Abstract
The occurrence of Clostridium difficile infections in patients that do not fulfill the classical risk factors prompted us to investigate new risk factors of disease. The goal of this study was to characterize strains and associated antimicrobial resistance determinants of C. difficile isolated from swine raised in Ohio and North Carolina. Genotypic approaches used include PCR detection, toxinotyping, DNA sequencing, and pulsed-field gel electrophoresis (PFGE) DNA fingerprinting. Thirty-one percent (37/119) of isolates carried both tetM and tetW genes. The ermB gene was found in 91% of isolates that were resistant to erythromycin (68/75). Eighty-five percent (521/609) of isolates were toxin gene tcdB and tcdA positive. A total of 81% (494/609) of isolates were positive for cdtB and carry a tcdC gene (a toxin gene negative regulator) with a 39-bp deletion. Overall, 88% (196/223) of pigs carry a single C. difficile strain, while 12% (27/223) of pigs carried multiple strains. To the best of our knowledge, this is the first report of individual pigs found to carry more than one strain type of C. difficile. A significant difference in toxinotype profiles in the two geographic locations was noted, with a significantly (P < 0.001) higher prevalence of toxinotype V found in North Carolina (84%; 189/224) than in Ohio (55%; 99/181). Overall, the study findings indicate that significant proportions of C. difficile in swine are toxigenic and often are associated with antimicrobial resistance genes, although they are not resistant to drugs that are used to treat C. difficile infections.
INTRODUCTION
Clostridium difficile is one of the leading causes of nosocomial diarrhea and has increasingly been reported as an emerging community-associated pathogen (17, 35). Each case of C. difficile-associated disease has been estimated to result in more than $3,600 in excess health care costs, and these costs may exceed $1 billion annually in the United States alone (14, 17). Although elderly hospitalized patients receiving antibiotics are the main group at risk of infection, an increasing number of C. difficile infections (CDI) in younger populations with no previous contact either with the hospital environment or with antibiotics has been documented (17). The rising rates of CDI have largely been attributed to the presence of the hypervirulent epidemic strain of C. difficile, NAP1/027, but are not limited to this strain. The occurrence of CDI in patients that do not fulfill the classical risk factors prompts for the investigations of new risk factors.
C. difficile is also associated with disease in animal species, including swine, calves, and horses (11, 23, 30). Extensive work has been conducted to describe C. difficile disease in swine; however, published data on the epidemiology and molecular characterization of C. difficile from swine herds is limited (9, 30, 33, 35, 37). Scientific data on the occurrence of phenotypically and genetically similar strains from swine and humans are very limited.
The emergence of hypervirulent and epidemic strains has increased the interest in C. difficile typing and stimulated the application of newer genotype-based methods, such as PCR ribotyping (3, 32), amplified fragment length polymorphism (AFLP) (18), multilocus sequence typing (MLST) (16), multilocus variable-number tandem-repeat analysis (MLVA) (34), and surface layer protein A gene sequence typing (slpAST) (10). Other methods have been used to characterize changes in the pathogenicity locus, which is the chromosomal region that carries the key toxin genes that render strains virulent or, in some cases, hypervirulent. The pathogenicity locus is a 19.6-kb chromosomal segment found in pathogenic strains of C. difficile that carry genes encoding enterotoxin or toxin A (tcdA), cytotoxin, or toxin B (tcdB) and accessory genes, including tcdC, tcdD, and tcdE. Variations in the pathogenicity locus sequence can be detected and provide the basis for the commonly used classification scheme known as toxinotyping (24).
There is little published data available on the antimicrobial susceptibility of C. difficile isolated from pigs or for animals in general (20, 33). The overall hypothesis of the study is that toxigenic strains of C. difficile are found in high prevalence in swine and are phenotypically and genotypically similar to isolates of human origin. The current study was conducted to investigate and characterize C. difficile of swine origin. The objectives were to investigate the epidemiology and develop baseline data of C. difficile in swine at farm and slaughter, to investigate whether swine could be sources of the organism to other pigs in the farm and to the processing continuum, and to assess the public health significance of C. difficile strains of porcine origin using phenotypic and genotypic analyses.
