Abstract
Wound debridement samples and contralateral (healthy) skin swabs acquired from 26 patients attending a specialist foot clinic were analyzed by differential isolation and eubacterium-specific PCR-denaturing gradient gel electrophoresis (DGGE) in conjunction with DNA sequencing. Thirteen of 26 wounds harbored pathogens according to culture analyses, with Staphylococcus aureus being the most common (13/13). Candida (1/13), pseudomonas (1/13), and streptococcus (7/13) were less prevalent. Contralateral skin was associated with comparatively low densities of bacteria, and overt pathogens were not detected. According to DGGE analyses, all wounds contained significantly greater eubacterial diversity than contralateral skin (P < 0.05), although no significant difference in total eubacterial diversity was detected between wounds from which known pathogens had been isolated and those that were putatively uninfected. DGGE amplicons with homology to Staphylococcus sp. (8/13) and S. aureus (2/13) were detected in putatively infected wound samples, while Staphylococcus sp. amplicons were detected in 11/13 noninfected wounds; S. aureus was not detected in these samples. While a majority of skin-derived DGGE consortial fingerprints could be differentiated from wound profiles through principal component analysis (PCA), a large minority could not. Furthermore, wounds from which pathogens had been isolated could not be distinguished from putatively uninfected wounds on this basis. In conclusion, while chronic wounds generally harbored greater eubacterial diversity than healthy skin, the isolation of known pathogens was not associated with qualitatively distinct consortial profiles or otherwise altered diversity. The data generated support the utility of both culture and DGGE for the microbial characterization of chronic wounds.
INTRODUCTION
Chronic wounds occur in approximately 2% of the population in developed countries (20), where they cause considerable morbidity and mortality (23, 24, 34). In 2008, over 200,000 patients in the United Kingdom were affected, associated with an economic burden of c. £3 billion (34).
Common forms of chronic wounds include pressure sores and diabetic/venous ulcers. Risk factors for the development of diabetic ulcers include conditions associated with neuropathy and/or venous insufficiency, which restrict oxygen supply and impair the transport and integration of leukocytes and macrophages into tissues, leading to ischemic necrosis and subsequent ulceration (2, 12, 43). This creates a portal of entry for bacteria and thus, increases susceptibility to infections. Infection can then lead to further tissue damage and impaired healing by exacerbating the inflammatory state (15, 26, 37).
Clinical laboratory investigations of chronic wound infections commonly rely upon bacterial isolation by culture, which most efficiently detects numerically dominant organisms amenable to growth on laboratory media. While this is a useful and well-established approach for the detection of many common pathogenic bacteria associated with wound infections, it may underestimate microbial diversity (45). Thus, various culture-independent methods have been assessed in a limited number of studies as potential adjuncts or replacements of culture for the microbial characterization of chronic wounds (11, 18, 23). Culture-independent investigations of the bacterial diversity utilizing pyrosequencing (11), PCR-denaturing gradient gel electrophoresis (DGGE) (8), other DNA fingerprinting techniques (39), and quantitative PCR (qPCR) (31) have generally identified a greater range of bacteria than traditional culture techniques, and taxa not previously detected in wounds have been reported. For example, Hill et al. (23) used 16S rRNA gene clone sequence analysis and culture to assess the microbial composition of a chronic venous leg ulcer. Acinetobacter sp. was detected by culture in both swabs and tissue samples; swab samples yielded Proteus sp. and Candida tropicalis, whereas Staphylococcus epidermidis was only isolated from tissue samples. Importantly, however, molecular analysis of the same samples identified clones that were closely related to the cultured organisms, together with species that had not been isolated from the samples (Morganella morganii, Bacteroides ureolyticus, Enterococcus faecalis, and Peptostreptococcus octavius). A study conducted by Davies et al. (8), which assessed the microbiota of healing and nonhealing chronic venous leg ulcers, also reported greater eubacterial diversity according to PCR-DGGE than culture. Of the sequences obtained in the Davies study, 40% were organisms which had not been isolated from the same samples using culture (8). These are among a limited number of reports in regard to the ability of molecular techniques to identify a distinct range of organisms in wounds and also illustrate the importance of sampling techniques and sample sites for the outcome of analyses (23).
While it is clear that culture-independent methods may provide deeper characterization of microbial diversity, the role that taxa thus identified play in infection remains poorly understood. This contrasts with isolation methods, where the pathogenicity of prominent culturable organisms, such as Staphylococcus aureus and Pseudomonas aeruginosa, has been well documented (3, 10, 19, 29).
The current study used clinical diagnostic isolation techniques in combination with eubacterium-specific PCR-DGGE to compare bacterial consortia associated with chronic wound debridement samples with those from healthy skin (17, 21), to assess the utility of PCR-DGGE in addition to culture, and to determine whether samples from which pathogens could be isolated were otherwise compositionally distinct.
MATERIALS AND METHODS
Chemicals and media.
Unless otherwise stated, the chemicals used were obtained from Sigma (Poole, Dorset, United Kingdom). Dehydrated bacteriological medium was obtained from Oxoid (Basingstoke, Hampshire, United Kingdom) and prepared according to instructions supplied by the manufacturer.
Ethical approval.
This study was reviewed by the North Manchester Research Ethics Committee and Central Manchester University Hospital Research and Development department (reference number 09/H1006/41, protocol number 1.0).
Collection of chronic wound tissue and contralateral skin swabs.
