Abstract
Using in situ hybridization, we describe, for the first time, the profiles of expression of serotonin receptors (Htr/5-HTR) along the dorsal–ventral axis of mouse hippocampus. cRNA probes for most Htrs, excluding Htr6, were used. All hippocampal subregions and the entorhinal cortex cells providing input into the hippocampus were examined. The study shows that some, but not all, Htrs are expressed in the cells of the hippocampal circuitry. At both the subfield and the cell type levels, a somewhat overlapping pattern is observed. Four serotonin receptors, Htr1a, Htr2a, Htr2c and Htr7, display an expression pattern that changes along the dorsal–ventral axis of the hippocampus. Given the proposed functional differentiation of the hippocampus along its long axis, with the dorsal pole more involved in cognitive functions and the ventral pole more involved in mood and anxiety, our results suggest that serotonin receptors enriched in the ventral pole probably contribute to mood- and anxiety-related behaviours.
Keywords: serotonin, hippocampus, antidepressant
1. Introduction
Serotonin (5-HT) is an ancient, evolutionarily conserved molecule, synthesized in the brain stem, capable of binding to 14 or more distinct receptors scattered throughout the brain. Major classes of serotonin receptors (Htrs) diverged approximately 750 Ma and are found in a wide diversity of species ranging from single-cell eukaryotes to man [1]. It was first proposed in the 1950s that distinct classes of receptors existed when it was found that the effects of 5-HT could be blocked in part by morphine and in part by dibenzyline [1,2]. Historically, the receptors have been visualized in tissue using radiolabelled pharmacological tools. In the 1970s, initial studies used [3H]5-HT, [3H] lysergic acid diethylamide (LSD) and various other radioligands to identify distinct classes of receptors [1,3] and by the mid-1980s it was proposed that at least three families of Htrs existed: Htr1, Htr2 and Htr3 [1,4]. This classification was solely based on assumed functional segregations, as radioligand binding, which suggested diversity within the Htr1 family, was still not convincing to many in the field [1]. With the increasing use of radioligand binding, distinct subtypes were described in the Htr1 family (named at the time Htr1a, 1b, 1c, 1d and 1e). Further functional studies suggested that Htr1c was more similar to the Htr2 family, suggesting diversity of Htr2 subtypes as well [1]. Another receptor, found in the late 1980s in the gastrointestinal tract, heart and brain, was named Htr4 [1,5].
Htr1a was cloned in the late 1980s [6,7], and in the following years most known and several previously unknown Htr receptors (named at the time Htr1e, 1f, 2f, 5a, 5b, 6 and 7) were cloned based on sequence homology [1]. In the early 1990s, the Serotonin Club Receptor Nomenclature Committee proposed a new nomenclature system, based on operational, structural and transductional information, giving us the current identity of 14 distinct receptors: Htr1a, 1b, 1d, 1e, 1f, 2a, 2b, 2c, 3, 4, 5a, 5b, 6 and 7 [1,8]. It has also been demonstrated that the Htr3 receptor consists of multiple subunits (referred to as Htr3a and Htr3b) in heteromeric (Htr3a/3b) or homomeric (Htr3a) combinations.
Since then, generation of mutant mice has helped in revealing functional role(s) of several Htrs. Altered behaviour has been observed in genetic knockouts for several different Htrs. For example, Htr1b-deficient mice display enhanced aggressive behaviour [9]. Htr1a-deficient mice have increased anxiety [10], whereas Htr2a-deficient mice display decreased anxiety [11]. Htr5a-deficient mice display increased locomotor activity, but no changes in anxiety behaviour [12]. Htr4 knockout mice display attenuated feeding and locomotor activity in response to stress [13].
The current knowledge of Htr expression patterns in the brain is somewhat limited. Although very informative for gross expression patterns, labelling the intact tissue with radioligands fails to provide a cellular level of resolution. Furthermore, there are conflicting results in the literature with regard to expression patterns, in part due to the fact that several of the readily available antibodies for serotonin receptors are suboptimal for immunostaining. For example, several Htr1a antibodies that are widely used label brain tissue from Htr1a knockout mice similarly to wild-type littermates (R. Hen 2000, unpublished data). In order to provide a framework to better understand how serotonin may modulate behaviour, we sought to determine the expression pattern of all Htrs at the level of neuronal cell type. Here, we chose to focus on Htr expression in cell types throughout the hippocampus, a brain structure required for normal anxiety behaviour [14,15] and also thought to be very important for the behavioural response to selective serotonin reuptake inhibitors (SSRIs) [16], a class of widely used antidepressants that function by increasing serotonin neurotransmission. We describe for the first time Htr expression profiles along the dorsal–ventral axis of the hippocampus at the cellular level, using in situ hybridization (ISH) with cRNA probes for most Htrs, excluding only Htr6.
