakravorty S, et al. 2010. Rapid universal identification of bacterial pathogens from clinical cultures by using a novel sloppy molecular beacon melting temperature signature technique. J. Clin. Microbiol. 48:258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chakravorty S, et al. 2011. Rapid detection of fluoroquinolone-resistant and heteroresistant Mycobacterium tuberculosis by use of sloppy molecular beacons and dual melting-temperature codes in a real-time PCR assay. J. Clin. Microbiol. 49:932–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen X, et al. 2011. Rapid detection of isoniazid, rifampin, and ofloxacin resistance in Mycobacterium tuberculosis clinical isolates using high-resolution melting analysis. J. Clin. Microbiol. 49:3450–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Choi GE, et al. 2010. High-resolution melting curve analysis for rapid detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis clinical isolates. J. Clin. Microbiol. 48:3893–3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Friedrich SO, et al. 2011. Suitability of Xpert MTB/RIF and genotype MTBDRplus for patient selection for a tuberculosis clinical trial. J. Clin. Microbiol. 49:2827–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garcia-Sierra N, et al. 2011. Pyrosequencing for rapid molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 49:3683–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Garcia de Viedma D, et al. 2002. New real-time PCR able to detect in a single tube multiple rifampin resistance mutations and high-level isoniazid resistance mutations in Mycobacterium tuberculosis. J. Clin. Microbiol. 40:988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Helb D, et al. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J. Clin. Microbiol. 48:229–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hillemann D, Rusch-Gerdes S, Richter E. 2007. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J. Clin. Microbiol. 45:2635–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoek KG, Van Rie A, van Helden PD, Warren RM, Victor TC. 2011. Detecting drug-resistant tuberculosis: the importance of rapid testing. Mol. Diagn. Ther. 15:189–194 [DOI] [PubMed] [Google Scholar]
- 17. Hofmann-Thiel S, et al. 2009. Mechanisms of heteroresistance to isoniazid and rifampin of Mycobacterium tuberculosis in Tashkent, Uzbekistan. Eur. Respir. J. 33:368–374 [DOI] [PubMed] [Google Scholar]
- 18. Huang Q, et al. 2011. Multiplex fluorescence melting curve analysis for mutation detection with dual-labeled, self-quenched probes. PLoS One 6:e19206 doi:10.1371/journal.pone.0019206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kiet VS, et al. 2010. Evaluation of the MTBDRsl test for detection of second-line-drug resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 48:2934–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kocagoz T, Saribas Z, Alp A. 2005. Rapid determination of rifampin resistance in clinical isolates of Mycobacterium tuberculosis by real-time PCR. J. Clin. Microbiol. 43:6015–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lacoma A, et al. 2008. GenoType MTBDRplus assay for molecular detection of rifampin and isoniazid resistance in Mycobacterium tuberculosis strains and clinical samples. J. Clin. Microbiol. 46:3660–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leung ET, et al. 2011. Rapid and simultaneous detection of Mycobacterium tuberculosis complex and Beijing/W genotype in sputum by an optimized DNA extraction protocol and a novel multiplex real-time PCR. J. Clin. Microbiol. 49:2509–2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ling DI, Zwerling AA, Pai M. 2008. GenoType MTBDR assays for the diagnosis of multidrug-resistant tuberculosis: a meta-analysis. Eur. Respir. J. 32:1165–1174 [DOI] [PubMed] [Google Scholar]
- 24. Luo T, et al. 2011. Multiplex real-time PCR melting curve assay to detect drug-resistant mutations of Mycobacterium tuberculosis. J. Clin. Microbiol. 49:3132–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marin M, Garcia de Viedma D, Ruiz-Serrano MJ, Bouza E. 2004. Rapid direct detection of multiple rifampin and isoniazid resistance mutations in Mycobacterium tuberculosis in respiratory samples by real-time PCR. Antimicrob. Agents Chemother. 48:4293–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ong DC, Yam WC, Siu GK, Lee AS. 2010. Rapid detection of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis by high-resolution melting analysis. J. Clin. Microbiol. 48:1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ramirez MV, et al. 2010. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J. Clin. Microbiol. 48:4003–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Riska PF, Jacobs WR, Jr, Alland D. 2000. Molecular determinants of drug resistance in tuberculosis. Int. J. Tuberc. Lung Dis. 4:S4–S10 [PubMed] [Google Scholar]
- 29. Skenders GK, Holtz TH, Riekstina V, Leimane V. 2011. Implementation of the INNO-LiPA Rif. TB(R) line-probe assay in rapid detection of multidrug-resistant tuberculosis in Latvia. Int. J. Tuberc. Lung Dis. 15:1546–1552 [DOI] [PubMed] [Google Scholar]
- 30. Tan Y, et al. 2011. The beginning of the rpoB gene in addition to the RRDR might be needed for identifying RIF/Rfb cross resistance in multidrug-resistant Mycobacterium tuberculosis isolates from southern China. J. Clin. Microbiol. 50:81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van Deun A, et al. 2009. Mycobacterium tuberculosis strains with highly discordant rifampin susceptibility test results. J. Clin. Microbiol. 47:3501–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van Rie A. 2011. A single Xpert MTB/RIF test of sputum for diagnosis of tuberculosis and multidrug resistance shows high sensitivity and specificity and reduces diagnosis and treatment delays. Evid. Based Med. 16:174–175 [DOI] [PubMed] [Google Scholar]
- 33. Vassall A, et al. 2011. Rapid diagnosis of tuberculosis with the Xpert MTB/RIF assay in high burden countries: a cost-effectiveness analysis. PLoS Med. 8:e1001120 doi:10.1371/journal.pmed.1001120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Viveiros M, et al. 2005. Direct application of the INNO-LiPA Rif.TB line-probe assay for rapid identification of Mycobacterium tuberculosis complex strains and detection of rifampin resistance in 360 smear-positive respiratory specimens from an area of high incidence of multidrug-resistant tuberculosis. J. Clin. Microbiol. 43:4880–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Williams DL, et al. 1998. Contribution of rpoB mutations to development of rifamycin cross-resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1853–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williamson DA, et al. 2011. Clinical failures associated with rpoB mutations in phenotypically occult multidrug-resistant Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 16:216–220 [DOI] [PubMed] [Google Scholar]
- 37. Wright A, et al. 2009. Epidemiology of antituberculosis drug resistance 2002-07: an updated analysis of the Global Project on Anti-Tuberculosis Drug Resistance Surveillance. Lancet 373:1861–1873 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.