A total of 26 wound tissue debridement samples from chronic diabetic foot wounds (defined as distal to the medial and lateral malleoli, with a known duration greater than 4 weeks) and 26 contralateral skin swabs were obtained from patients with diabetic chronic foot wounds attending a specialist foot clinic in Manchester, United Kingdom, between 2 February 2010 and 2 February 2011 as follows. Wound tissue samples were taken from the wound bed and surrounding tissue by the attending clinician using a sterile scalpel and placed in sterile 0.85% (wt/vol; 5 ml) saline for transportation. Skin swabs of an area of intact contralateral skin measuring 40 cm2 were also collected using Dual Amies transport swabs (Duo Transwab; MWE, Wiltshire, United Kingdom). Swabs were moistened with sterile saline and then used to thoroughly scrub skin sites within the contralateral area. All samples were transported to the laboratory and processed within 3 h of collection, as detailed in the following section.
Semiquantitative culture and differential bacteriological identification.
Tissue samples were dissected with a sterile scalpel in the laboratory, weighed, and homogenized using a sterile tissue pulper (VWR, Leicestershire, United Kingdom) in 3 ml of sterile saline. Dual Amies swabs were aseptically separated with sterile scissors, with one swab archived at −80°C for bacterial DNA extraction in 1.5-ml microcentrifuge tubes. The remaining skin swabs and samples of homogenized tissue were streaked for isolation onto four quadrants of Health Protection Agency (HPA)-recommended agars and grown under appropriate atmospheres as shown in Table 1 to isolate clinically relevant organisms according to standardized methods (25). Residual samples of homogenized tissue were archived at −80°C for bacterial DNA extraction. Bacterial species isolated from the four quadrants were reported as scant (<10 colonies), light (first quadrant), moderate (second quadrant), or heavy (third-fourth quadrant) growth according to methods outlined and validated by Angel et al. (1) and Healy and Freedman (22). Bacterial identification was based upon colony morphology, Gram staining, catalase reaction, latex coagulase reaction tests, Lancefield group reaction to identify β-hemolytic streptococci (Prolex Streptococcal grouping latex kits; Pro-Lab Diagnostics, Cheshire, United Kingdom), and subculture onto brilliance UTI medium (25, 40).
Table 1.
Bacteriological agars used in the study
| Medium | Incubation conditions | Target bacterial groupa |
|---|---|---|
| Cysteine lactose electrolyte-deficient agar | 5% CO2, 37°C | Enterobacteriacea and Pseudomonads |
| 5% Horse blood agar with vancomycin plus 5-μg metronidazole disc | Anaerobic, 37°C | Gram-negative anaerobes |
| 5% Horse blood agar with neomycin plus 5-μg metronidazole disc | Anaerobic, 37°C | Gram-positive anaerobes |
| 5% Horse blood agar | 5% CO2, 37°C | Streptococci (Lancefield groups A, C, and G), Pasteurella spp., S. aureus, Vibrio spp., and Aeromonas spp. |
Commonly isolated wound-associated pathogens which may be indicative of infection (25).
Chronic wound tissue and skin swab bacterial community profiling using PCR-DGGE.
DNA was extracted from archived macerated tissue samples and swab samples using a DNeasy blood and tissue kit (Qiagen Ltd., West Sussex, United Kingdom) in accordance with the manufacturer's instructions. The V2-V3 region of the 16S rRNA gene was amplified using the eubacterium-specific primers HDA1 (with additional GC clamp) (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGG GAC TCC TAC GGG AGG CAG CAG T-3′) and HDA2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′) (44). The reaction mixture was as follows: Red Taq DNA polymerase ready mix (25 μl), HDA primers (2 μl of each [5 μM]), NANOpure water (16 μl), and extracted DNA template. The reactions were performed in 0.2-ml DNA-free PCR tubes with a T-Gradient DNA thermal cycler (Biometra, Germany). The thermal amplification program was as follows: 94°C (4 min), followed by 30 thermal cycles of 94°C (30 s), 56°C (30 s), and 68°C (60 s). The final cycle incorporated a 7-min chain elongation step (68°C). Positive and negative controls (5 μl of microbial DNA extracted from saliva and NANOpure water, respectively) were run concurrently with each reaction run.
Polyacrylamide electrophoresis was done using 30% and 60% denaturing concentrations using the DCode Universal Mutation Detection System (Bio-Rad, Hemel Hempstead, United Kingdom) according to the manufacturer's recommendations for perpendicular DGGE. Parallel denaturing gradient gels of 10% acrylamide–bisacrylamide (37:1:5) (Sigma, Poole, Dorset, United Kingdom) were cast with a linear gradient of urea and formamide ranging from 60% at the base to 30% at the top (100% of the denaturants corresponds to 7 M urea and 40% formamide). The gels were left to equilibrate at room temperature overnight in the tank containing 7 liters of 1× Tris-acetate buffer solution. Gel electrophoresis was carried out at 140 V at a constant 60°C for 5.5 h (16, 30). All gels were stained using SYBR Gold stain (Molecular Probes, Leiden, The Netherlands) for 30 min with occasional agitation, after which they were transferred to a UV transilluminator (UVP, Upland, CA), visualized under UV light at 312 nm, and photographed using a Canon D60 digital single lens reflex (DSLR) camera (Canon, Surrey, United Kingdom).
Construction of dendrograms.
Gel images were processed and aligned (with the aid of the positive controls of saliva as internal controls used on each gel) using Adobe Photoshop Elements version 7 and analyzed using the BioNumerics Fingerprint package (Applied Maths, Belgium). Bands were detected automatically and then checked manually. Dendrograms were constructed by cluster comparison using the unweighted pair group method with arithmetic mean (UPGMA) algorithm (28).
Diversity indices were determined for each individual sample and also for each specimen group (i.e., the presence or absence of cultured wound pathogens) and using the Shannon-Weaver index of diversity (H′) with the following equation, where s is the number of species and Pi is the proportion of species in the sample i (13, 16):
The resultant indices were compared between wound and skin samples and between the presence or absence of cultured wound pathogen cohorts by the Mann-Whitney U test performed with SPSS version 16 (SPSS, Chicago, IL).