2. Methods
(a). Animals
All mice were adult C57BL/6 males. For all probes, sections from at least five distinct mice were used. The procedures described herein were conducted in accordance with National Institutes of Health regulations and approved by the Institutional Animal Care and Use Committees of Columbia University and the New York State Psychiatric Institute.
(b). Tissue preparation
Mice were deeply anaesthetized by ketamine (120 mg kg−1) plus xylazine (8 mg kg−1) and perfused with 4 per cent paraformaldehyde (PFA) solution in phosphate buffered saline (PBS). Brains were removed and post-fixed overnight by the same fixative. Then the brains were cryoprotected overnight in diethylpyrocarbonate (DEPC)-treated PBS containing 20 per cent sucrose, frozen in methylbutane cooled by dry ice and cut on a cryostat at 20 µm thickness.
(c). In situ hybridization
The sequences for all probes used in this manuscript are listed in table 1. For ISH, we used a modification of methods previously described [17]. Briefly, sections were treated with 4 per cent PFA in PBS for 20 min, followed by washing twice with PBS for 5 min and treatment with 40 µg ml−1 of Proteinase K (Roche) for 30 min at room temperature. Following this, the sections were washed with PBS and fixed with 4 per cent PFA in PBS for 15 min to inactivate proteinase. After acetylation with 0.25 per cent acetic anhydride (Sigma) in 1 per cent triethanolamine (Sigma) solution for 10 min, prehybridization was carried out for 5 h at room temperature in hybridization buffer, consisting of 50 per cent formamide (Roche), 5×SSC (saline sodium citrate buffer), 5×Denhardts (Sigma), 0.25 mg ml−1 yeast tRNA (Ambion) and 0.4 mg ml−1 Salmon Sperm DNA (Stratagene). After removing the prehybridization buffer, hybridization buffer containing digoxigenin (DIG)-labelled cRNA probe was added, and hybridization was performed at 60°C overnight. After hybridization, sections were washed with 5×SSC at 60°C for 5 min, 2×SSC at 60°C for 5 min, 0.2×SSC/50 per cent formamide at 60°C for 30 min, and 0.2×SSC at room temperature for 10 min. After a stringent wash, they were then incubated with blocking buffer (1% blocking reagent; Roche) for 60 min, followed by incubation with alkaline phosphatase-conjugated anti-DIG antibody (1 : 5000 dilution; Roche) for 90 min at room temperature. After unbound antibody was removed with two 30 min washes in MABT buffer (100 mM maleic acid, 150 mM NaCl, 0.1% Tween-20, pH 7.5), the sections were incubated with freshly prepared nitroblue tetrazolium chloride/5-bromo-4-chloro-3-indolylphosphate p-toluidine salt (NBT/BCIP) colour substrate (Roche) for up to 16 h at room temperature, after which the reaction was stopped by immersion into PBS. After ISH staining, the sections were counterstained with Nuclear Fast Red (Vectastain).
Table 1.
List of probe position of Htrs and marker genes. Alphanumerals correspond to NCBI accession number. Probe position can be referred to the length of cDNA and the position of coding region
| gene | probe | coding region | length of cDNA | accession no. |
|---|---|---|---|---|
| Htr1a | 2114..4089 | 493..1758 | 4356 | NM_008308 |
| Htr1b | 1..1161 | 1..1161 | 1161 | NM_010482 |
| Htr1d | 4..2275 | 972..2096 | 2908 | NM_008309 |
| Htr1f | 65..1610 | 380..1480 | 2578 | NM_008310 |
| Htr2a | 973..2787 | 1094..2509 | 2972 | NM_172812 |
| Htr2b | 63..2073 | 328..1767 | 2073 | NM_008311 |
| Htr2c | 2516..3444 | 691..2070 | 4751 | NM_008312 |
| Htr3a | 1239..2017 | 61..1524 | 2035 | NM_013561 |
| Htr3b | 24..2029 | 207..1520 | 2420 | NM_020274 |
| Htr4 | 214..1289 | 214..1380 | 4667 | NM_008313 |
| Htr5a | 363..1340 | 503..1576 | 5576 | NM_008314 |
| Htr5b | 140..1513 | 312..1424 | 2061 | NM_010483 |
| Htr7 | 30..1478 | 108..1454 | 3095 | NM_008315 |
| GAD65 | 603..1414 | 337–2094 | 5625 | NM_008078 |
| GAD67 | 281..821 | 234–2015 | 3231 | NM_008077 |
| calretinin | 12..1460 | 70..885 | 1462 | NM_007586 |
(d). Double fluorescent in situ hybridization
For double fluorescent ISH (FISH) between serotonin receptors and other genes, sections were incubated with DIG-labelled serotonin receptor cRNA probe and fluorescein isothiocyanate (FITC)-labelled marker gene probe. After a stringent wash, sections were incubated with peroxidase-conjugated anti-DIG antibody (1 : 1000; Roche) and labelled with Cy3 by using the tyramide signal amplification (TSA) system (PerkinElmer, USA). Followed by quenching with 1 per cent H2O2, sections were incubated with peroxidase-conjugated anti-FITC antibody (1 : 1500; Roche) and labelled with FITC by the TSA system.
