Abstract
In this article, the first isolation of Mycobacterium kyorinense specimens in Brazil is described. M. kyorinense is a recently identified species, with a few strains reported only in Japan. The Brazilian isolates were initially identified as Mycobacterium celatum by PCR restriction enzyme pattern analysis (PRA) with hsp65. However, biochemical tests indicated the same profile of M. kyorinense and distinguished them from M. celatum and Mycobacterium branderi. The sequencing of the hsp65, rpoB, and 16S rRNA genes allowed the accurate identification of isolates as M. kyorinense.
TEXT
Mycobacterium kyorinense is a nonpigmented slowly growing mycobacterium that was first isolated from sputum of a patient with pneumonia. The species was described in 2009, and to date a few strains have been isolated, all from patients in Japan (5, 9). All patients were immunocompetent, and most of them had a history of pulmonary disease. M. kyorinense was considered a possible cause of clinically significant respiratory disease. The sequences of 16S rRNA genes, hsp65, and rpoB were identical in the strains tested but different from those of the two phylogenetically most related species, Mycobacterium celatum and Mycobacterium branderi (1, 4). Biochemical tests can also distinguish M. kyorinense from M. celatum and M. branderi (1, 4, 5, 8).
The aim of this work is to report the first isolation of M. kyorinense in Brazil and, to our knowledge, its first isolation outside Japan and to characterize its hsp65 restriction profile by PCR restriction enzyme pattern analysis (PRA).
This study includes two pulmonary specimens (HF1629 and HF1836) that were initially identified as M. celatum in 2007 by PRA with hsp65 in the National Reference Laboratory for Tuberculosis of the Centro de Referência Professor Hélio Fraga/ENSP/FIOCRUZ. These isolates were obtained from two separate expectorated sputum samples from a patient from Rio de Janeiro State, Brazil, according to microbiological diagnostic criteria for nontuberculous mycobacterial lung disease according to the American Thoracic Society guidelines (3).
At admission to the health authority, the patient presented with complete destruction of the left lung by fibrotic lesions, which over the course of the disease extended to the right lung. Treatment with rifampin, isoniazid, pyrazinamide, and ethambutol temporarily improved the patient′s condition, but after less than 1 year, symptoms reappeared, leading to death.
The DNA extraction protocol used in this work was developed in our laboratory and is being introduced here. Briefly, one loopful of bacteria grown on Lowenstein-Jensen medium was mixed in 300 μl of 1% Triton X-100 and 100 μl of acid-washed glass beads with a diameter of 106 μm. The cells were shaken vigorously in a Vortex mixer for 1 min and incubated at room temperature for 8 min. After this incubation, 50 μl of mixture was placed in a microtube with 100 μl of lysis buffer (equal amounts of 15% Chelex 100, 0.5% Tween 20, and Tris-EDTA). The mixture was heated to 100°C for 20 min and shaken vigorously in a vortex mixer for 1 min, followed by centrifugation at 13,000 × g for 8 min. The supernatant was then transferred to a microtube and stored at 4°C.
Amplification and digestion of hsp65 fragments were performed as described by Telenti et al. (7), and the resulting restriction digest pattern was compared to those in the PRAsite database (http://app.chuv.ch/prasite/index.html). The full rpoB gene was amplified using 20 primers (see Table S1 in the supplemental material), and the 16S rRNA gene was amplified using the MicroSEQ full-gene 16S rRNA gene PCR kit (Life Technologies). All sequences obtained were compared on the basis of similarity to those in the GenBank database by using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The hsp65 441-bp fragment, the almost complete 16S rRNA gene, and the full rpoB gene sequences of strain HF1629 (the older of the two isolates) were used to construct a concatenated neighbor-joining tree with MEGA version 5 (2, 6).
Cleavage of the hsp65 PCR product obtained from strains HF1629 and HF1836 with BstEII produced fragments of about 230, 130, and 80 bp, and with HaeIII, the digestion resulted in fragments of about 125, 105, 80, 40, 35 and 25 bp (see Fig. S1 in the supplemental material). The closest match in the PRAsite database was the pattern of M. celatum type 2 (BstEII, 235, 130, and 85 bp; HaeIII, 130, 105, and 80 bp) with a score of 9, but very distinct from M. celatum type 1 (BstEII, 235 and 210 bp; HaeIII, 130, 80, and 60 bp). Thus, the isolates were initially identified as M. celatum type 2, because the M. kyorinense profile was not available in PRAsite. To remedy this, the novel profile has been deposited in the PRAsite database.
