Abstract
Hepatitis C virus (HCV) infection typically leads to antibody response within weeks after primary infection. Here, we describe the case of a child infected with HCV by mother-to-child transmission who remained persistently seronegative despite the presence of high levels of circulating HCV RNA.
CASE REPORT
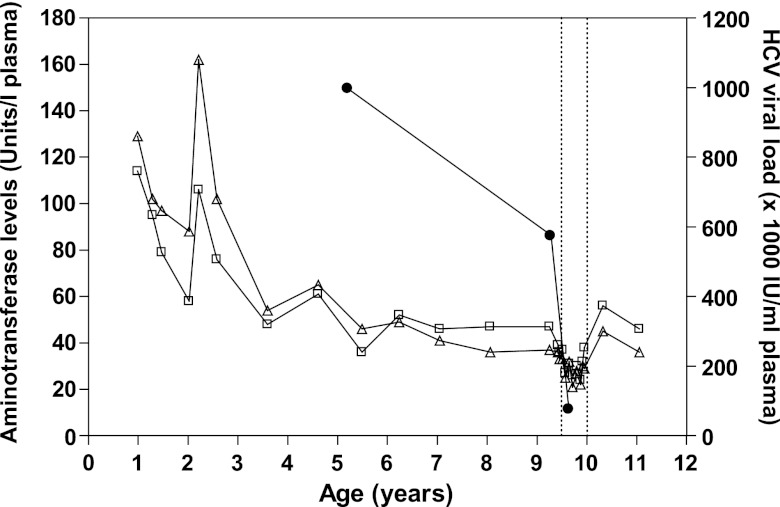
The female subject of this case report was born in 1999 by cesarean section to a woman infected with hepatitis C virus (HCV) genotype 1a. There was no evidence of human immunodeficiency virus type 1 (HIV-1) infection in the child or the mother, a former injection drug user who stopped using drugs in 1998 (Table 1). The subject was diagnosed with hepatitis C following two positive HCV RNA PCR tests (Cobas Amplicor HCV, version 2.0; Roche Diagnostics, Montreal, Quebec, Canada) at 10 and 12 months of age. The subject also tested positive for HCV RNA at 1.47, 2.04, 2.58, 3.59, 4.61, 5.49, 6.24, 7.04, 8.06, 9.25, 10.3, and 11.5 years of age using the same technique. In sharp contrast, testing for anti-HCV antibodies using anti-HCV microparticle enzyme immunoassay (MEIA) (AxSYM HCV, version 3.0; Abbott Laboratories, Saint-Laurent, Quebec, Canada) was repeatedly negative (tested 10 times between 1.47 and 10.3 years of age). At age 11.1 years, testing using EIA-1/EIA-2 (Monolisa anti-HCV plus [Bio-Rad Laboratories, Hercules, FL] and HCV ELISA [enzyme-linked immunosorbent assay] test system, version 3.0 [Ortho-Clinical Diagnostics, Raritan, NJ]) yielded indeterminate results. However, samples from the same time point tested negative using the INNO-LIA HCV score assay (Innogenetics, Ghent, Belgium). Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were regularly monitored between the ages of 12 months and 11 years, and HCV plasma RNA levels were quantified in 3 separate instances over this time period using the COBAS Amplicor HCV monitor assay, version 2.0 (Roche Diagnostics) (Fig. 1). At age 9.42 years, persistently elevated (2 times normal) ALT and AST levels justified the introduction of combination treatment with PEGylated interferon alpha-2b (50 μg per week) and ribavirin (600 mg per day) (Pegetron; Schering-Plough, Kenilworth, NJ). The HCV plasma viral load was 997,000 IU per ml, 576,000 IU per ml, and 76,000 IU per ml at age 5.20 years, 9.28 years, and 9.64 years, respectively. Treatment was stopped after 30 weeks at age 10.0 years in the absence of sustained virological response. Liver histology scored according to Metavir before the beginning of treatment showed stage 1 fibrosis and absence of inflammation.
Table 1.
Clinical characteristics of study subjects
| Characteristic | Manner or value |
|---|---|
| Possible modes of HCV infection (mother) | Injection drug use, blood transfusion |
| Gestational age (wk) | 35.7 |
| Birth wt (g) | 2,560 |
| Mode of delivery | Cesarean sectiona |
| Apgar score (1 min) | 5 |
| Apgar score (5 min) | 7 |
| Maternal HCV viral load at delivery | Not determinedb |
| Maternal HCV viral load in 2010 | 1.0 × 107 UI per ml plasmac |
Membrane rupture >24 h; cephalo-pelvic disproportion.
