Abstract
Plant growth promotion is a multigenic process under the influence of many factors; therefore an understanding of these processes and the functions regulated may have profound implications. Present study reports microarray analysis of Arabidopsis thaliana plants inoculated with Pseudomonas putida MTCC5279 (MTCC5279) which resulted in significant increase in growth traits as compared with non-inoculated control. The gene expression changes, represented by oligonucleotide array (24652 genes) have been studied to gain insight into MTCC5279 assisted plant growth promotion in Arabidopsis thaliana. MTCC5279 induced upregulated Arabidopsis thaliana genes were found to be involved in maintenance of genome integrity (At5g20850), growth hormone (At3g23890 and At4g36110), amino acid synthesis (At5g63890), abcissic acid (ABA) signaling and ethylene suppression (At2g29090, At5g17850), Ca+2 dependent signaling (At3g57530) and induction of induced systemic resistance (At2g46370, At2g44840). The genes At3g32920 and At2g15890 which are suggested to act early in petal, stamen and embryonic development are among the downregulated genes. We report for the first time MTCC5279 assisted repression of At3g32920, a putative DNA repair protein involved in recombination and DNA strand transfer in a process of rapid meiotic and mitotic division.
Keywords: Induced systemic resistance, Plant growth promoting bacteria
Introduction
Plants are in continuous contact with soil micro biota, pathogenic or beneficial in nature. Beneficial free-living root colonizing bacteria, directly or indirectly supporting plant growth and yield are collectively termed as plant growth promoting rhizobacteria (PGPR).1 In past years several mechanisms of plant growth promotions by PGPR including (1) bacterial production of phytohormone like indole-3-acetic acid,2,3 gibrellins4 cytokinin5 (2) triggering induced systemic resistance (ISR)6 (3); breakdown of plant produced ethylene by bacterial 1-aminocyclopropane-1-carboxylate (ACC) deaminase7 (4); suppression of plant pathogens8 and (5) increased availability of soil nutrients9 to plants have been discussed. In addition to plant growth promotion, PGPR also increases plant tolerance against abiotic environmental factors like drought, extreme temperature, high soil salinity, nutrient deficiency and metal toxicity.10 Bacterial ACC deaminase activity and regulation of ACC production is considered to be an important beneficial mechanism for abiotic stressed plants.11
Plant-microbe interactions with PGPRs commonly result in a phenomenon called Induced systemic resistance (ISR) which prepares the host plant to resist a wide range of stresses and plant pathogens.12–14 However mechanism of ISR is very diverse. These mechanisms may be, ethylene and jasmonic acid (JA) or salicylic acid (SA) dependent.15,16 Pseudomonas fluorescens has been reported to induce ISR, independent of SA and dependent of JA but could also induces ISR even in ethylene insensitive mutant Arabdiopsis thaliana plants.17 Moreover some specific strains trigger ISR by SA signaling indicates that the pathways for ISR are not universal and are very much dependent on plant species and bacterial partners. Most of the studies investigating plant-microbe interaction mechanisms are worked out with bacterial strains with traits like niche specificity, PGPR and biocontrol activity etc.18–20 However reports of elucidating mechanisms of PGPR with abiotic stress tolerant bacterial strains is scanty. Hence the current study was taken up with the objective of to (1) understand the plant phsiological alteration under the influence of a abiotic stress tolerant plant growth promoting rhizobacteria Pseudomonas putida MTCC5279 (MTCC5279), (2) microarray analysis was performed for a broader understanding of the molecular determinants involved in plant growth promotion, which can serve as a viable strategy for promoting plant growth.