(e). Sections
Sections were cut in a variety of orientations to allow for visualization of either the whole hippocampus (coronal) or the dorsal–ventral axis (sagittal and horizontal). In addition to §3, the electronic supplementary material figure S1 provides a complete overview of which section type was used in each figure, as well as a cartoon of the dorsal–ventral axis.
(f). Image acquisition
Bright field images were taken with an Axioplan-2 upright microscope (Zeiss, Thornwood, NY). Fluorescent images were taken by a Zeiss Axioplan-2 microscope equipped with a charge-coupled device (CCD) camera.
(g). Probe generation
For generating probes, the plasmids containing mouse cDNA were used as listed in the table. DIG- or FITC-labelled cRNA probes were transcribed by appropriate RNA polymerase. A mixture of glutamine acid decarboxylase (GAD) 65 and 67 cRNA probes were used to mark gamma-aminobutyric acid (GABA)ergic inhibitory neurons, and a calretinin cRNA probe was used to mark mossy cells.
3. Results
(a). Htr mRNA expression in the mouse hippocampus
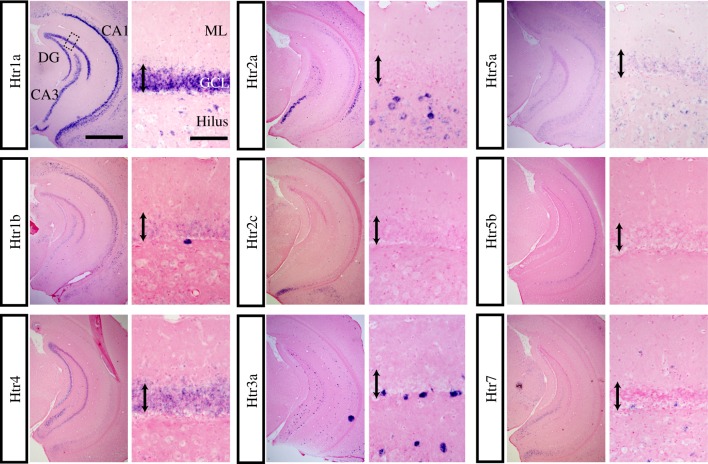
To investigate the precise expression patterns of Htrs in the hippocampus, we first asked which receptors were present in this structure. By ISH, we detected the following Htr mRNA expression: Htr1a, 1b, 1f, 2a, 2c, 3a, 4, 5a, 5b and 7. Htr1d mRNA was not found in the hippocampus, but was expressed in other regions, such as piriform cortex (data not shown). We were unable to detect Htr2b and 3b expression anywhere in the central nervous system. We did detect Htr1f mRNA in CA1 (data not shown), however, its level was very low as previously reported [18]. Figure 1 shows the regional Htr mRNA expression in the ventral hippocampus in coronal sections (left panel of each pair).
Figure 1.
Regional Htr mRNA expression in the ventral hippocampus and cellular expression in the dentate gyrus (DG). ISHs are shown for the indicated Htr probe. Sections are coronal and are through the ventral hippocampus. For each probe, the left panel is an overview of the whole hippocampus. In the Htr1a pictures, the DG and the CA3 and CA1 subfields, as well as the molecular layer (ML), granule cell layer (GCL) and hilus, are labelled (scale bar, 1 mm). The right panels are a higher magnification of the dentate gyrus granule cell layer indicated by dotted box in top left panel (scale bar, 100 µm). The double-headed arrow indicates the granule cell layer.