MATERIALS AND METHODS
Porcine fecal samples were collected from farms in North Carolina (n = 5) and Ohio (n = 3). Details of collection, isolation, and antimicrobial resistance testing are described in a previous publication (33). Briefly, swine fecal samples were collected from a cohort of 30 pigs at different stages of production, including farrowing, nursery, and finishing stages in the two geographic regions. C. difficile isolates were compared using antimicrobial resistance profiles, antimicrobial resistance gene detection, toxin gene detection, restriction fragment length polymorphism (RFLP) toxinotyping, and pulsed-field gel electrophoresis (PFGE). Fecal samples were collected from a total of 251 piglets and 68 sows at farrowing. Piglets were identified and again sampled at the nursery stage. All bacterial isolation was completed at North Carolina State University using standard protocols (38). Antimicrobial resistance testing was also completed at North Carolina State University using Epsilometric test strips. One isolate per animal was selected for testing. The isolates were tested against an antimicrobial panel consisting of ampicillin (Amp; MIC, 2 μg/ml [range, 0.016 to 256 μg/ml]), ciprofloxacin (Cip; MIC, 8 μg/ml [range, 0.002 to 32 μg/ml]), erythromycin (Ery; MIC, 2 μg/ml [0.016 to 256 μg/ml]), metronidazole (Met; MIC, 16 μg/ml [range, 0.016 to 256 μg/ml]), tetracycline (Tet; MIC, 4 μg/ml [range, 0.016 to 256 μg/ml]), and vancomycin (Van; MIC, 4 μg/ml [range, 0.016 to 256 μg/ml]).
All isolates were further confirmed as C. difficile by the amplification of the tpi gene. Primers were tpi specific (Table 1) and generated a 230-bp amplified fragment specific for C. difficile (16). PCR was done using Illustra PuReTag Ready-To-Go PCR beads (GE Healthcare, Buckinghamshire, United Kingdom). Each 25-μl reaction mix contained 2.5 U of PureTaq DNA polymerase; 10 mM Tris-HCl (pH 9.0 at room temperature); 50 mM KCl; 1.5 mM MgCl; 200 μM dATP, dCTP, dGTP, and dTTP; and stabilizers, including bovine serum albumin (BSA). Thermocycler running conditions included an initial denaturation step at 95°C for 15 min and then 30 cycles of a denaturation step at 95°C for 1 min, primer annealing at 54°C for 1 min, and an extension step at 72°C for 1 min. The final extension time was 7 min at 72°C. The reaction mix was then held at 4°C. Reference isolates of human origin were received from the Centers for Disease Control and Prevention (CDC) and had been previously characterized by PFGE NAP (North American pulsed-field) typing and toxinotyping. Strains used included a NAP1/toxinotype III strain (CDC4118), an unnamed/toxinotype XIV strain (CDC7167), a NAP5/toxinotype O strain (CDC5496), and a NAP7/toxinotype V strain (CDC5127).
Table 1.
List of primers used in this study
| Gene or fragment | Primer |
Amplicon (bp) | Reference | |
|---|---|---|---|---|
| Name | Sequence (3′-5′) | |||
| Gene | ||||
| tpi | tpi-F | AAAGAAGCTACTAAGGGTACAAA | 230 | 13 |
| tpi-R | CATAATATTGGGTCTATTCCTAC | |||
| tcdB | tcdB-F | GGAAAAGAGAATGGTTTTATTAA | 160 | 13 |
| tcdB-R | ATCTTTAGTTATAACTTTGACATCTTT | |||
| tcdA | tcdA-F | AGATTCCTATATTTACATGACAATAT | 369 (A+B+)/110 (A−B+)a | 13 |
| tcdA-R | GTATCAGGCATAAAGTAATATACTTT | |||
| tcdC | Tim2- | GCACCTCATCACCATCTTCAA | 345 | 28 |
| Struppi2 | TGAAGACCATGAGGAGGTCAT | |||
| cdtB | cdtBpos | CTTAATGCAAGTAAATACTGAG | 500 | 31 |
| cdtBrev | AACGGATCTCTTGCTTCAGTC | |||
| tetM | TETMd | TGGAATTGATTTATCAACGG | 1,000 | 27 |
| TETMr | TTCCAACCATACAATCCTTG | |||
| tetW | WRC1 | CATCTCTGTGATTTTCAGCTTTTCTCTCCC | 457 | 27 |
| WRC2 | AGTCTGTTCGGGATAAGCTCTCCGCCG | |||
| ermB | E5 | CTCAAAACTTTTTAACGAGTG | 711 | 29 |
| E6 | CCTCCCGTTAAATAATAGATA | |||
| Fragment | ||||
| B1 | B1C | AGAAAATTTTATGAGTTTAGTTAATAGAAA | 21 | |
| B2N | CAGATA ATGTAGGAAGTA AGTCTATAG | |||
| A3 | A3C | TATTGATAGCACCTGATTT ATATACAAG | 21 | |
| A4N | TTATCAAACATATATTTAGCCATATATC | |||
The amplicon size for toxin A-positive (A+) strains was 369 bp. However, toxin A-negative (A−) strains showed a smaller size, 110 bp.