HCV RNA positive at delivery using qualitative PCR (Cobas Amplicor HCV, version 2.0).
Measured using COBAS Amplicor HCV Monitor assay version 2.0.
Fig 1.
Clinical parameters measured in the study subject. Alanine aminotransferase (ALT; open squares), aspartate aminotransferase (AST; open triangles), and plasma HCV RNA levels (closed circles) were measured as described in the text. Vertical dashed lines indicate the beginning and end of PEGylated interferon alpha-2b and ribavirin treatment.
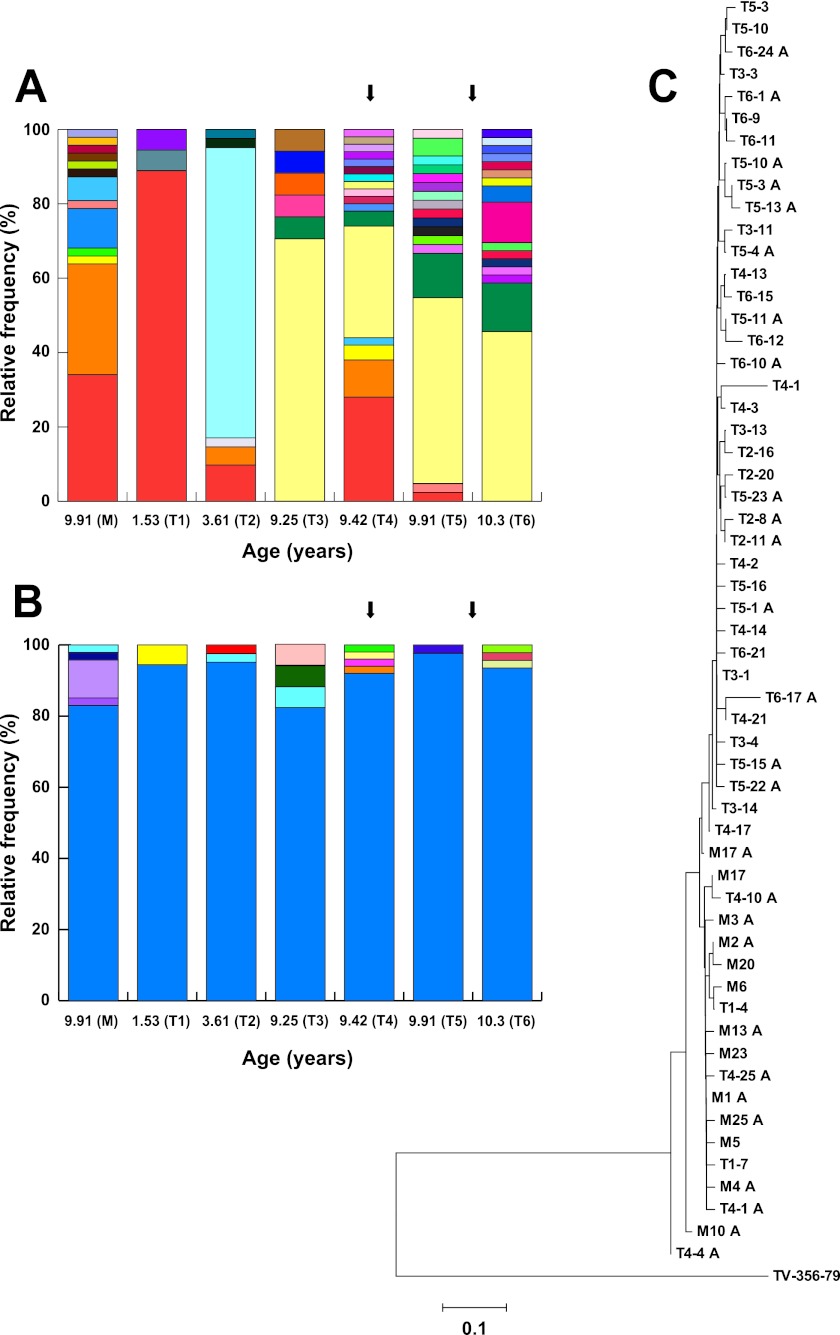
Six serum samples obtained from the subject at ages 1.53 years, 3.61 years, 9.25 years, 9.42 years (initiation of treatment), 9.91 years, and 10.3 years (T1 to T6) and one sample obtained from her mother when the subject was 9.91 years of age (M) were used for analysis of quasispecies evolution based on the sequence of the first hypervariable region of the HCV E2 envelope protein (HVR1), which is subjected to selective pressures exerted by the host's humoral and cellular immune responses. The evolution of the variant spectrum or viral quasispecies profile based on this particular region of the HCV genome was associated with the spontaneous clearance of HCV infection and the transition from acute to chronic hepatitis C (11), with the response to interferon treatment (13), with the extent of liver injury (19), and with the onset of immune responses in children infected by mother-to-child transmission (22). Briefly, HCV RNA was extracted from serum using a QIAamp viral RNA minikit (Qiagen, Mississauga, Ontario, Canada) and then reverse transcribed and amplified based on previously published protocols (4, 11, 13, 28). The PCR products were visualized on 1.7% agarose gels, purified using an agarose gel extraction kit (Feldan-bio, Quebec, Quebec, Canada), and cloned in pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA). A total of 261 independent recombinants (17 to 50 recombinants per time point) were randomly selected and sequenced unidirectionally using an ABI 3730xl automated DNA sequencer (Applied Biosystems, Foster City, CA) at the Centre de Recherche du Centre Hospitalier de l'Université Laval, Québec, Quebec, Canada. Sequences were visualized and edited using Chromas, version 1.45 (Technelysium, Southport, Australia), and were aligned using ClustalX, version 1.81 (34) (GenBank accession numbers JQ808144 to JQ808404). Based on nucleic acid sequences, the quasispecies profile revealed a main variant (variant M1A) representing 88.9% of the total number of clones isolated from the subject at the first time point (1.53 years of age) (Fig. 2A). This variant was identified in the subject at 3.61, 9.42, and 9.91 years of age, indicative of long-term longitudinal persistence. In addition, variant M1A was also found in the mother, in whom it represented 34.0% of the sequences obtained (Fig. 2A). This