In the CA1 pyramidal cell layer, Htr1a mRNA was strongly expressed, Htr1b and 5b were moderately expressed, and Htr2a was weakly expressed. In the CA3 pyramidal cell layer, Htr1a, 1b, 2a, 2c, 4 and 7 mRNA were all expressed. In the dentate granule cell layer, Htr1a and 4 were strongly expressed and 1b was weakly expressed. Htr5a mRNA expression was weak and ubiquitous. Higher magnification pictures (right panel of each pair) show cellular level Htr mRNA expression in the dentate gyrus. In the molecular layer, only Htr7 mRNA was expressed. In the hilus, Htr1a, 2a, 3a and 7 mRNA were expressed. In the subgranular zone (the border between molecular layer and hilus), Htr1a, 1b, 3a, 4 and 7 were expressed. In sum, different combinations of Htrs are expressed in different cell types throughout the hippocampus. Based on these results, we chose to further study the expression profiles of Htr1a, 1b, 2a, 2c, 3a, 4, 5a, 5b and 7.
(b). Htr expression along the dorsal–ventral axis of the hippocampus
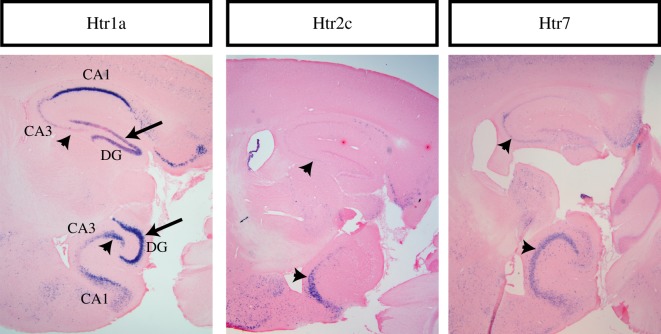
While inspecting the expression patterns of Htrs in the hippocampus, we noticed that a few receptors have a very distinct distribution of expression along the dorsal–ventral axis. In CA1, Htr1a expression is very prominent dorsally and slightly less so ventrally. In contrast, in the dentate gyrus and CA3, Htr1a expression increases gradually across the dorsal–ventral axis, so it is very prominent ventrally but much less so dorsally. Likewise, Htr2c and Htr7 show much more prominent expression in ventral CA3 than in dorsal CA3. This can be seen in sagittal sections containing both dorsal and ventral hippocampus (figure 2).
Figure 2.
Htr expression along the dorsal–ventral axis of the hippocampus. Sagittal sections showing both dorsal and ventral hippocampus for indicated probes. In each panel, dorsal hippocampus is on the top and ventral hippocampus at the bottom. Arrowheads indicate CA3 whereas full arrows indicate dentate gyrus. In the Htr1a picture, the DG and the CA3 and CA1 subfields are also labelled.
(c). Htr mRNA expression in hilar cells
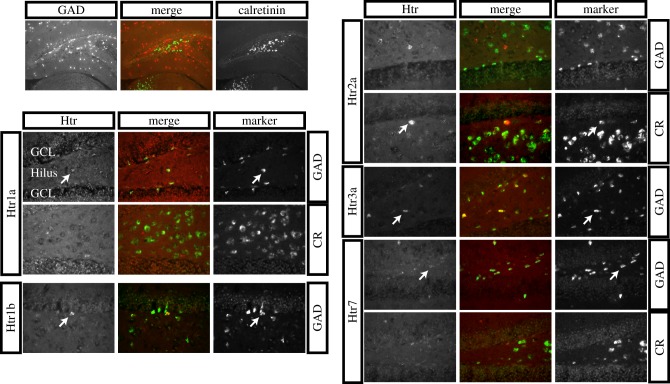
We next turned to an in-depth analysis of expression patterns within the hilus. In the hilus, there are two types of cells: GABAergic inhibitory neurons and excitatory mossy cells. Hilar GABAergic inhibitory neurons terminate throughout the dentate gyrus where they inhibit the tone. Mossy cells terminate in the inner third of the molecular layer of the dentate gyrus and form excitatory synapses with granule cells. Glutamate decarboxylase is a marker of GABAergic cells and calretinin is a marker of mossy cells, and their expression is mutually exclusive in the hilus (figure 3, top left).
Figure 3.