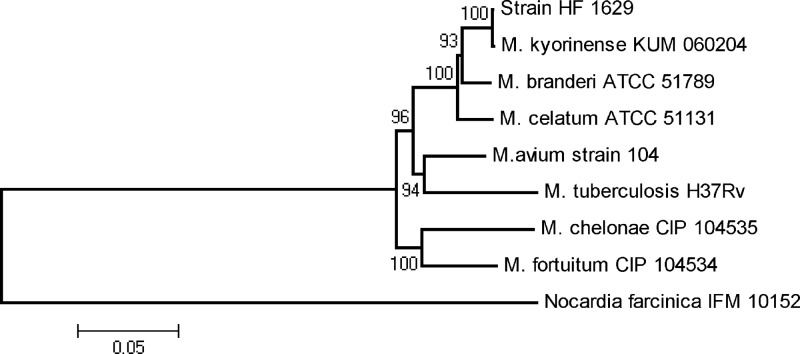
Sequences of hsp65, rpoB, and 16S rRNA genes from HF1629 and HF1836 were obtained to further explore the identity of the isolates. First, the sequences of these three genes were identical in HF1629 and HF1836, indicating that those two isolates belong to the same species. A BLAST search showed that the hsp65 441-bp fragment is 100% identical to the M. kyorinense type strain KUM060204 sequence. Alignment of the full rpoB sequence from the Japanese and Brazilian isolates (unpublished data) showed them to be 100% identical. The 16S rRNA gene sequence displaying the highest identity score was that of M. kyorinense type strain KUM060204, with 4 mismatches (99.73%). M. celatum type strain ATCC 51131 showed 98.28% identity (25 mismatches in 1,450 bases), and M. branderi type strain ATCC 51789 showed 97.9% identity (31 mismatches in 1,475 bases). The concatenated phylogenetic tree revealed that the HF1629 strain was tightly clustered (with 100% bootstrap value) to the type strain of M. kyorinense, but distinct from those of M. celatum and M. branderi or all other sequences tested (Fig. 1).
Fig 1.
Phylogenetic tree computed from the concatenation of 16S rRNA gene, rpoB, and hsp65 sequences by the neighbor-joining method and with Kimura's two-parameter model as the substitution model. The significance of branches is indicated by bootstrap values calculated for 1,000 replicates. Nocardia farcinica sequence was used as an outgroup.
To complete the identification of HF1629 and HF1836 as M. kyorinense, we carried out microbiological and biochemical tests. Isolates presented slow growth, and pigment was not observed. The biochemical results agree with those obtained for M. kyorinense type strain KUM060204 (see Table S2 in the supplemental material). The arylsulfatase activity and tellurite reduction assays distinguish M. kyorinense from M. celatum, and the heat-stable catalase test allows M. kyorinense to be distinguished from M. branderi (1, 4, 5, 8).
The results of molecular, microbiological, and biochemical analyses of two Brazilian mycobacterial isolates from a patient with lung disease have allowed their identification as M. kyorinense. Comparison of genetic similarity and phylogenetic analysis show that these isolates are very closely related to M. kyorinense, with only a slight difference from the 16S rRNA gene sequence of this species' type strain. Thus, all results confirm the identification of our isolates as M. kyorinense.
As we used the American Thoracic Society (ATS) microbiologic criteria that are important to make a diagnosis of nontuberculous mycobacterial (NTM) lung disease and there are M. kyorinense disease reports previously reported in Japan, we speculate that M. kyorinense infection is also the cause of the pulmonary deterioration in the Brazilian patient.
Nucleotide sequence accession numbers.
The 16S rRNA, hsp65, and rpoB gene sequences from the oldest isolate of this study have been deposited in GenBank for strain HF1629 under accession no. JN634643, JN974461, and JQ744020, respectively. All accession numbers of sequences retrieved from GenBank are available in the supplemental material.
Supplementary Material
ACKNOWLEDGMENT
We thank the Health Ministry of Brazil for financial support.
Footnotes
Published ahead of print 18 April 2012
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Butler WR, et al. 1993. Mycobacterium celatum sp. nov. Int. J. Syst. Bacteriol. 43:539–548 [DOI] [PubMed] [Google Scholar]
- 2. Devulder G, Pérouse de Montclos M, Flandrois JP. 2005. A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55:293–302 [DOI] [PubMed] [Google Scholar]
- 3. Griffith DE, et al. 2007. ATS Mycobacterial Diseases Subcommittee; American Thoracic Society; Infectious Disease Society of America. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 175:367–416 [DOI] [PubMed] [Google Scholar]
- 4. Koukila-Kähkölä P, et al. 1995. Mycobacterium branderi sp. nov., a new potential human pathogen. Int. J. Syst. Bacteriol. 45:549–553 [DOI] [PubMed] [Google Scholar]
- 5. Okazaki M, et al. 2009. Mycobacterium kyorinense sp. nov., a novel, slow-growing species, related to Mycobacterium celatum, isolated from human clinical specimens Int. J. Syst. Evol. Microbiol. 59:1336–1341 [DOI] [PubMed] [Google Scholar]
- 6. Tamura K, et al. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Telenti A, et al. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vincent V, Gutierres MC. 2007. Mycobacterium: laboratory characteristics of slowly growing mycobacteria, p 573–588 In Murray PR, Baron EJ, Landry ML, Jorgensen JH, Pfaller MA. (ed), Manual of clinical microbiology, 9th ed American Society for Microbiology, Washington, DC [Google Scholar]
- 9. Wada H, Yamamoto M, Okazaki M, Watanabe T, Goto H. 2009. Isolation of Mycobacterium kyorinense in a patient with respiratory failure. Ann. Intern. Med. 150:568–570 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.