suggests that this particular viral variant was transmitted from the mother to the child during pregnancy or childbirth. Four additional maternal variants (M2A, M3A, M13A, and M17A) were also found in the child. M2A was found at 3.61 (4.88%) and 9.42 years (10.0%), M3A (4%) and M17A (2%) were found at 9.42 years (4.00% and 2.00%, respectively), and M13A was found at 9.91 years (2.38%), also suggestive of vertical transmission (Fig. 2A). In contrast, neither of two other minor variants found in the child at 1.5 years of age (T14, 5.56%, and T17, 5.56%) nor any other variant subsequently identified in the child were observed in the mother. Examination of the quasispecies profile based on amino acid sequences revealed extensive longitudinal conservation and persistence of the HVR1 peptide, with >82.4% representation at all time points in the child and 83.0% representation in the mother (i.e., the large majority of mutations were synonymous nucleotide substitutions) (Fig. 2B). To examine the evolution of viral sequences during the follow-up period, phylogenies based on Kimura 2-parameter distance matrices and the neighbor-joining method were constructed using MEGA4 (33). The resulting tree exhibits low bootstrap values and extensive cross-grouping of variants isolated from different time points, indicative of a lack of directional divergence of nucleotide sequences (Fig. 2C). Sequences originating from the mother (M) did not cluster separately from sequences derived from the subject. Finally, the mean frequencies of nonsynonymous substitutions per nonsynonymous site (dN) and of synonymous substitutions per synonymous site (dS) and the dN/dS ratio were computed based on HVR1 sequences according to the Nei-Gojobori method with the Jukes-Cantor correction (26). The resulting value of dN/dS (0.22) was compatible with nondirectional genetic drift and an absence of selective pressure on HVR1, even during interferon treatment. Taken together, these results are consistent with impairment or absence of host immune responses directed against HCV.
Fig 2.
Longitudinal analysis of HCV quasispecies structure in the study subject and her mother. Each color represents an identical HCV HVR1 variant in longitudinal analysis. A total of 261 sequences were analyzed as described in the text. M, mother, n = 47; T1, 1.53 years, n = 18; T2, 3.61 years, n = 41; T3, 9.25 years, n = 17; T4, 9.42 years, n = 50; T5, 9.91 years, n = 42; T6, 10.3 years, n = 46. (A) Analysis of the variant profile based on HVR1 nucleic acid sequences. (B) Analysis of the variant profile based on HVR1 amino acid sequences. Arrows indicate the beginning and end of PEGylated interferon alpha-2b and ribavirin treatment. (C) Phylogenetic analysis of HVR1 nucleic acid sequences. The TV-356-79 reference sequence (GenBank accession no. AY385797 [4]) was used as the outgroup.