PCR was done to identify toxins of the pathogenicity locus region (tcdA, tcdB, and tcdC), binary toxins (cdtB), and antimicrobial resistance genes (tetW, tetM, and ermB) (Table 1). Clostridium difficile DNA was extracted using the Qiagen DNeasy tissue kit (Qiagen, Valencia, CA). PCR-RFLP toxinotyping was adopted from previously described methods (24, 25). Briefly, the amplifications of B1 and A3 fragments of the pathogenicity locus region were done using the primers as described in Table 1. PCR was done using Illustra PuReTag Ready-To-Go PCR beads (GE Healthcare, Buckinghamshire, United Kingdom) as described above. Thermocycler running conditions were 93°C for 3 min and then 35 cycles of 51°C (A3) or 57°C (B1) for 8 min, followed by 93°C for 3 s. The run is completed by 47°C for 10 min and a hold at 4°C. The B1-amplified products were then cut with two restriction enzymes, AccI and HincII, and the A3-amplified products were cut with the EcoRI restriction enzyme. All restrictions were then visualized on 1% agarose gels with 0.5× Tris-borate-EDTA (TBE) buffer. Known types of restriction patterns are then determined using the published tables (24).
DNA fingerprinting was done using PFGE. A total of 105 isolates were genotyped, including 101 swine isolates and four control strains provided by the CDC (Atlanta, GA). Swine isolates were selected as representatives from each farm and each toxinotype found within each farm. The protocol used was a modified version of the previously reported methods (12). Fresh, isolated single colonies were added to 2 ml of cell suspension buffer (CSB). The cells were resuspended in 300 μl of Gram-positive lysis buffer and dispensed into the plug molds. Plugs were digested using SmaI. The standard markers, Salmonella enterica BAA-664 plugs, were also digested using XbaI at the same time. PFGE parameters used were a gradient of 6, run time of 18 h, included angle of 120, initial switch time of 5 s, and final switch time of 40 s. After the run was complete, the gel was stained using 5 μl/ml of ethidium bromide in distilled water and photographed using Gel Doc 2000 (Bio-Rad). Images were analyzed using Bionumerics 4.0 (applied Math) using a Dice similarity coefficient and a unweighted-pair group method using average linkages (UPGMA) dendrogram type. Position tolerance settings included optimization at 1.50% and position tolerance at 2.0%.
The characterization of the toxin regulator gene tcdC was conducted by amplification and DNA sequencing. tcdC fragments were amplified using the PCR protocol and gel electrophoresis protocol as described above. PCR products were extracted from the gel using the QIAquick gel extraction kit (Qiagen, Valencia, CA). Sequencing was done using a Beckman Coulter GenomeLab dye terminator cycle sequencing with quick start kit and Beckman Coulter CEQ 8000 genetic analysis system (Beckman Coulter, Fullerton, CA). All sequencing results were analyzed and confirmed using the basic local alignment search tool (BLAST).
RESULTS
C. difficile prevalence.
C. difficile was isolated from 73% (n = 183) of farrowing piglets, with a significantly higher prevalence in Ohio (87.5%) than North Carolina (64%). C. difficile was also isolated from 47% (n = 39) of sows, with no significant difference being noted between the two geographic regions. One pig was found to be positive at the nursery (33).
Antimicrobial resistance genes.
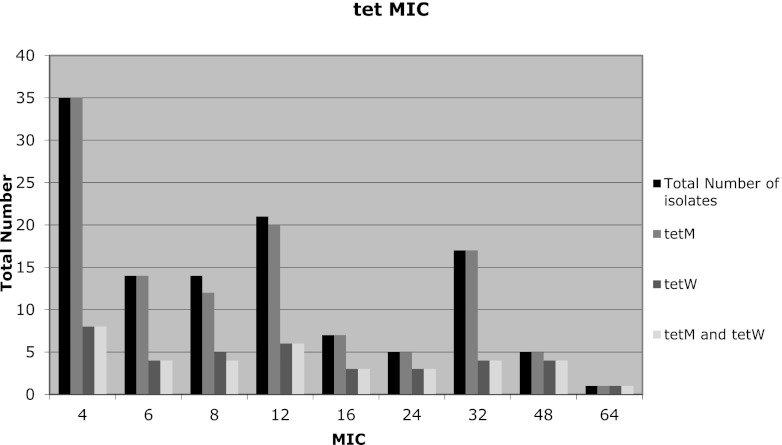
Of the 119 isolates found to be resistant to tetracycline, 97% (116/119) and 32% (38/119) of the isolates were positive for the tetM and tetW genes, respectively. In addition, 31% (37/119) of the tetracycline-resistant isolates were carried both tetM and tetW genes (Fig. 1). The ermB gene was found in 91% of the isolates that were found to be resistant to erythromycin (68/75). The majority (97%; 66/68) of isolates with a MIC of >256 μg/ml were found to carry the ermB gene. A total of 88.6% (39/44) isolates that were resistant to both erythromycin and tetracycline tested positive for both the ermB and tetM genes.