DGGE band excision and reamplification for sequence identification.
Replicated DGGE bands (visualized on a UV transilluminator), i.e., those which were present across several samples, and unique bands were excised using a sterile scalpel and placed in nuclease-free tubes with 20 μl NANOpure water. The bands were then stored at 4°C for 24 h before being archived at −80°C. Before sequence analysis, the tubes were vortexed for 30 s and then centrifuged (MSE Microcentaur; Sanyo, Loughborough, United Kingdom) for 10 min at 14,462 × g. Extracts (5 μl) were then used as templates for PCR using the protocol outlined above for bacterial community profiling. PCR products derived from excised DGGE bands were purified using a QIAquick PCR purification kit (Qiagen Ltd., West Sussex, United Kingdom) in accordance with the manufacturer's instructions. PCR products were sequenced using the non-GC clamp (reverse) HDA primer. The sequencing reaction was as follows: 94°C (4 min) followed by 25 cycles of 96°C (30 s), 50°C (15 s), and 60°C (4 min). Once chain termination was complete, sequencing was carried out in a Perkin-Elmer ABI 377 sequencer. DNA sequences were compiled using CHROMAS-LITE (Technelysium Pty. Ltd., Australia). Sequence matching was undertaken using the Basic Local Alignment Search Tool bioinformatics program (http://www.ncbi.nlm.nih.gov/blast) to mine the prokaryotic database for matching sequences.
PCA.
To produce graphical representations in which clusters can be differentiated, principal component analysis (PCA) was used. Similarity matrix data derived from UPGMA algorithms of DNA fingerprints of chronic wound tissue and intact-skin swabs were utilized to determine principal component data. Briefly, similarity matrix data of correlated variables were reduced using factor analysis (SPSS; SPSS, Inc., Chicago, IL) in which variances between groups, i.e., the different band position for each sample when compared to another, are maximized to produce three overall uncorrelated variables (principal components). The first principal component accounts for the greatest degree of variance in the overall group, with each succeeding component representing the remaining variances. The resulting principal component data were plotted on a 3-axis scatter plot.
RESULTS
Semiquantitative bacterial counts and identification of bacteria from wound and skin samples.
Data in Tables 2 and 3 show semiquantitative growth data from chronic wound tissue samples and intact-skin swabs, respectively. Tissue samples were grouped based on the isolation or absence of pathogenic wound organisms (1, 22, 25). From the organisms cultured, S. aureus was the only overt pathogen identified using the defined assessment parameters (1, 22, 25). Intact-skin swabs produced comparatively low numbers of skin-associated bacteria, with no sample producing moderate to heavy growth of any bacterial isolate.
Table 2.
Microbial characterization of chronic wounds by semiquantitative culture
| Bacterial groupa | Characteristic in isolateb from patient: |
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | |
| S. aureus | 3+c | 2+d | 2+ | 2+ | 2+c | 3+c | 2+c | 1+c | 1+d | 1+ | 2+d | 1+c | 2+c | |||||||||||||
| CNS | 3+c | 3+c | 3+d | 2+ | 2+c | 2+d | 3+d | 3+c | 2+d | 3+d | 3+ | 3+ | 1+d | 1+d | 3+c | 1+d | 2+d | 3+d | 1+ | 3+d | ||||||
| E. coli | 1+ | 3+ | 3+ | |||||||||||||||||||||||
| Coliform | 2+d | 3+c | 2+ | 1+ | 3+ | 3+c | 3+d | |||||||||||||||||||
| GDS | 1+d | 2+c | 2+c | 1+ | ||||||||||||||||||||||
| GBS | 1+ | 2+ | ||||||||||||||||||||||||
| GGS | 2+c | |||||||||||||||||||||||||
| Corynebacterium spp. | 2+ | 2+ | 2+ | |||||||||||||||||||||||
| Pseudomonas spp. | 1+ | |||||||||||||||||||||||||
| Micrococcus spp. | 2+ | |||||||||||||||||||||||||
| Anaerobes (GPC) | 1+d | |||||||||||||||||||||||||
| Candida spp. | 2+ | |||||||||||||||||||||||||
CNS, coagulase-negative staphylococci; GBS, β-hemolytic streptococci (Lancefield group B); GDS, Enterococcus faecalis (Lancefield group D); GGS, β-hemolytic streptococci (Lancefield group G); GPC, Gram-positive cocci.
Shading indicates isolation of wound pathogens from tissue according to established culture-based criteria. 1+, light growth; 2+, moderate growth; 3+; heavy growth (see Materials and Methods for definitions). Blank cells, none detected.
Genus identified on DGGE sequence analysis from bands derived from the sample or aligned sequenced bands.
Species identified on DGGE sequence analysis from bands derived from the sample or aligned sequenced bands.
Table 3.
Microbial characterization of contralateral skin sites by semiquantitative culture
| Bacterial groupa | Characteristic in isolateb from patient: |
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | |
| CNS | Sd | Sd | Sc | Sd | Sd | S | 1+c | S | Sd | Sd | 1+d | 1+d | 1+d | 1+d | 1+d | 1+d | 1+d | 1+d | 1+c | 1+d | Sc | S | S | |||
| Corynebacterium spp. | S | S | S | 1+ | ||||||||||||||||||||||
| Micrococcus spp. | S | 1+ | ||||||||||||||||||||||||
CNS, coagulase-negative staphylococci. Additional bacterial groups shown in Table 2 were not detected.
Shading indicates skin samples from contralateral sites of patients with wounds whose tissue samples cultured pathogens, defined according to established culture-based criteria. S, scant growth; 1+, light growth (see Materials and Methods for definitions).