Double FISH between Htr mRNA and hilar cell marker mRNA in the hilus. All panels show ISHs, and the merge is the overlay of the double FISH. Top left panels provide an overview of the hippocampus showing the non-overlapping GAD and calretinin patterns of labelling in the hilus. The other panels show the double FISH for the indicated Htr probes, with either GAD or calretinin (CR) as indicated. Arrows indicate dually labelled cells. The granule cell layer (GCL) and hilus are indicated in the first Htr1a panel.
To address which hilar cells express which Htr mRNA, we performed double FISH using a Htr probe and either GAD or calretinin to mark GABAergic and mossy cells, respectively. In this experiment, since the signals for several Htrs were weak, we only performed a qualitative analysis. Hilar cells with Htr1a, 1b, 3a and 7 mRNA were colabelled with GAD mRNA but not with calretinin mRNA suggesting interneuron expression, and cells with Htr2a mRNA were colabelled with calretinin but not with GAD mRNA suggesting mossy cell expression. Therefore, in the hilus there does not appear to be any overlap in expression of Htrs between cell types.
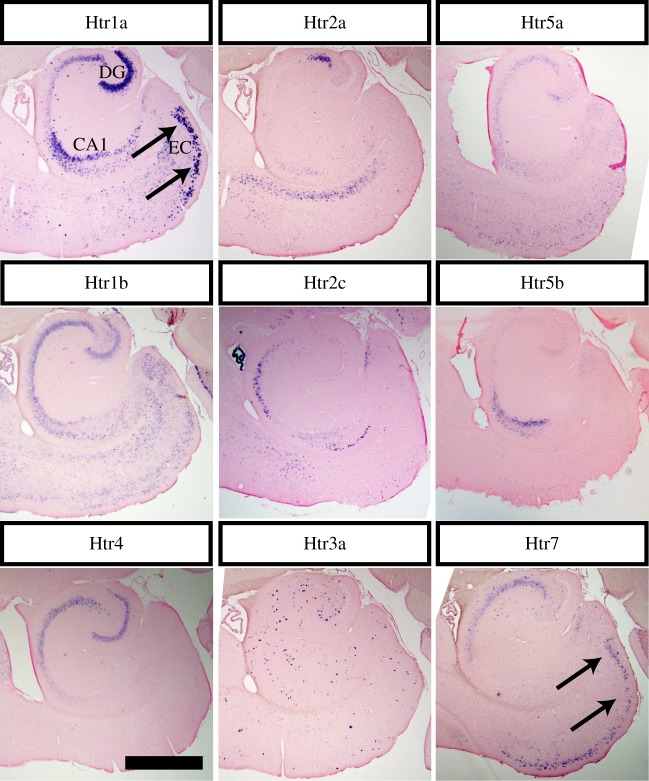
(d). Htr mRNA expression in the entorhinal cortex
We next examined Htr expression profiles in the cells that provide one of the main inputs into the hippocampus. Pyramidal cells in layer II of the entorhinal cortex that terminate in the outer two thirds of the molecular layer of the dentate gyrus, provide major inputs to the dentate gyrus from outside of the hippocampus. To assess Htr mRNA expression, we performed ISH using horizontal sections containing the entorhinal cortex (figure 4). Htr1a and 7 mRNA were strongly expressed in the layer II pyramidal cells in the entorhinal cortex (arrows); Htr1b and 5a mRNA were weakly expressed; Htr3a mRNA signals were scattered in the entorhinal cortex.
Figure 4.
Htr mRNA expression in the entorhinal cortex. The indicated Htr probes are visualized in horizontal sections across the entorhinal cortex (EC). Arrows indicate prominent labelling for Htr1a and Htr7. Limited expression is seen for other Htrs. Scale bar, 1 mm.
4. Discussion
This study first examined the expression of serotonin receptors across the distinct subfields of the hippocampus (see figure 5). Htr1a and 4 were highly expressed in granule cells of the dentate gyrus and Htr1a, 2a, 2c, 4 and 7 were also expressed in CA3 pyramidal cells. CA1 pyramidal cells expressed Htr1a, 1b and 5b. In entorhinal cortex, there was a prominent expression of Htr1a and 7 in layer II pyramidal cells. Double FISH analysis revealed that Htr1a, 1b, 3a and 7 were expressed in GAD+ interneurons of the hilus, whereas Htr2a was found in mossy cells of the hilus.
Figure 5.