Fig 1.
Distribution of tet resistance genes through MIC ranges of isolates of swine origin. The total number of isolates found to be resistant to tetracycline and therefore tested by PCR for the presence of tet genes was 200. The total number with a MIC of ≥4 was 119.
Toxin gene PCR detection and sequencing.
Toxin gene detection using PCR was completed on a total of 609 isolates of porcine origin, 346 from North Carolina and 263 from Ohio. These isolates originated from the 223 total pigs that were positive for C. difficile, characterizing up to 3 colonies per pig. Eighty-five percent (521/609) of isolates tested were tcdB and tcdA positive. A small proportion, 1.1% (7/609), was tcdB positive and tcdA negative. A total of 81% (494/609) were positive for cdtB and also carried a tcdC gene with a 39-bp deletion. A tcdC gene with no noted deletion was also found in 5.6% (34/609) of the isolates.
Toxin gene profiles were assessed based on PCR and DNA sequencing, and four profile types were noted. The majority of isolates, 80% (487/609), carried genes encoding enterotoxin, cytotoxin, and the binary toxins (tcdB, tcdA, and cdtB), and these strains carried the corresponding downregulator gene, tcdC, with a 39-bp deletion (profile type 1). The second group consisted of 5.6% (34/609) of the isolates, which were positive for the toxin genes on the pathogenicity locus region (tcdB and tcdA) with no deletion of the downregulator gene, tcdC, and negative for the binary toxin marker gene cdtB (profile type 2). An additional two profiles, including a nontoxigenic one, were detected, as shown in Table 2.
Table 2.
Description and prevalence of toxin profile types
| Toxin gene profilea | Profile type | % Prevalence (no. positive/total no.) |
|---|---|---|
| A+B+, cdtB+, and 39-bp deletion in tcdC gene | 1 | 80 (487/609) |
| A+B+, lacking cdtB, and no deletion in tcdC gene | 2 | 5.6 (34/609) |
| A−B+, cdtB+, and 39-bp deletion in tcdC gene | 3 | 1.1 (7/609) |
| A−B−, lacking cdtB and tcdC | Nontoxigenic | 13 (81/609) |
A+ and A− represent the presence and absence of tcdA, and B+ and B− represent the presence and absence of tcdB, respectively.
The heterogeneity of toxin gene profiles among C. difficile strains isolated from individual pigs was assessed. Overall, 88% (196/223) of pigs carried only one type of C. difficile, and 12% (27/223) of the pigs carried more than one type of C. difficile. The majority of pigs, 73% (162/223), carried only the profile type 1 strain. A total of 11.2% (25/223) of pigs carried only a nontoxigenic strain of C. difficile, and a small percentage (4.0%; 9/223) carried only isolates that were profile type 2. The majority of pigs, 6.3% (14/223), that carried more than one strain carried one nontoxigenic isolate and one profile type 1 isolate. A few pigs, 4.0% (9/223), carried both type 1 and type 2. A small group of pigs, 1.8% (4/223), carried both profile type 1 and profile type 3.
Geographic comparison of C. difficile isolates.
Differences were noted between C. difficile isolates of swine origin from North Carolina and Ohio. A significantly larger number of isolates from North Carolina (88%; 306/346) were profile type 1, tcdB positive, tcdA positive, and cdtB positive, and they carried a tcdC gene with a 39-bp deletion compared to the tcdC gene of the Ohio isolates (69%; 181/263) (P < 0.001). North Carolina (8.1%; 28/346) was also found to have a significantly higher prevalence of profile type 2 isolates than Ohio (2.3%; 6/263) (P < 0.005). There was a significantly higher prevalence of nontoxigenic (profile type 4) C. difficile strains in Ohio (29%; 75/263) than in North Carolina (1.7%; 6/346) (P < 0.001).
PCR-RFLP toxinotyping.
Toxinotyping was done on 324 isolates; at least one isolate was selected per pig, with additional isolates selected when variations in toxin gene carriage were detected. More than one isolate was selected only from the pigs with heterogenicity found during toxin gene detection. All toxinotypes found were types that had been reported previously. The most common toxinotype found in the isolates was toxinotype V (89%; 289/324). Other toxinotypes found included toxinotype O (8.9%; 29/324) and toxinotype V-like (1.9%; 6/324). Most pigs (73%; 162/223) were found to carry only toxinotype V. Ten percent (25/223) of pigs were found to carry nontoxigenic isolates. A relatively small proportion of the pigs (4.0%; 9/223) carried only toxinotype 0. Overall, 12% of pigs carried more than one toxinotype. Toxinotype V and nontoxigenic strains were found in 6.3% (14/223) of pigs, toxinotypes V and O were found in 4.0% (9/223) of pigs, and toxinotypes V and V-like were found in 1.8% (4/223) of pigs (Table 3).