No apparent abnormalities in IgG profiles were noted in this particular subject, and the presence of antibody responses against childhood vaccinations (hepatitis A virus [HAV], hepatitis B virus [HBV], tetanus, mumps, and rubella) was confirmed (data not shown). To characterize HCV-specific cell-mediated immune responses, enzyme-linked immunospot assay (ELISpot) was performed using the subject's peripheral blood mononuclear cells (PBMC) obtained at age 10.3 years and pools of overlapping peptides representing the core protein, NS3, and NS5B of HCV-1a (H77; BEI Resources, American Type Culture Collection, Manassas VA), as previously described (8). Purified recombinant core protein and NS3 (Feldan Bio) were also used as antigens. The CEF (cytomegalovirus, Epstein-Barr virus, and influenza virus) peptide pool (NIH AIDS Research and Reference Reagents Program) and phorbol myristate acetate (PMA)-ionomycin were used as positive controls. Spots were counted using an S4 immunospot analyzer (CTL Technologies Ltd., Shaker Heights, OH), and data were expressed as spot-forming units (SFU) per 106 PBMC. The subject responded very well to simulation by PMA-ionomycin but only exhibited comparatively modest responses, below the generally accepted threshold of positivity (50 SFU per 106 PBMC). These responses were directed against NS3 pool 5 (5 SFU per 106 PBMC), NS3 pool 6 (5 SFU per106 PBMC), NS5B pool 10 (5 SFU per 106 PBMC), purified core protein (5 SFU per 106 PBMC), and purified NS3 protein (10 SFU per 106 PBMC) (data not shown).
The presence of multiple risk factors for HCV infection and elevated HCV viral load in the mother have been associated with mother-to-child transmission in a number of studies (5, 25) but not in others (7; reviewed in reference 20). In addition, elective caesarean section was not shown to be effective to prevent vertical HCV transmission (10). In infants with chronic hepatitis C, HVR1 variability increases with the development of immunity, consistent with mounting selective pressures being exerted on HCV gene products by host humoral and cell-mediated immune responses (12, 22). In the case reported herein, no HCV-specific antibody responses were detected, and comparatively few cells producing gamma interferon (IFN-γ) in response to HCV antigens were found in peripheral blood. These observations are compatible with reduced selective pressure on HVR1 and with variations in HVR1 sequences being largely limited to synonymous substitutions caused by the high error rate of the viral RNA-dependent RNA polymerase. Seronegative HCV infection in immunocompetent subjects has been reported previously, usually in the context of early reports where viral population analysis was not performed (9, 21, 31, 32). However, to our knowledge, it was never observed in the context of mother-to-child transmission. In contrast, seronegative HCV infection was reported in patients with X-linked agammaglobulinemia (XLA) or common variable immunodeficiency (CVID) (16), two diseases that have a strong impact on humoral immunity. Viral variants isolated from XLA patients showed low variability in HVR1 (1 or 2 amino acid residues of the 27 amino acid residues that comprise HVR1), similar to the subject examined in the present study (i.e., the variant spectrum remains relatively similar between time points spread out over a long period of time). However, variants isolated from CVID patients typically present with comparatively more amino acid substitutions (i.e., ∼10) (16). Seronegative HCV infection and stable variant spectra were also observed in subjects with hepatitis C who are coinfected with HIV-1, a T-lymphotropic virus that is well known to interfere with the initiation and maintenance of humoral and cell-mediated immune responses (3, 4, 6, 17, 24, 28). George et al. (17) reported that CD4+ T cell counts were higher in HCV- and HIV-coinfected subjects who were HCV seropositive than in coinfected subjects who were HCV seronegative despite detectable HCV RNA levels, suggestive of a relationship between the absence of HCV-specific antibody production and HIV-associated impairment of cell-mediated immunity.