Genus identified on DGGE sequence analysis from bands derived from the sample or aligned sequenced bands.
Species identified on DGGE sequence analysis from bands derived from the sample or aligned sequenced bands.
DGGE analysis of the microbial diversity of chronic wound tissue samples and contralateral skin swabs.
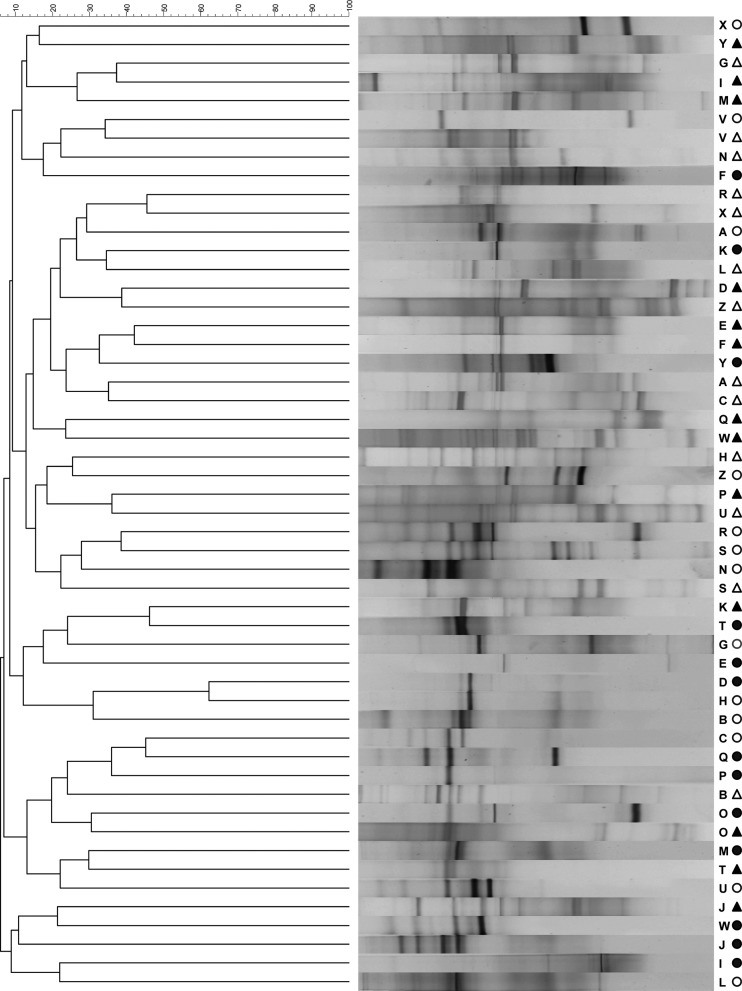
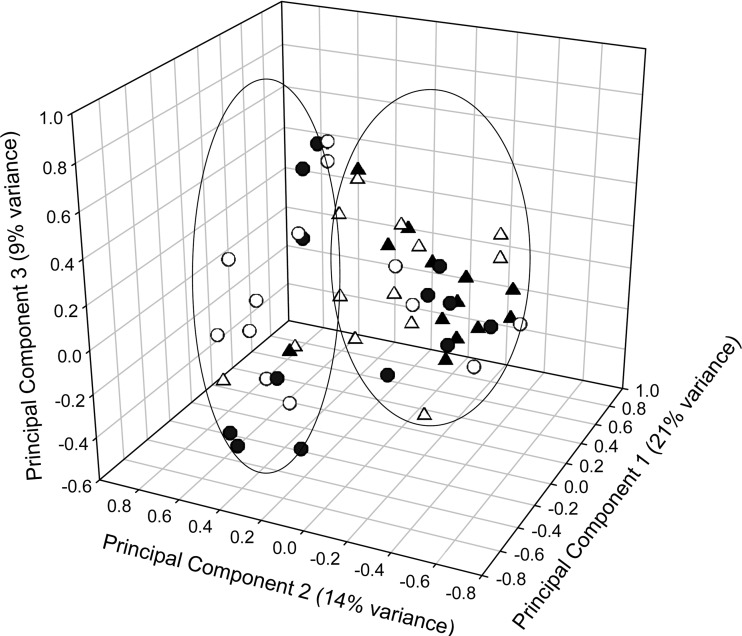
A UPGMA dendrogram was constructed to compare the overall eubacterial DNA fingerprints of wound and skin communities derived from chronic wound tissue samples and contralateral intact-skin swabs. The similarity scores ranged from 10 to 60%, with the average similarity score below 50%, indicating that, generally, skin surface and wounds were colonized with divergent consortial profiles. However, clustering can be seen for a minority of DGGE profiles derived from wound and skin consortia in the results in Fig. 1. This is explored further via principal component analysis of similarity matrix scores (Fig. 2) in which two major clusters are apparent. While one of these represents a combination of skin and wound profiles, the other is composed primarily of skin-derived consortial profiles. Comparisons between diversity indices derived from individual wound and skin samples (Table 4) revealed marked differences in eubacterial diversity between all the wound and skin samples and those where no pathogens were cultured (P < 0.05), although significant differences were not detected between wound and skin cohorts where pathogens were isolated.
Fig 1.
A UPGMA dendrogram for patients A to Z, showing percentage matching of wound DGGE fingerprints. Closed triangles, wound debridement samples from which pathogens were isolated; open triangles, wound debridement samples from which pathogens were not isolated; closed circles, contralateral skin samples from individuals with wounds from which pathogens were isolated; open circles, contralateral skin samples from individuals with wounds from which pathogens were not isolated.
Fig 2.
Principal component analysis of DGGE fingerprints of chronic wound samples and intact-skin swabs (patients A to Z). See legend to Fig. 1 for key to symbols.