Table 3.
Toxinotyping results from isolates of swine origina
| Toxinotype (n = 223) | No. (%) from: |
||
|---|---|---|---|
| OH | NC | Total | |
| V | 55 (56) | 107(85) | 162 (73) |
| Nontoxigenic | 25 (26) | 0 | 25 (10) |
| V and Nontoxigenic | 14 (14) | 0 | 14 (6.3) |
| O | 1 (1) | 8 (6) | 9 (4.0) |
| V and O | 2 (2) | 7 (6) | 9 (4.0) |
| V and V-like | 0 | 4 (3) | 4 (1.8) |
This is a comparison at the pig level, with the total number of pigs found positive for C. difficile being 223 (126 from NC and 97 from OH).
Further, we determined the extent of geographic variations of toxinotypes. Toxinotype V was found more commonly in North Carolina (84%; 189/224) than in Ohio (55%; 99/181) (P < 0.001). Swine in North Carolina were also found to carry a significantly higher prevalence of toxinotype O (10%; 23/224) than Ohio (3.3%; 6/181) (P < 0.001). In return, a higher prevalence of nontoxigenic strains of C. difficile (41%; 75/181) was found in Ohio than in North Carolina (2.7%; 6/224) (P < 0.001).
PFGE genotyping.
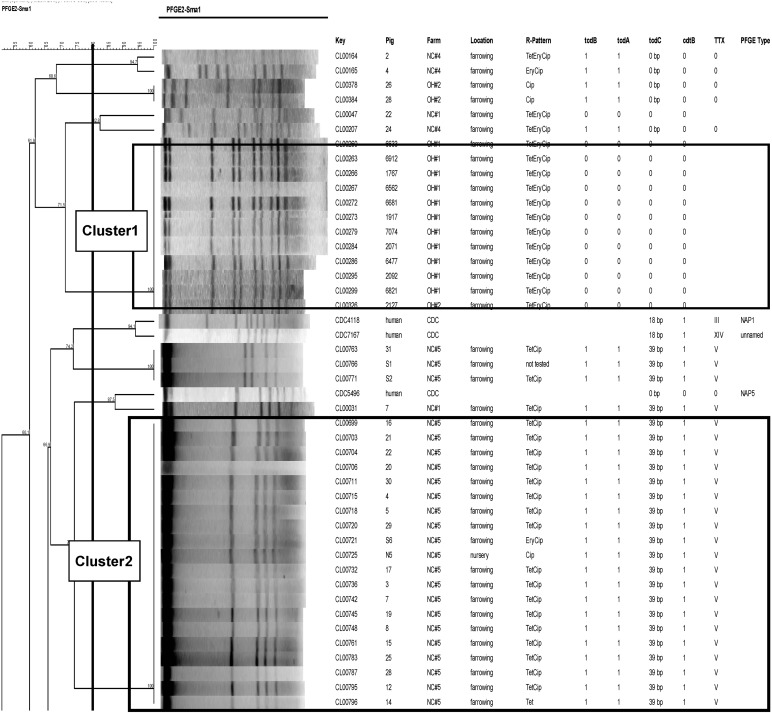
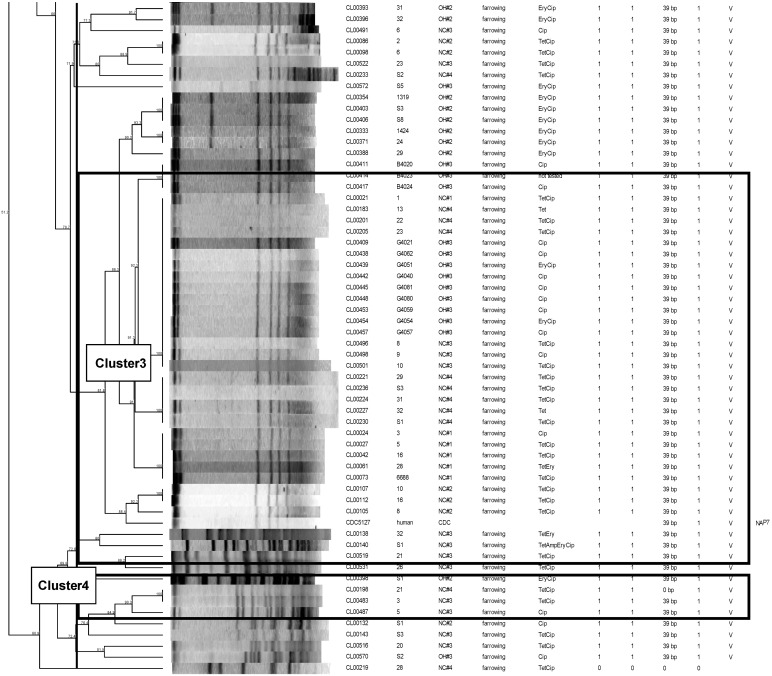
A total of 101 isolates of swine origin were genotyped using PFGE (Fig. 2). The selection was done systematically to represent each state and farm, as well as toxin gene carriage variations noted among farms. Using a breakpoint threshold of 80% similarity, a total of 20 genotypic clusters were identified. A large portion of the isolates of swine origin, 33% (35/105), were found to have 86.3% or more genetic similarity (cluster 3). The isolates in this large cluster were all toxinotype V strains but were from multiple different farms, and they included isolates from both geographic regions (North Carolina and Ohio). A second cluster (cluster 2), containing 20 isolates of swine origin, was found to have 100% similarity. This cluster contained all toxinotype V isolates, all of which were from North Carolina farm 5, and included 11 piglet strains and 1 isolate from the nursery age group. Another cluster of 12 isolates with 100% genetic similarity was also noted (cluster 1). This cluster contained all nontoxigenic strains, all from two farms in Ohio (11 from one farm and 1 from a second farm). Three isolates, all toxinotype V and from one farm in North Carolina, were found to be 88.4% similar to one of the reference strains, the human CDC NAP7 (cluster 4). Overall, isolates were found to cluster within toxinotypes but not geographic origin, although multiple clusters per toxinotype were found.
Fig 2.
PFGE dendrogram showing the extent of clonal relatedness among C. difficile isolates. TTX, toxinotype.
DISCUSSION
Community-associated infections are a rising concern in the epidemiology of C. difficile. This study identified a potential food animal source of such infections. Parallel to the findings of this study, recent investigations reported that as many as 47.6% of litters and 90% of herds are infected with C. difficile (29, 36). In the Netherlands, patients infected with ribotype 078 were younger (67.4 versus 73.5 years) and had community-associated disease more frequently (17.5% versus 6.7%) than patients infected with ribotype 027 (27). Similarly to the findings reported here, previous researchers have found genetic relatedness among strains isolated from humans and pigs, suggesting a possible epidemiological relationship (4, 13).