HCV quasispeciation profiles are strongly influenced by HIV-1 infection (23, 29). In the presence of low CD4+ T cell counts and/or suboptimal antiretroviral therapy (ART), the HCV variant spectrum becomes homogenous as a result of reduced immune selective pressure exerted on HCV antigens. Conversely, when HIV-1 replication is suppressed by ART, the variant spectrum rapidly evolves and diversifies (4, 28). In the present case, CVID, HIV infection, and XLA (subject is female) were all excluded as possible causes of persistent HCV seronegativity and absence of selective pressure. Regardless, these diseases are associated with defective immune responses against entire groups of pathogens and not specific ones, such as what seems to be observed in this case. Also, subjects who suffer from XLA do not usually exhibit major defects in T cell-mediated immune responses (14). Alternative explanations could include (i) unique biological properties of the HCV strain infecting this particular subject, leading to an unusually efficient viral avoidance of host immune responses, (ii) impairment of the generation of antibody responses by HCV via direct infection of T cell and/or B cell subsets (1, 2, 15, 30), and (iii) the presence in the child of a putative genetic polymorphism or defect that would affect humoral immune responses directed against HCV in particular. Finally, since the subject could have been infected in utero, this could represent a possible case of neonatal immune tolerance specific to HCV leading to the absence of significant generation of long-term humoral and cell-mediated immune responses as a result of T cell and B cell anergy and/or clonal deletion. Immune tolerance to HCV was recently hypothesized as a possible cause for low HVR1 diversity in immunocompetent children who were infected with HCV during the perinatal period (18). This situation might be analogous to that observed in hepatitis B virus infection, where mother-to-child transmission leads to neonatal tolerance, long-term seronegativity, and chronic viral persistence (27).
ACKNOWLEDGMENTS
The work was supported by grants from the Canadian Institutes of Health Research (CIHR) to H.S. (grant HOP-98831) and N.H.S. (grant MOP-106468) and by an infrastructure grant from le Réseau SIDA et Maladies Infectieuses of le Fonds de Recherche du Québec-Santé (FRQS). M.Q.-V. was the recipient of the Gabriel-Marquis Scholarship, Université de Montréal, and was supported by la Fondation de l'Hôpital Sainte-Justine, FRQS, and CIHR. N.H.S. is the recipient of a Chercheur Boursier (Junior-2) salary award from FRQS.
Footnotes
Published ahead of print 25 April 2012
REFERENCES
- 1. Azzari C, et al. 2008. Higher risk of hepatitis C virus perinatal transmission from drug user mothers is mediated by peripheral blood mononuclear cell infection. J. Med. Virol. 80:65–71 [DOI] [PubMed] [Google Scholar]
- 2. Azzari C, et al. 2000. Vertical transmission of HCV is related to maternal peripheral blood mononuclear cell infection. Blood 96:2045–2048 [PubMed] [Google Scholar]
- 3. Bharti AR, et al. 2011. Clinical variables identify seronegative HCV co-infection in HIV-infected individuals. J. Clin. Virol. 52:328–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Canobio S, et al. 2004. Differing patterns of liver disease progression and hepatitis C virus (HCV) quasispecies evolution in children vertically coinfected with HCV and human immunodeficiency virus type 1. J. Clin. Microbiol. 42:4365–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ceci O, et al. 2001. Vertical transmission of hepatitis C virus in a cohort of 2,447 HIV-seronegative pregnant women: a 24-month prospective study. J. Pediatr. Gastroenterol. Nutr. 33:570–575 [DOI] [PubMed] [Google Scholar]
- 6. Chamie G, et al. 2007. Factors associated with seronegative chronic hepatitis C virus infection in HIV infection. Clin. Infect. Dis. 44:577–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conte D, Fraquelli M, Prati D, Colucci A, Minola E. 2000. Prevalence and clinical course of chronic hepatitis C virus (HCV) infection and rate of HCV vertical transmission in a cohort of 15,250 pregnant women. Hepatology 31:751–755 [DOI] [PubMed] [Google Scholar]
- 8. Drouin C, Lamarche S, Bruneau J, Soudeyns H, Shoukry NH. 2010. Cell-mediated immune responses directed against hepatitis C virus (HCV) alternate reading frame protein (ARFP) are undetectable during acute infection. J. Clin. Virol. 47:102–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Durand F, Beauplet A, Marcellin P. 2000. Evidence of hepatitis C virus viremia without detectable antibody to hepatitis C virus in a blood donor. Ann. Intern. Med. 133:74–75 [DOI] [PubMed] [Google Scholar]