Table 4.
Eubacterial diversity indices and proportions of skin amplicons also detected in woundsa
| Patienta | Shannon-Weaver diversity indexb |
Shared ampliconsc | |
|---|---|---|---|
| Wound | Skin | ||
| A | 0.583 | 0.901 | 4/11 |
| B | 0.583 | 0.410 | 0/5 |
| C | 1.166 | 0.328 | 3/4 |
| D | 0.510 | 0.328 | 0/4 |
| E | 0.364 | 0.164 | 0/2 |
| F | 0.219 | 0.819 | 0/10 |
| G | 0.656 | 0.492 | 2/7 |
| H | 0.874 | 0.246 | 2/3 |
| I | 0.656 | 0.410 | 0/5 |
| J | 0.801 | 0.492 | 1/6 |
| K | 0.510 | 0.410 | 0/5 |
| L | 0.583 | 0.655 | 0/8 |
| M | 0.729 | 0.328 | 1/4 |
| N | 0.364 | 0.573 | 0/6 |
| O | 0.364 | 0.328 | 2/4 |
| P | 0.801 | 0.082 | 0/1 |
| Q | 0.219 | 0.246 | 0/3 |
| R | 0.437 | 0.492 | 2/6 |
| S | 0.510 | 0.819 | 3/10 |
| T | 0.146 | 0.328 | 1/4 |
| U | 0.801 | 0.410 | 1/5 |
| V | 0.510 | 0.328 | 2/3 |
| W | 1.239 | 0.410 | 0/5 |
| X | 0.364 | 0.328 | 0/4 |
| Y | 0.583 | 0.492 | 0/6 |
| Z | 0.729 | 0.492 | 1/6 |
Shaded rows indicate wounds which harbored pathogens based on established culture criteria.
Diversity indices were compared between wound and skin and between infected and noninfected cohorts by the Mann-Whitney U test. Significant differences were found between diversity indices of wound and skin samples and between wound and skin samples when grouped into noninfected cohorts (P < 0.05). No significant difference was found between wound and skin when grouped into infected cohorts.
Proportion of bands present in intact-skin DGGE analysis found in chronic wound DGGE analysis.
Comparisons of 16S DNA sequence data between chronic wound tissue samples and contralateral intact-skin swabs for each patient.
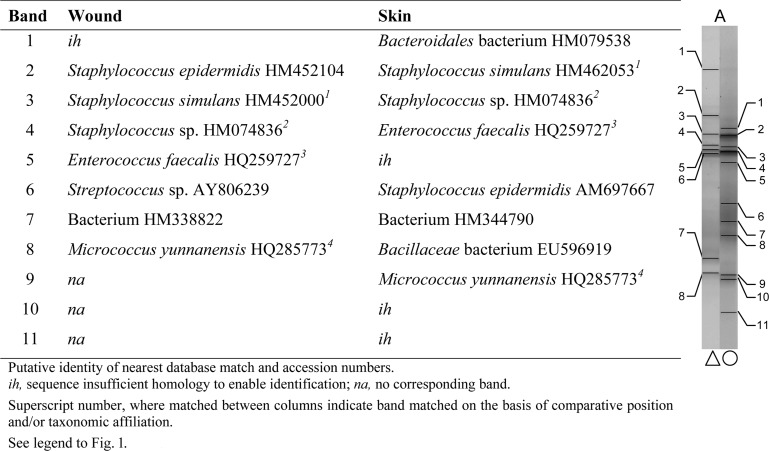
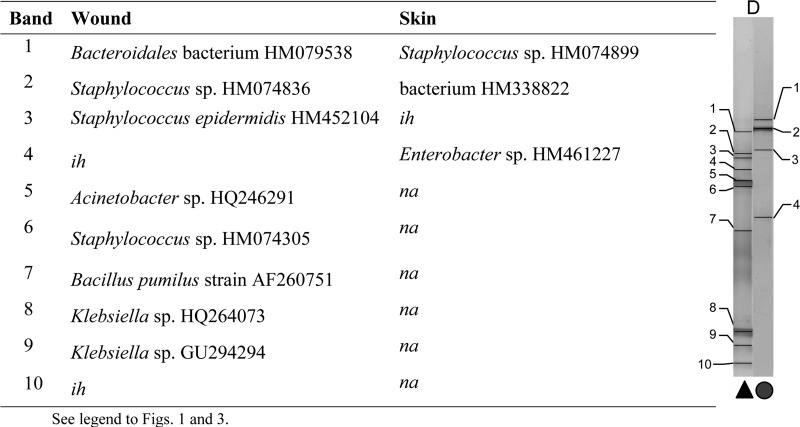
Comparisons were made between bands with matching positions or taxonomic affiliation and sequences across the wound and the contralateral control skin swab for each patient. Examples are given from patients A and G (no pathogens isolated), presented in Fig. 3 and 4, and patients D and I (wound pathogens isolated), presented in Fig. 5 and 6. In general, greater proportions of skin-associated bacteria were detected in contralateral wound sites (two or more correlated bands in 8 samples) where no wound pathogens were cultured than in wounds where pathogens were cultured (no correlated bands in 11 samples).
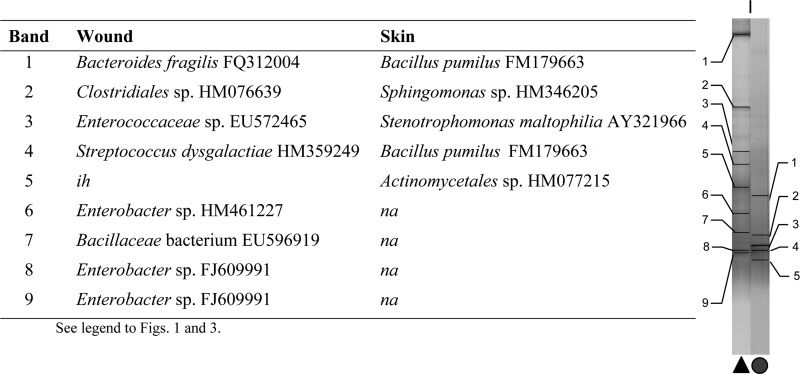
Fig 3.