There is little published data available on the antimicrobial susceptibility of C. difficile isolated from pigs and other food animals (20, 33), and there is even less on antimicrobial resistance genes. Based on this study, the majority of C. difficile isolates (80.5%) have a MIC of >32 μg/ml for ciprofloxacin. This finding is in agreement with previous reports, which have noted that the older fluoroquinolones, such as ciprofloxacin, have moderate to poor activity against C. difficile (7). One recent study found all isolates of swine origin to be sensitive to metronidazole and vancomycin, 98% were sensitive to chloramphenicol, and 90% were sensitive to tetracycline (20). Previous studies have reported that the majority of C. difficile strains with tetracycline resistance have the tetM gene, which is carried on a conjugative transposon (19). The tetW gene has the second largest host range of the tetracycline resistance genes, second only to tetM. In this study, the tetM gene was found in 97% of resistant isolates and the tetW gene was found in 32% of tetracycline-resistant isolates of swine origin. There did not appear to be a combined benefit to carrying both genes, as isolates that carried both tetM and tetW genes had a wide range of MICs within the resistant zone.
The ermB gene has been found to encode macrolide lincosamide and streptogramin resistance and has been shown to result in high levels of resistance (15, 22). This gene was found in 91% of erythromycin-resistant isolates in this study. Of those isolates found to carry this gene, 97% were found to have a MIC of >256 μg/ml, which is consistent with previous reports. This gene appears to play a significant role in high-level erythromycin resistance in C. difficile isolated from swine. A large portion of the isolates that were resistant to both erythromycin and tetracycline were found to carry both the ermB and the tetM genes. A link between these two resistance genes has previously been described in C. difficile isolates of human origin, where they are associated with a Tn916-like element (31). Further studies to characterize these elements in swine and to determine if they are linked would be noteworthy.
Toxin gene results for these swine are similar to results previously published, and all combinations of genes have been previously reported for food animals (20, 23, 33). This study is the first known report of heterogeneity in C. difficile strains carried within individual pigs, although this has been found previously in humans at a rate of approximately 7% (5). The findings clearly imply that the characterization of a single isolate from a subject (i.e., pig) could be misleading, as pigs may carry multiple strains at the same time. Such carriage of multiple strains by one pig could promote the horizontal transfer of resistance genes and toxin genes between strain types.