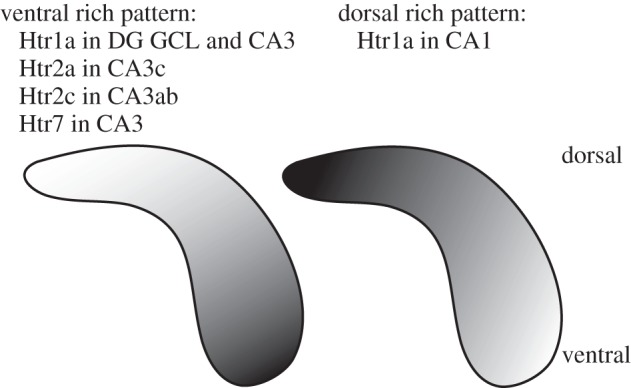
Summary of Htr expression along the dorsal–ventral axis of hippocampus. The diagram illustrates the gradient of dorsal–ventral expression for Htr1a, Htr2a, Htr2c and Htr7 in the indicated subfields.
We did not look for coexpression of different Htrs at the cellular level, but qualitatively, according to their ubiquitous expression within the same cell layer, it appears probable that dentate gyrus granule cells coexpress Htr1a and 4, and entorhinal projection neurons coexpress Htr1a and 7. This unique coexpression of different receptors suggests the existence of synergistic signal transduction within these regions, since downstream signalling is thought to be different: stimulation of Htr1a results in decreased cyclic adenosine monophosphate (cAMP) levels and distinct alterations in ion channels, while stimulation of Htr4 or 7 increases cAMP levels.
The expression of Htrs was also investigated along the dorsal–ventral axis of the hippocampus. Htr1a showed a very unique expression pattern in which it was most prominent in CA1 dorsally, but much more prominent ventrally in CA3 and dentate gyrus. Htr2a, 2c and 7 also showed much more prominent expression in ventral CA3 than in dorsal CA3 (see figure 5).
Due to their participation in different circuitries, it has been suggested that the dorsal and ventral hippocampus are distinct structures [19,20]. The dorsal dentate gyrus receives inputs from lateral and caudomedial entorhinal cortex and medially located cells of the medial septal nucleus [16,21]. Outputs of the dorsal hippocampus are towards the mammillary complex, dorsal lateral septum and lateral entorhinal cortex. In contrast, the ventral dentate gyrus receives inputs from the rostromedial entorhinal cortex and laterally located cells of the medial septal nucleus, where the outputs of the ventral hippocampus are towards the prefrontal cortex, amygdala, nucleus accumbens, hypothalamus, medial entorhinal cortex, bed nucleus of stria terminalis and rostral and ventral lateral septum [16,21]. These different circuitries suggest that the dorsal hippocampus may be more important for learning and memory, and the ventral hippocampus more involved in emotion [16,19–21]. Some lesion studies have supported this hypothesis [22–25].
Given that there is a functional segregation along the dorsal–ventral axis of hippocampus [19], it is possible that Htr1a, 2c and 7 in distinct subfields may represent populations of the same receptor serving very different functions. This could be particularly meaningful for the study of Htr1a, as it suggests an important role in cognition for this receptor in CA1, but a more limbic role in regulating stress, emotion and affect in the dentate gyrus. Interestingly, it has been shown that the antidepressant agomelatine is capable of having distinct effects on neurogenesis in the dorsal and ventral hippocampus [26]. Subfield (or dorsal–ventral)-specific receptor knockout models should test this idea.
Radioligands have widely been used in previous experiments to label serotonin receptors in brain tissue. The rich pharmacology available for serotonin receptors has allowed for relatively specific labelling of the different receptors, providing most of the current knowledge of Htr expression patterns. However, the radioligand binding technique fails to provide a sufficient resolution to differentiate between cell types within the hippocampal circuitry. For example, it is nearly impossible to differentiate hilar interneurons from dentate gyrus granule cells at the low resolution of the autoradiographic technique. This being said, the ISH studies also suffer from a major limitation: they do allow visualization of mRNAs at the cellular as well as the regional level, but not the receptors themselves. Receptor localization within cells cannot be determined. Given that some Htrs are known to have a distinct localization in axonal and somatodendritic compartments, a combination of techniques will be required to achieve a detailed and complete description of their distribution.
The present study nevertheless suggests that several Htrs are expressed in mouse hippocampus, and that each of these receptors has a unique pattern of expression. A framework is thus provided for better understanding the respective contributions of distinct Htrs to hippocampal functions, ranging from learning and memory to emotional affect. Specifically, the enrichment of Htr1a, 2a, 2c and 7 in the ventral hippocampus suggests that these receptors play a role in mood- and anxiety-related behaviour, as well as in the antidepressant and anxiolytic effects of SSRIs.