Results
Plant growth promotion by Pseudomonas putida MTCC5279
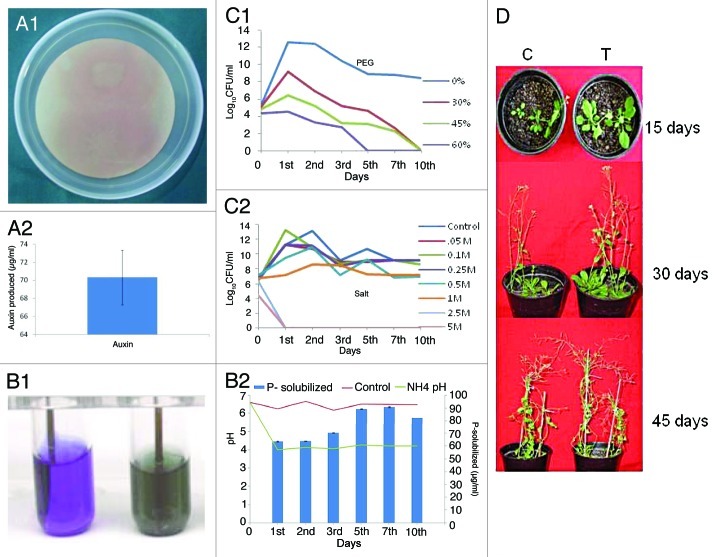
MTCC5279 was isolated from desert regions of Rajasthan, India and identified by fatty acid methyl ester analysis by CABI Biosciences and 16SrRNA sequencing as Pseudomonas putida and submitted to Microbial Type Culture Collection at IMTECH, Chandigarh; India. The strain was characterized for its abiotic stress tolerance and plant growth promotional attributes summarized in Figure 1. MTCC5279 shows growth in presence of up to 60% PEG (Fig. 1C1) and 500 mM NaCl (Fig. 1C2), produced Auxin, 70.23 µg ml−1 (Fig. 1A2) and solubilized insoluble tri-calcium phosphate (TCP) (Fig. 1B1 and B2). The PGPR activity of MTCC5279 was determined using Arabidopsis thaliana (Col-0) as host plant. Inoculation with MTCC5279 resulted in significant increase in vegetative growth of Arabidopsis thaliana plants (Fig. 1D). Significant increase in plant height by 43.2%, number of branches by 91.3% and dry weight by 79.8% was noted in MTCC5279 inoculated Arabidopsis thaliana in comparison to non-inoculated plants (Table 1). Significant increase in number of branches per plant resulted in higher silique formation (110.2% as compared with control), which ultimately gave better seed yield of about 32.3% in comparison to uninoculated control. Colonization in terms of mean colony-forming units (CFU) mg−1 of the MTCC5279 treated Arabidopsis thaliana plants showed 5.6 × 105 CFU mg−1 in rhizosphere and 7.0 × 105 CFU mg−1 in phyllosphere after 45 d of inoculation, whereas control plant roots did not show any colonization.
Figure 1. Auxin production by Pseudomonas putida MTCC5279. (A) Auxin production in Pseudomonas putida MTCC5279 was performed as per the protocol of Bric et al.63 Phosphate solubilization by Pseudomonas putida MTCC5279. Pseudomonas putida MTCC5279 was inoculated in NBRI-BPB and NBRIP media and P-solubilization at different time interval was performed as described by Mehta and Nautiyal. (B1 and 2).64 Abiotic stress tolerance of Pseudomonas putida MTCC5279 was performed by growing the culture in presence of different concentration of polyethylene glycol (PEG-6000) and salt (NaCl) and CFU ml-1 was determined at different time interval (C1 and 2).65 Effect of Pseudomonas putida MTCC5279 inoculation on the growth of Arabidopsis thaliana.
Table 1. Plant growth promoting effect of Pseudomonas putida MTCC5279 using Arabidopsis thaliana as a host plant after 45 d of inoculation.
| Plant growth parameters |
Control |
Treated |
CD at |
|
|---|---|---|---|---|
| 5% | 1% | |||
| Root length (cm) |
2.46 ± 0.15 |
2.64 ± 0.21 |
0.29 |
0.40 |
| Shoot length (cm) |
23.10 ± 0.71 |
33.08 ± 1.24 |
3.64 |
5.13 |
| No. of branches |
5.79 ± 0.13 |
11.08 ± 0.72 |
1.92 |
2.71 |
| No. of siliques |
29.75 ± 1.75 |
62.54 ± 3.46 |
11.62 |
16.39 |
| Fresh weight(mg) |
630.00 ± 39.83 |
1245.00 ± 114.79 |
240.62 |
339.56 |
| Dry weight (mg) |
156.66 ± 8.43 |
281.66 ± 21.35 |
47.97 |
67.69 |
| Seed weight (mg/plant) | 5.10 ± 0.43 | 6.75 ± 0.56 | 0.82 | 1.12 |
Microarray analysis of Arabidopsis thaliana leaves after inoculation with Pseudomonas putida MTCC5279
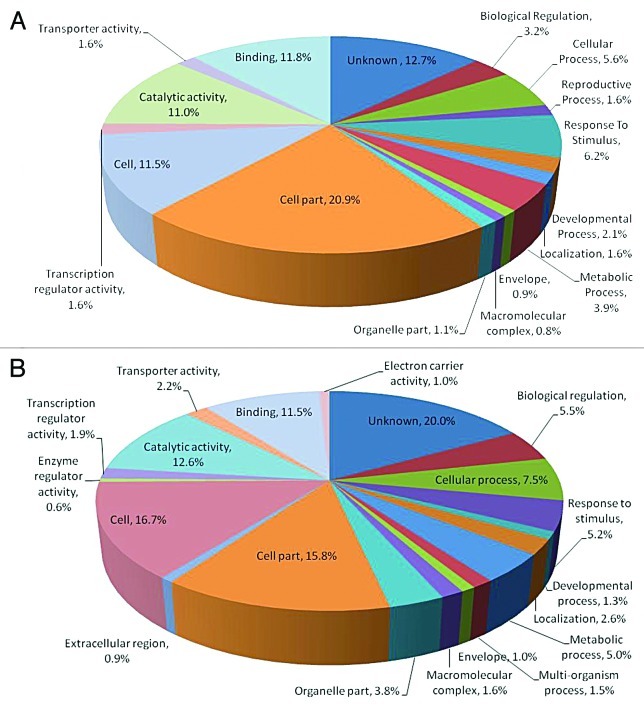