Characterization of major taxa in wound and skin samples based on dominant PCR amplicons and matched bands derived from DGGE gels (patient A; no pathogens isolated).
Fig 4.
Characterization of major taxa in wound and skin samples based on dominant PCR amplicons and matched bands derived from DGGE gels (patient G; no pathogens isolated).
Fig 5.
Characterization of major taxa in wound and skin samples based on dominant PCR amplicons and matched bands derived from DGGE gels (patient D; pathogens isolated).
Fig 6.
Characterization of major taxa in wound and skin samples based on dominant PCR amplicons and matched bands derived from DGGE gels (patient I; pathogens isolated).
Corroboration of isolation data by DGGE sequence analyses.
DGGE-derived sequence identities were compared to isolation data for each patient and between sample cohorts. Sequence analyses of DGGE amplicons suggested the presence of Staphylococcus sp. in 8/13 and S. aureus in 2/13 wound samples from which S. aureus was cultured. PCR amplicons with homology to coagulase-negative staphylococci were detected in 8/13 and Staphylococcus sp. in 3/13 of the wound samples where no pathogens were isolated (data not shown). For both tissue and skin samples, the most prevalent genera were Staphylococcus and Bacillus; the latter were not detected by culture. Additionally, a greater number of obligate anaerobic organisms were identified in both the skin and tissue isolates by DGGE than by culture. According to PCR-DGGE analyses, of the 22 genera identified in the wound tissue samples and 21 genera in skin swabs, four were unique to the wounds (Klebsiella sp., Abiotrophia sp., Escherichia coli, and Peptoniphilus sp.) and three were unique to the intact-skin swabs (Kocuria rhizophila, Morexellaceae sp., and Rhodocyclaceae sp.).
DISCUSSION
The etiology of chronic wounds commonly relates to underlying pathologies; initiation is often associated with primary tissue damage, which creates a portal of entry for microorganisms in which complex microbial communities can develop and infection may occur (15, 26, 37). The progression and chronicity of wounds can be correlated with infection which, from a microbiological perspective, is dependent upon the types of bacteria present and their relationship with the host immune responses. Wound infection is commonly defined according to the presence of pathogens and colonization densities exceeding ≥106 organisms per gram of tissue and the development of significant tissue damage and clinical signs of infection (32, 36, 37).
There has been considerable speculation regarding the potential etiological role of the taxonomically diverse microbial populations which commonly develop within chronic wounds (14, 27, 37, 38) and the role that culture-independent techniques could play in research and diagnosis (42). The current study investigated the relationship between the isolation of overt pathogens from wounds, as defined by established culture methods, and eubacterial diversity, assessed using PCR-DGGE. These techniques were also used to compare the bacterial composition of wounds and contralateral healthy skin sites. Semiquantitative culture, often used clinically as an indication of infection severity (22, 25, 35, 41), was adopted, providing a means by which culturable pathogenic organisms could be detected and a relevant comparator for PCR-DGGE.
Of the 26 chronic wound tissue samples investigated, common wound pathogens could be isolated from 13; S. aureus was detected in all of these, in addition to enteric species and various representatives of the regional skin microbiota. Additionally, 2 of 13 samples (E and M) were also associated with Candida sp., Pseudomonas sp., and hemolytic group G streptococci. Coliforms were isolated from seven samples, of which six also harbored skin and/or enteric flora, indicating putative colonization or contamination. A moderate growth of coliforms was noted for patient sample Z, which was not considered to be associated with infection based on clinical details. A scant culture of E. coli was also isolated from sample A. While E. coli and other coliforms may be considered pathogenic and thus significant in specific wound cultures, such as gastrointestinal surgical wound sites, in the context of the current study, sample A was classified as not harboring wound pathogens due to the low numbers of E. coli isolated and the wound type from which it was isolated.
Comparisons of the bacteriological composition of wounds from which pathogens had or had not been isolated using eubacterium-specific PCR-DGGE detected S. aureus in 2/13 and Staphylococcus sp. in 8/13 of wound samples associated with pathogens. Several sequences obtained using DGGE analysis could not be identified to species level, an observation which is commonly associated with this method. Isolation methods detected a more limited range of taxa than DGGE from both wound tissue and skin swabs. Additionally, a greater number of obligate anaerobic organisms were identified in both the skin and tissue isolates using the PCR-based technique than the culture, despite the fact that validated anaerobic isolation methods were used. Interestingly, a higher proportion of those taxa present on the contralateral (control) skin sites occurred in wounds which did not harbor overt pathogens than in those from which pathogens had been isolated. Within the 13 tissue samples where pathogens had been isolated, 11 produced no bands (i.e., PCR amplicons) which matched to the contralateral skin swab profile, whereas all tissue samples where no pathogens had been isolated were associated with least one or more matching skin swab bands. Previous diversity profiling studies of the human skin microbiota by Gao et al. (17) and Grice et al. (21) suggest that while there is comparatively little interindividual compositional similarity in healthy skin microbiotas, high levels of conservation between the contralateral skin sites in individuals can be demonstrated (17, 21). On this basis, contralateral intact-skin samples may provide an insight into the normal microbiota of the site and thus, the microbial composition of skin prior to wounding.