Acknowledgements
We thank Dr Yuichiro Yanagawa for GAD65 and GAD67 cDNA plasmids and Dr Laurent Descarries for thorough reading and editing of the manuscript. K.F.T. was supported by NARSAD 2008 Young Investigator Award. B.A.S. is a Charles H. Revson Senior Fellow in Life Sciences. R.H. was supported by grants from NARSAD, the New York Stem Cell Initiative (NYSTEM), the National Institutes of Health (R01 MH068542), and the Hope for Depression Research Foundation (HDRF).
References
- 1.Hannon J., Hoyer D. 2008. Molecular biology of 5-HT receptors. Behav. Brain. Res. 195, 198–213 10.1016/j.bbr.2008.03.020 (doi:10.1016/j.bbr.2008.03.020) [DOI] [PubMed] [Google Scholar]
- 2.Gaddum J. H., Picarelli Z. P. 1957. Two kinds of tryptamine receptor. Br. J. Pharmacol. Chemother. 12, 323–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peroutka S. J., Snyder S. H. 1979. Multiple serotonin receptors: differential binding of [3H]5-hydroxytryptamine, [3H]lysergic acid diethylamide and [3H]spiroperidol. Mol. Pharmacol. 16, 687–699 [PubMed] [Google Scholar]
- 4.Bradley P. B., Engel G., Feniuk W., Fozard J. R., Humphrey P. P., Middlemiss D. N., Mylecharane E. J., Richardson B. P., Saxena P. R. 1986. Proposals for the classification and nomenclature of functional receptors for 5-hydroxytryptamine. Neuropharmacology 25, 563–576 10.1016/0028-3908(86)90207-8 (doi:10.1016/0028-3908(86)90207-8) [DOI] [PubMed] [Google Scholar]
- 5.Villalon C. M., den Boer M. O., Heiligers J. P., Saxena P. R. 1990. Mediation of 5-hydroxytryptamine-induced tachycardia in the pig by the putative 5-HT4 receptor. Br. J. Pharmacol. 100, 665–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobilka B. K., Frielle T., Collins S., Yang-Feng T., Kobilka T. S., Francke U., Lefkowitz R. J., Caron M. G. 1987. An intronless gene encoding a potential member of the family of receptors coupled to guanine nucleotide regulatory proteins. Nature 329, 75–79 10.1038/329075a0 (doi:10.1038/329075a0) [DOI] [PubMed] [Google Scholar]
- 7.Fargin A., Raymond J. R., Lohse M. J., Kobilka B. K., Caron M. G., Lefkowitz R. J. 1988. The genomic clone G-21 which resembles a beta-adrenergic receptor sequence encodes the 5-HT1A receptor. Nature 335, 358–360 10.1038/335358a0 (doi:10.1038/335358a0) [DOI] [PubMed] [Google Scholar]
- 8.Humphrey P. P., Hartig P., Hoyer D. 1993. A proposed new nomenclature for 5-HT receptors. Trends Pharmacol. Sci. 14, 233–236 10.1016/0165-6147(93)90016-D (doi:10.1016/0165-6147(93)90016-D) [DOI] [PubMed] [Google Scholar]
- 9.Saudou F., Amara D. A., Dierich A., LeMeur M., Ramboz S., Segu L., Buhot M. C., Hen R. 1994. Enhanced aggressive behavior in mice lacking 5-HT1B receptor. Science 265, 1875–1878 10.1126/science.8091214 (doi:10.1126/science.8091214) [DOI] [PubMed] [Google Scholar]
- 10.Ramboz S., Oosting R., Amara D. A., Kung H. F., Blier P., Mendelsohn M., Mann J. J., Brunner D., Hen R. 1998. Serotonin receptor 1A knockout: an animal model of anxiety-related disorder. Proc. Natl Acad. Sci. USA 95, 14 476–14 481 10.1073/pnas.95.24.14476 (doi:10.1073/pnas.95.24.14476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisstaub N. V., et al. 2006. Cortical 5-HT2A receptor signaling modulates anxiety-like behaviors in mice. Science 313, 536–540 10.1126/science.1123432 (doi:10.1126/science.1123432) [DOI] [PubMed] [Google Scholar]
- 12.Grailhe R., Waeber C., Dulawa S. C., Hornung J. P., Zhuang X., Brunner D., Geyer M. A., Hen R. 1999. Increased exploratory activity and altered response to LSD in mice lacking the 5-HT(5A) receptor. Neuron 22, 581–591 10.1016/S0896-6273(00)80712-6 (doi:10.1016/S0896-6273(00)80712-6) [DOI] [PubMed] [Google Scholar]