To obtain a global picture of the genes differentially expressed on colonization of MTCC5279, microarray analysis was performed in Arabidopsis thaliana using 25K OciChip™. After filtration 24,652 probes were retained, which were used for downstream statistical and biological analysis and the comparison across samples was performed by using the median absolute deviation (MAD). To determine the differentially expressed candidate genes multivariate outlier detection was also performed. This approach provides fold-change value, 520 genes were found to be overexpressed and 364 were repressed using 3-fold change as a criterion. The functional classification of the differentially expressed genes were performed using Genowiz™, a gene expression analysis and tracking tool that enables to determine biological significance of differentially expressed genes and facilitates expression pattern analysis and gives an insight into metabolic pathways. Gene ontology data shows that in a biological process 15.58%, 13.62% in cellular component and 13.88% in molecular process were getting upregulated while the downregulated genes in terms of biological function, molecular function and cellular component were 21.30%, 13.62% and 18.20% respectively.
Pathway analysis data showed that the genes involved in glyoxylate and dicarboxylate metabolism, fructose and mannose metabolism, oxidative phosphorylation, carbon fixation, glycerolipid metabolism, lysine biosynthesis, glycine, serine and threonine metabolism, glycan biosynthesis and metabolism, one carbon pool by folate, biosynthesis of secondary metabolites, ribosome, SNARE interactions in vesicular transport, inositol phosphate metabolism and behavior are upregulated in treated with respect to control. Genes involved in butanoate metabolism, galactose metabolism, starch and sucrose metabolism, pentose and glucuronate interconversions, oxidative phosphorylations, nucleotide metabolism, histidine, tryptophan, glutathione, alanine and aspartate metabolism and phenyl propanoid biosynthesis, styrene degradation, caprolactam degradation, proteasome, and phosphatidyl inositol signaling system were downregulated in treated plants when compared with control plants.
Functional characterization of majority of the genes getting differentially expressed by MTCC5279 showed that genes upregulated in biological process were mainly involved in cellular process, 5.6%; biological regulation 3.2%; response to stimulus 6.2% and metabolic process 3.8% and 20.8% in cell part and 11.45% in cell in cellular process and 10.96% in catalytic activity and 11.7% in binding in terms of molecular function. Downregulated genes on functional characterization in terms of biological process showed 5.47% in biological regulation, 7.49% in cellular process, 5.15% in response to stimulus and 4.99% involved in metabolic process. Among cellular process 15.79% in cell part and 16.6% in cell and in molecular function class catalytic activity (12.57%) and genes responsible for binding (11.57%) were downregulated.
At gene level the genes getting upregulated by the colonization of MTCC5279 have shown 3- to 17-fold change factor in their expression value (supplementary data). The majority of genes getting upregulated shows similarity to the plant growth related traits such as carbohydrate metabolism (At1g05030, 5.71-fold), nucleoside transferase (At1g63730, 6.59-fold), kinases (At1g06730, 5.32-fold; At1g53730, 4.69-fold), transcription factor (At1g12890, 5.83-fold; At1g01260, 7.66-fold), defense and abiotic stress related genes, induction of amino acid biosynthetic pathway and nutrient uptake (At5g63890, 3.89-fold), hormone synthesis (At3g23890, 3.66-fold), ABA signaling and ethylene suppression (At2g29090, 3.17-fold), signal transduction through calcium signaling components (At3g57530, 3.60-fold) induction of induced systemic resistance (At2g46370, 3.87-fold) and induction in the mitotic and meiotic division (At5g20850, 3.02-fold change).