Primary colonizers of wounds are reportedly often members of the autochthonous skin microbiota due to their proximity to the tissue injury (4, 5). However, delayed healing may enable adventitious bacteria to proliferate and thus compete against autochthonous species. Additionally, since the microbiota of healthy skin is likely to be water and nutrient limited (6), hydration and nutrient availability may have a marked influence on the microbial composition of wounds. The restricted nutrient and moisture content of healthy skin may limit the proliferation of fastidious organisms and thus select for Gram-positive bacteria, such as coagulase-negative staphylococci, corynebacteria, and propionibacteria (6, 7). In contrast, the comparatively nutrient-rich environment of a wound may facilitate the growth of a wider variety of organisms, including S. aureus, P. aeruginosa, streptococci, enterobacteriaceae, and other facultative anaerobic species (6, 9). The transition from healthy skin to colonized/infected wound may therefore be associated with the clonal expansion of bacteria not normally associated with health. In most cases, wounds were colonized by more diverse microbial communities than healthy skin, but the overall eubacterial diversity of wounds which harbored pathogens and those from which no pathogens were isolated did not differ significantly. It could be agued that this observation highlights the utility of culture as a straightforward means of detecting wound pathogens. However, it also indicates the need for further investigations of potential associations between microbial profiles and clinical outcome, because the contribution of altered microbial colonization to causality remains poorly understood, in contrast to the involvement of overtly pathogenic species, which are more readily detectable by culture (25). Previous analysis of chronic wound diversity by Dowd et al. (11) using pyrosequencing, DGGE, and rRNA gene shotgun sequencing identified several genera and species not isolated upon culture, a method which in general failed to identify the primary bacterial populations of the wounds tested (11). Importantly, sequence analyses of DGGE amplicons within the current study indicated that wounds which did not harbor pathogens were associated with a greater proportion of normal skin bacteria than were infected wounds.
This study provides a cross-sectional assessment of the bacterial diversity of wounds which were initially assessed according to the presence or absence of culture-defined pathogenic species. It also provides an opportunity to compare isolation methods to a culture-independent DNA profiling technique. In some cases, pathogens detected by isolation were not detected by PCR-DGGE, and conversely, bacterial diversity indicated on DGGE gels was not readily detectable by culture. Both DGGE and culture have distinct characteristics: culture is relatively simple to implement, cost effective, and can detect numerically dominant culturable pathogens. It is, however, limited by the culturability of target bacteria. Conversely, DGGE can be used to analyze complex communities while facilitating the identification of unculturable organisms which may only represent as little as 1% of the total bacterial population (33). Due to the length of the DNA sequences that DGGE produces, however, the taxonomic associations made may not be categorical.
Analysis of the microbiotas of wound and contralateral skin sites indicated that, in general, healthy skin-associated organisms were underrepresented in wounds from which pathogens were cultured but that significant alterations in total eubacterial diversity were not detected. Therefore, while DGGE is a useful tool for the reproducible culture-independent profiling of bacterial consortia, data presented in the current investigation also highlight the utility of culture. On this basis, the two analytical approaches are complementary.
ACKNOWLEDGMENTS
This work was partially supported by grants from the BBSRC and ConvaTec.
Footnotes
Published ahead of print 2 May 2012
REFERENCES
- 1. Angel DE, Lloyd P, Carville K, Santamaria N. 2011. The clinical efficacy of two semi-quantitative wound-swabbing techniques in identifying the causative organism(s) in infected cutaneous wounds. Int. Wound J. 8:176–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bass MJ, Phillips LG. 2007. Pressure sores. Curr. Probl. Surg. 44:101–143 [DOI] [PubMed] [Google Scholar]
- 3. Bourke WJ, O'Connor CM, FitzGerald MX, McDonnell TJ. 1994. Pseudomonas aeruginosa exotoxin A induces pulmonary endothelial cytotoxicity: protection by dibutyryl-cAMP. Eur. Respir. J. 7:1754–1758 [DOI] [PubMed] [Google Scholar]
- 4. Bowler PG, Duerden BI, Armstrong DG. 2001. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 14:244–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brook I, Frazier EH. 1990. Aerobic and anaerobic bacteriology of wounds and cutaneous abscesses. Arch. Surg. 125:1445–1451 [DOI] [PubMed] [Google Scholar]
- 6. Chiller K, Selkin B, Murakawa G. 2001. Skin microflora and bacterial infections of the skin. J. Invest. Dermatol. Symp. Proc. 6:170–174 [DOI] [PubMed] [Google Scholar]
- 7. Cogen AL, Nizet V, Gallo RL. 2008. Skin microbiota: a source of disease or defence? Br. J. Dermatol. 158:442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Davies CE, et al. 2004. Use of 16S ribosomal DNA PCR and denaturing gradient gel electrophoresis for analysis of the microfloras of healing and nonhealing chronic venous leg ulcers. J. Clin. Microbiol. 42:3549–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davies CE, et al. 2001. Use of molecular techniques to study microbial diversity in the skin: chronic wounds reevaluated. Wound Repair Regen. 9:332–340 [DOI] [PubMed] [Google Scholar]