The major toxinotype, toxinotype V, which was found commonly in the present study, has also been reported previously (1, 2, 24). Toxinotype V-like has been reported recently (8). However, to the best of our knowledge, this is the first study indicating its occurrence among isolates of swine origin. Toxinotypes V and O have also been reported in humans (8, 26). The common occurrence of toxinotype V in the present study underscores a potential concern, as this type has been associated with community-associated disease and increasing prevalence in CDI in humans (6, 21, 26, 27). Further investigation is necessary, as some recent reports have found toxinotype V in asymptomatic people (21), and others found that there was no association between virulence attributes and clinical outcomes (28).
Previous studies have determined that PFGE has a high index of discrimination for C. difficile (12). PFGE results showed many samples clustering within toxinotypes as well as within farms. For example, a large cluster of 100% similar nontoxigenic isolates were mostly from one Ohio farm, although one isolate within that cluster was from a second Ohio farm. All clusters noted were found to carry only one toxinotype, although one toxinotype was found to split into separate clusters within the dendrogram, showing diversity within the toxinotype. This finding was expected, as toxinotyping only describes one small part of the genome and PFGE is descriptive of the total genomic DNA.
In comparisons of swine strains to control CDC isolates, clusters included isolates of swine and human origin. Although this is concerning for public health reasons, further information is needed to determine the epidemiological relationship of these isolates. As mentioned by previous studies which have found highly similar isolates in human and swine, it is possible that strains humans and strains in pigs are of a common source, but these findings do not show a direct causal or epidemiologic relationship among the isolates (4). Further research is needed to determine the potential transmission of swine isolates to humans, either through contaminated food, environmental contamination, or other exposure routes.
ACKNOWLEDGMENTS
This project was financially sponsored by the National Pork Board (grant 07-044).
We also thank Brandi Limbago of the Centers for Disease Control and Prevention for providing reference strains.
Footnotes
Published ahead of print 18 April 2012
REFERENCES
- 1. Avbersek J, et al. 2009. Diversity of Clostridium difficile in pigs and other animals in Slovenia. Anaerobe 15:252–255 [DOI] [PubMed] [Google Scholar]
- 2. Baker AA, Davis E, Rehberger T, Rosener D. 2010. Prevalence and diversity of toxigenic Clostridium perfringens and Clostridium difficile among swine herds in the midwest. Appl. Environ. Microbiol. 76:2961–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bidet P, Barbut F, Lalande V, Burghoffer B, Petit JC. 1999. Development of a new PCR-ribotyping method for Clostridium difficile based on ribosomal RNA gene sequencing. FEMS Microbiol. Lett. 175:261–266 [DOI] [PubMed] [Google Scholar]
- 4. Debast SB, et al. 2009. Clostridium difficile PCR ribotype 078 toxinotype V found in diarrhoeal pigs identical to isolates from affected humans. Environ. Microbiol. 11:505–511 [DOI] [PubMed] [Google Scholar]
- 5. Eyre DW, et al. 2012. Clostridium difficile mixed infection and reinfection. J. Clin. Microbiol. 50:142–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goorhuis A, et al. 2008. Clostridium difficile PCR ribotype 078: an emerging strain in humans and in pigs? J. Clin. Microbiol. 46:1157–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang H, Weintraub A, Fang H, Nord CE. 2009. Antimicrobial resistance in Clostridium difficile. Int. J. Antimicrob. Agents 34:516–522 [DOI] [PubMed] [Google Scholar]
- 8. Jamal WY, et al. 2009. Correlation of multidrug resistance, toxinotypes and PCR ribotypes in Clostridium difficile isolates from Kuwait. J. Chemother. 21:521–526 [DOI] [PubMed] [Google Scholar]
- 9. Jones MA, Hunter D. 1983. Isolation of Clostridium difficile from pigs. Vet. Rec. 112:253. [DOI] [PubMed] [Google Scholar]
- 10. Kato H, Yokoyama T, Arakawa Y. 2005. Typing by sequencing the slpA gene of Clostridium difficile strains causing multiple outbreaks in Japan. J. Med. Microbiol. 54:167–171 [DOI] [PubMed] [Google Scholar]
- 11. Keel MK, Songer JG. 2007. The distribution and density of Clostridium difficile toxin receptors on the intestinal mucosa of neonatal pigs. Vet. Pathol. 44:814–822 [DOI] [PubMed] [Google Scholar]