- 13.Compan V., et al. 2004. Attenuated response to stress and novelty and hypersensitivity to seizures in 5-HT4 receptor knock-out mice. J. Neurosci. 24, 412–419 10.1523/JNEUROSCI.2806-03.2004 (doi:10.1523/JNEUROSCI.2806-03.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deacon R. M., Bannerman D. M., Rawlins J. N. 2002. Anxiolytic effects of cytotoxic hippocampal lesions in rats. Behav. Neurosci. 116, 494–497 10.1037/0735-7044.116.3.494 (doi:10.1037/0735-7044.116.3.494) [DOI] [PubMed] [Google Scholar]
- 15.File S. E., Gonzalez L. E., Andrews N. 1996. Comparative study of pre- and postsynaptic 5-HT1A receptor modulation of anxiety in two ethological animal tests. J. Neurosci. 16, 4810–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuels B. A., Hen R. 2011. Neurogenesis and affective disorders. Eur. J. Neurosci. 33, 1152–1159 10.1111/j.1460-9568.2011.07614.x (doi:10.1111/j.1460-9568.2011.07614.x) [DOI] [PubMed] [Google Scholar]
- 17.Ma J., Matsumoto M., Tanaka K. F., Takebayashi H., Ikenaka K. 2006. An animal model for late onset chronic demyelination disease caused by failed terminal differentiation of oligodendrocytes. Neuron Glia Biol. 2, 81–91 10.1017/S1740925X06000056 (doi:10.1017/S1740925X06000056) [DOI] [PubMed] [Google Scholar]
- 18.Amlaiky N., Ramboz S., Boschert U., Plassat J. L., Hen R. 1992. Isolation of a mouse ‘5HT1E-like’ serotonin receptor expressed predominantly in hippocampus. J. Biol. Chem. 267, 19 761–19 764 [PubMed] [Google Scholar]
- 19.Fanselow M. S., Dong H. W. 2010. Are the dorsal and ventral hippocampus functionally distinct structures. Neuron 65, 7–19 10.1016/j.neuron.2009.11.031 (doi:10.1016/j.neuron.2009.11.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moser M. B., Moser E. I. 1998. Functional differentiation in the hippocampus. Hippocampus 8, 608–619 (doi:10.1002/(SICI)1098-1063(1998)8:6<608::AID-HIPO3>3.0.CO;2-7) [DOI] [PubMed] [Google Scholar]
- 21.Sahay A., Hen R. 2007. Adult hippocampal neurogenesis in depression. Nat. Neurosci. 10, 1110–1115 10.1038/nn1969 (doi:10.1038/nn1969) [DOI] [PubMed] [Google Scholar]
- 22.McHugh S. B., Deacon R. M., Rawlins J. N., Bannerman D. M. 2004. Amygdala and ventral hippocampus contribute differentially to mechanisms of fear and anxiety. Behav. Neurosci. 118, 63–78 10.1037/0735-7044.118.1.63 (doi:10.1037/0735-7044.118.1.63) [DOI] [PubMed] [Google Scholar]
- 23.Moser E., Moser M. B., Andersen P. 1993. Spatial learning impairment parallels the magnitude of dorsal hippocampal lesions, but is hardly present following ventral lesions. J. Neurosci. 13, 3916–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moser M. B., Moser E. I., Forrest E., Andersen P., Morris R. G. 1995. Spatial learning with a minislab in the dorsal hippocampus. Proc. Natl Acad. Sci. USA 92, 9697–9701 10.1073/pnas.92.21.9697 (doi:10.1073/pnas.92.21.9697) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kjelstrup K. G., Tuvnes F. A., Steffenach H. A., Murison R., Moser E. I., Moser M. B. 2002. Reduced fear expression after lesions of the ventral hippocampus. Proc. Natl Acad. Sci. USA 99, 10 825–10 830 10.1073/pnas.152112399 (doi:10.1073/pnas.152112399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soumier A., et al. 2009. Mechanisms contributing to the phase-dependent regulation of neurogenesis by the novel antidepressant, agomelatine, in the adult rat hippocampus. Neuropsychopharmacology 34, 2390–2403 10.1038/npp.2009.72 (doi:10.1038/npp.2009.72) [DOI] [PubMed] [Google Scholar]