- 10. Dinges MM, Orwin PM, Schlievert PM. 2000. Exotoxins of Staphylococcus aureus. Clin. Microbiol. Rev. 13:16–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dowd S, et al. 2008. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 8:43 doi:10.1186/1471-2180-8-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eaglstein WH, Falanga V. 1997. Chronic wounds. Surg. Clin. North Am. 77:689–700 [DOI] [PubMed] [Google Scholar]
- 13. Edwards ML, Lilley AK, Timms-Wilson TH, Thompson IP, Cooper I. 2001. Characterisation of the culturable heterotrophic bacterial community in a small eutrophic lake (Priest Pot). FEMS Microbiol. Ecol. 35:295–304 [DOI] [PubMed] [Google Scholar]
- 14. Edwards RH, Keith G. 2004. Bacteria and wound healing. Skin Soft Tissue Infect. 17:91–96 [DOI] [PubMed] [Google Scholar]
- 15. Fazli M, et al. 2009. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 47:4084–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gafan GP, et al. 2005. Statistical analyses of complex denaturing gradient gel electrophoresis profiles. J. Clin. Microbiol. 43:3971–3978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gao Z, Tseng C-h, Pei Z, Blaser MJ. 2007. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. U. S. A. 104:2927–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gontcharova V, Youn E, Sun Y, Wolcott RD, Dowd SE. 2010. A comparison of bacterial composition in diabetic ulcers and contralateral intact skin. Open Microbiol. J. 4:8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gorbet MB, Sefton MV. 2005. Endotoxin: the uninvited guest. Biomaterials 26:6811–6817 [DOI] [PubMed] [Google Scholar]
- 20. Gottrup F. 2004. A specialized wound-healing center concept: importance of a multidisciplinary department structure and surgical treatment facilities in the treatment of chronic wounds. Am. J. Surg. 187:38S–43S [DOI] [PubMed] [Google Scholar]
- 21. Grice EA, et al. 2008. A diversity profile of the human skin microbiota. Genome Res. 18:1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Healy B, Freedman A. 2006. Infections. BMJ 332(7545):838–841 doi:10.1136/bmj.332.7545.838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hill KE, et al. 2003. Molecular analysis of the microflora in chronic venous leg ulceration. J. Med. Microbiol. 52:365–369 [DOI] [PubMed] [Google Scholar]
- 24. Howell-Jones RS, et al. 2005. A review of the microbiology, antibiotic usage and resistance in chronic skin wounds. J. Antimicrob. Chemother. 55:143–149 [DOI] [PubMed] [Google Scholar]
- 25. HPA 2009. Investigation of skin and superficial and non-surgical wound swabs. UK standards for microbiology investigations, vol 11 V11 Issue 5 Health Protection Agency, London, United Kingdom [Google Scholar]
- 26. Jones SG, Edwards R, Thomas DW. 2004. Inflammation and wound healing: the role of bacteria in the immuno-regulation of wound healing. Int. J. Low. Extrem. Wounds 3:201–208 [DOI] [PubMed] [Google Scholar]
- 27. Kirketerp-Moller K, et al. 2008. The distribution, organization and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 46:2717–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ledder RG, et al. 2006. Individual microflora beget unique oral microcosms. J. Appl. Microbiol. 100:1123–1131 [DOI] [PubMed] [Google Scholar]
- 29. Lyczak JB, Cannon CL, Pier GB. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051–1060 [DOI] [PubMed] [Google Scholar]
- 30. McBain AJ, et al. 2003. Growth and molecular characterization of dental plaque microcosms. J. Appl. Microbiol. 94:655–664 [DOI] [PubMed] [Google Scholar]
- 31. Melendez JH, et al. 2010. Real-time PCR assays compared to culture-based approaches for identification of aerobic bacteria in chronic wounds. Clin Microbiol. Infect. 16:1762–1769 [DOI] [PubMed] [Google Scholar]
- 32. Murphy RC, Robson MC, Heggers JP, Kadowaki M. 1986. The effect of microbial contamination on musculocutaneous and random flaps. J. Surg. Res. 41:75–80 [DOI] [PubMed] [Google Scholar]
- 33. Muyzer G, de Waal EC, Uitterlinden AG. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Posnett J, Franks PJ. 2008. The burden of chronic wounds in the UK. Nurs. Times 104:44–45 [PubMed] [Google Scholar]
- 35. RNAO 2005. Assessment and management of foot ulcers for people with diabetes. Registered Nurses' Association of Ontario, Toronto, Ontario, Canada [Google Scholar]
- 36. Robson MC. 1979. Infection in the surgical patient: an imbalance in the normal equilibrium. Clin. Plast. Surg. 6:493–503 [PubMed] [Google Scholar]
- 37. Robson MC. 1997. Wound infection: a failure of wound healing caused by an imbalance of bacteria. Surg. Clin. North Am. 77:637–650 [DOI] [PubMed] [Google Scholar]
- 38. Robson MC, Stenberg BD, Heggers JP. 1990. Wound healing alterations caused by infection. Clin. Plast. Surg. 17:485–492 [PubMed] [Google Scholar]
- 39. Singh SK, et al. 2009. Detecting aerobic bacterial diversity in patients with diabetic foot wounds using ERIC-PCR: a preliminary communication. Int. J. Low. Extrem. Wounds 8:203–208 [DOI] [PubMed] [Google Scholar]
- 40. Steel KJ, Barrow GI, Feltham RKA, Cowan ST. 1993. Cowan and Steel's manual for the identification of medical bacteria. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 41. Stone ND, et al. 2008. Importance of bacterial burden among methicillin-resistant Staphylococcus aureus carriers in a long-term care facility. Infect. Control Hosp. Epidemiol. 29:143–148 [DOI] [PubMed] [Google Scholar]
- 42. Thomsen TR, et al. 2008. The bacteriology of chronic venous leg ulcer examined by culture-independent molecular methods. Wound Rep. Regen. 18:38–49 [DOI] [PubMed] [Google Scholar]
- 43. Vasconez LO, Schneider WJ, Jurkiewicz MJ. 1977. Pressure sores. Curr. Probl. Surg. 14:1–62 [DOI] [PubMed] [Google Scholar]
- 44. Walter J, et al. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilson MJ, Weightman AJ, Wade WG. 1997. Applications of molecular ecology in the characterization of uncultured microorganisms associated with human disease. Rev. Med. Microbiol. 8:91–102 [Google Scholar]