- 12. Killgore G, et al. 2008. Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein A gene sequence typing. J. Clin. Microbiol. 46:431–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koene MG, et al. 25 August 2011. Clostridium difficile in Dutch animals: their presence, characteristics and similarities with human isolates. Clin. Microbiol. Infect. doi:10.1111/j.1469-0691.2011.03651.x [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 14. Kyne L, Hamel MB, Polavaram R, Kelly CP. 2002. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin. Infect. Dis. 34:346–353 [DOI] [PubMed] [Google Scholar]
- 15. Leclercq R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482–492 [DOI] [PubMed] [Google Scholar]
- 16. Lemee L, et al. 2004. Multiplex PCR targeting tpi (triose phosphate isomerase), tcdA (toxin A), and tcdB (toxin B) genes for toxigenic culture of Clostridium difficile. J. Clin. Microbiol. 42:5710–5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McDonald LC, et al. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433–2441 [DOI] [PubMed] [Google Scholar]
- 18. Mohammadi T, Reesink HW, Pietersz RN, Vandenbroucke-Grauls CM, Savelkoul PH. 2005. Amplified-fragment length polymorphism analysis of Propionibacterium isolates implicated in contamination of blood products. Br. J. Haematol. 131:403–409 [DOI] [PubMed] [Google Scholar]
- 19. Mullany P, et al. 1990. Genetic analysis of a tetracycline resistance element from Clostridium difficile and its conjugal transfer to and from Bacillus subtilis. J. Gen. Microbiol. 136:1343–1349 [DOI] [PubMed] [Google Scholar]
- 20. Norman KN, et al. 2009. Varied prevalence of Clostridium difficile in an integrated swine operation. Anaerobe 15:256–260 [DOI] [PubMed] [Google Scholar]
- 21. Norman KN, et al. 2011. Prevalence and genotypic characteristics of Clostridium difficile in a closed and integrated human and swine population. Appl. Environ. Microbiol. 77:5755–5760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roberts MC, et al. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodriguez-Palacios A, et al. 2006. Clostridium difficile PCR ribotypes in calves, Canada. Emerg. Infect. Dis. 12:1730–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rupnik M, Avesani V, Janc M, von Eichel-Streiber C, Delmee M. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J. Clin. Microbiol. 36:2240–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rupnik M, et al. 1997. Characterization of polymorphisms in the toxin A and B genes of Clostridium difficile. FEMS Microbiol. Lett. 148:197–202 [DOI] [PubMed] [Google Scholar]
- 26. Rupnik M, Widmer A, Zimmermann O, Eckert C, Barbut F. 2008. Clostridium difficile toxinotype V, ribotype 078, in animals and humans. J. Clin. Microbiol. 46:2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 28. Sirard S, Valiquette L, Fortier LC. 2011. Lack of association between clinical outcome of Clostridium difficile infections, strain type, and virulence-associated phenotypes. J. Clin. Microbiol. 49:4040–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Songer JG, Anderson MA. 2006. Clostridium difficile: an important pathogen of food animals. Anaerobe 12:1–4 [DOI] [PubMed] [Google Scholar]
- 30. Songer JG, Post KW, Larson DJ, Jost BH, Glock RD. 2000. Infection of neonatal swine with Clostridium difficile. Swine Health Production 8:185–189 [Google Scholar]
- 31. Spigaglia P, Barbanti F, Mastrantonio P. 2007. Detection of a genetic linkage between genes coding for resistance to tetracycline and erythromycin in Clostridium difficile. Microb. Drug Resist. 13:90–95 [DOI] [PubMed] [Google Scholar]
- 32. Stubbs SL, Brazier JS, O'Neill GL, Duerden BI. 1999. PCR targeted to the 16S–23S rRNA gene intergenic spacer region of Clostridium difficile and construction of a library consisting of 116 different PCR ribotypes. J. Clin. Microbiol. 37:461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thakur S, Putnam M, Fry PR, Abley M, Gebreyes WA. 2010. Prevalence of antimicrobial resistance and association with toxin genes in Clostridium difficile in commercial swine. Am. J. Vet. Res. 71:1189–1194 [DOI] [PubMed] [Google Scholar]
- 34. van den Berg RJ, Schaap I, Templeton KE, Klaassen CH, Kuijper EJ. 2007. Typing and subtyping of Clostridium difficile isolates by using multiple-locus variable-number tandem-repeat analysis. J. Clin. Microbiol. 45:1024–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Waters EH, Orr JP, Clark EG, Schaufele CM. 1998. Typhlocolitis caused by Clostridium difficile in suckling piglets. J. Vet. Diagn. Investig. 10:104–108 [DOI] [PubMed] [Google Scholar]
- 36. Weese JS. 2010. Clostridium difficile in food–innocent bystander or serious threat? Clin. Microbiol. Infect. 16:3–10 [DOI] [PubMed] [Google Scholar]
- 37. Weese JS, Wakeford T, Reid-Smith R, Rousseau J, Friendship R. 2010. Longitudinal investigation of Clostridium difficile shedding in piglets. Anaerobe 16:501–504 [DOI] [PubMed] [Google Scholar]
- 38. Wilson KH, Kennedy MJ, Fekety FR. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15:443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]