- 10. European Paediatric Hepatitis C Virus Network 2005. A significant sex—but not elective cesarean section—effect on mother-to-child transmission of hepatitis C virus infection. J. Infect. Dis. 192:1872–1879 [DOI] [PubMed] [Google Scholar]
- 11. Farci P, et al. 2000. The outcome of acute hepatitis C predicted by the evolution of viral quasispecies. Science 288:339–344 [DOI] [PubMed] [Google Scholar]
- 12. Farci P, et al. 2006. Evolution of hepatitis C viral quasispecies and hepatic injury in perinatally infected children followed prospectively. Proc. Natl. Acad. Sci. U. S. A. 103:8475–8480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farci P, et al. 2002. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc. Natl. Acad. Sci. U. S. A. 99:3081–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fried AJ, Bonilla FA. 2009. Pathogenesis, diagnosis, and management of primary antibody deficiencies and infections. Clin. Microbiol. Rev. 22:396–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gabrielli A, et al. 1994. Active hepatitis C virus infection in bone marrow and peripheral blood mononuclear cells from patients with mixed cryoglobulinaemia. Clin. Exp. Immunol. 97:87–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaud U, Langer B, Petropoulou T, Thomas HC, Karayiannis P. 2003. Changes in hypervariable region 1 of the envelope 2 glycoprotein of hepatitis C virus in children and adults with humoral immune defects. J. Med. Virol. 69:350–356 [DOI] [PubMed] [Google Scholar]
- 17. George SL, et al. 2002. Hepatitis C virus viremia in HIV-infected individuals with negative HCV antibody tests. J. Acquir. Immune Def. Syndr. 31:154–162 [DOI] [PubMed] [Google Scholar]
- 18. Gismondi MI, et al. 2009. Evolution of hepatitis C virus hypervariable region 1 in immunocompetent children born to HCV-infected mothers. J. Viral Hepat. 16:332–339 [DOI] [PubMed] [Google Scholar]
- 19. Honda M, et al. 1994. Degree of diversity of hepatitis C virus quasispecies and progression of liver disease. Hepatology 20:1144–1151 [PubMed] [Google Scholar]
- 20. Indolfi G, Resti M. 2009. Perinatal transmission of hepatitis C virus infection. J. Med. Virol. 81:836–843 [DOI] [PubMed] [Google Scholar]
- 21. Luengrojanakul P, et al. 1994. Hepatitis C virus infection in patients with chronic liver disease or chronic renal failure and blood donors in Thailand. J. Med. Virol. 44:287–292 [DOI] [PubMed] [Google Scholar]
- 22. Manzin A, et al. 2000. Dominant role of host selective pressure in driving hepatitis C virus evolution in perinatal infection. J. Virol. 74:4327–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mao Q, et al. 2001. Human immunodeficiency virus seroconversion and evolution of hepatitis C virus quasispecies. J. Virol. 75:3259–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mazza C, et al. 1998. Prospective study of mother-to-infant transmission of hepatitis C virus (HCV) infection. Study Group for Vertical Transmission. J. Med. Virol. 54:12–19 [PubMed] [Google Scholar]
- 25. Murakami J, et al. 12 January 2012. Risk factors for mother-to-child transmission of hepatitis C virus: maternal high viral load and fetal exposure in the birth canal. Hepatol. Res. [Epub ahead of print]. doi:10.1111/j.1872-034X.2012.00968.x [DOI] [PubMed] [Google Scholar]
- 26. Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418–426 [DOI] [PubMed] [Google Scholar]
- 27. Ni YH. 2011. Natural history of hepatitis B virus infection: pediatric perspective. J. Gastroenterol. 46:1–8 [DOI] [PubMed] [Google Scholar]
- 28. Quesnel-Vallières M, Lemay M, Lapointe N, Martin SR, Soudeyns H. 2008. HCV quasispecies evolution during treatment with interferon alfa-2b and ribavirin in two children coinfected with HCV and HIV-1. J. Clin. Virol. 43:236–240 [DOI] [PubMed] [Google Scholar]
- 29. Roque-Afonso AM, et al. 2002. Influence of CD4 cell counts on the genetic heterogeneity of hepatitis C virus in patients coinfected with human immunodeficiency virus. J. Infect. Dis. 185:728–733 [DOI] [PubMed] [Google Scholar]
- 30. Sarhan MA, Pham TN, Chen AY, Michalak TI. 2012. Hepatitis C virus infection of human T lymphocytes is mediated by CD5. J. Virol. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmidt WN, et al. 1997. Surreptitious hepatitis C virus (HCV) infection detected in the majority of patients with cryptogenic chronic hepatitis and negative HCV antibody tests. J. Infect. Dis. 176:27–33 [DOI] [PubMed] [Google Scholar]
- 32. Silini E, et al. 1993. Virological features of hepatitis C virus infection in hemodialysis patients. J. Clin. Microbiol. 31:2913–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tamura K, Dudley J, Nei M, Kumar SS. 2007. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 34. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]