Validation of the microarray experiments through semiquantitative RT-PCR
To verify the GeneChip results, a semiquantitative RT-PCR analysis was performed on selected genes, including up- and downregulated genes identified in the microarray analysis. The upregulated genes selected for semiquantitative RT-PCR were of biological process, related to functions, DNA metabolic process in response to gamma radiation (At5g20850), histidinol dehydrogenase activity in response to UV (At5g63890), abcisic acid metabolic process responsive (At2g29090), induced systemic resistance responsive (At2g46370), At3g57530 responsible for calcium- and calmodulin-dependent protein kinase activity, At3g23890 in a DNA topological change in response to hormones, DEAD/H- box RNA helicase (At5g62190) involved in RNA metabolic process and At5g17850 involved in cation transport. At2g15890, At4g36110, At1g69490, At5g17220, At2g16660 were chosen among upregulated candidate genes based on prior report by Wang et al.20 Some of the downregulated genes chosen for revalidation are At3g32920, involved in SOS response; At1g74930 involved in regulation of transcription and At2g44840 involved in ET mediated signaling pathway. The RT-PCR experiments were performed using RNA from mock (control) and MTCC5279 supplemented (treated) plants grown for 45 d initially and after 15, 30 and 45 d of inoculation to get relative expression of the selected genes in an independent experiment from the microarray analysis. The PCR reaction products obtained using gene specific primers with 28 cycles of amplification were analyzed by gel electrophoresis. The results of RT-PCR analysis clearly showed increased expression in treated leaves, whereas the mRNA level of the selected downregulated genes were lower in MTCC5279 treated plants (Figure 4). As expected, transcript levels of the selected genes are in accordance with the microarray results and thus confirm the data from the microarray experiments. Results showed that among all the targeted eight genes two were significantly downregulated in MTCC 5279 treated Arabidopsis thaliana as compared with untreated control; however differences were more prominent at 45 d in At4g36110, At5g17850, At5g20850, At5g63890, At2g29090, At3g23890 genes respectively whereas two At2g15890 and At3g32920 were upregulated at 15 and 30 d followed by their downregulation at 45 d (Fig. 4C).
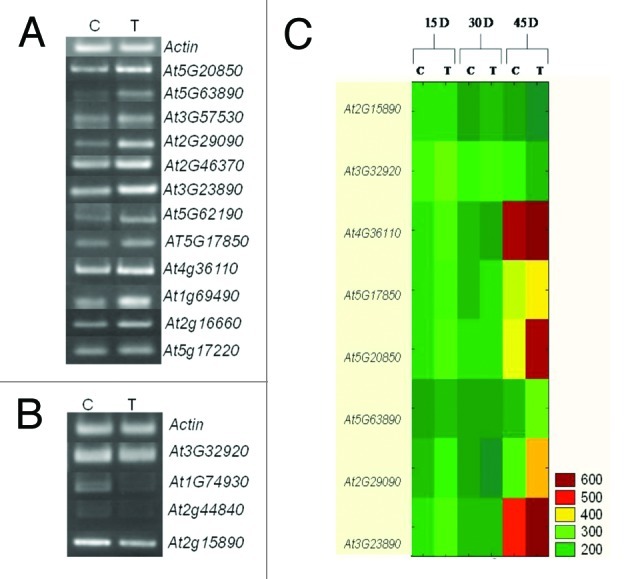
Figure 4. Semiquantitative RT-PCR analysis of genes showing significant differential expression. Increased (A) and decreased (B) expression in Arabidopsis thaliana plants colonized by Pseudomonas putida MTCC5279 after 6 weeks. (C) Relative expression of selected genes at different time intervals.
Discussion
Many plant-associated soil microorganisms interact with plant roots resulting in mutualistic and communalistic interactions, which plays a pivotal role on plant growth and development.21,22 However, plant growth and production is continually decreasing owing to abiotic and biotic stress factors, which is increasingly becoming a severe global problem for agriculture production.23,24 Therefore the methods to improve growth, development and stress tolerance in plants have become more and more important for crop production. One of the ways to evoke the above problem in crops is the exploitation of plant-associated microorganisms, which live in reciprocally beneficial relationships with plants.25 Pseudomonas putida MTCC5279 is an abiotic stress tolerant, auxin producing, P-solubilizing, potent plant growth promoting strain and has a potential to be exploited in P translocation in the plants, especially in P limiting and abiotic stress conditions as a further extension of the work. The present study presents a global view on gene expression and physiological processes involved in MTCC5279 induced PGP in Arabidopsis thaliana. The expression profiling of Arabidopsis thaliana response to MTCC5279 assisted changes has allowed us to identify transcripts differentially expressed confirming that plant growth promotion is a multigenic process. Some of the genes identified are similar to general plant growth promotional response while others may be specific to plant growth promotional response of PGPR to Arabidopsis thaliana. However, we have been able to characterize previously uncharacterized genes involved in this process. The genes identified in this study, induced or repressed (3.0- to 6.0-fold) suggest they have very important roles and are worthy of further functional analyses.
Many of the genes induced in MTCC5279 assisted plants reported for the first time are involved in maintenance of genome integrity concerned with DNA metabolic process, auxin responsive growth stimulus, induction of amino acid biosynthetic pathway and nutrient uptake, ABA signaling and induction of induced systemic resistance. The new report of downregulated gene has been so far suggested to act early in petal, stamen and embryonic development ending in dormancy. Other important gene associated with plant growth promotion which is being reported for the first time by MTCC5279 assisted repression is At3g32920. Role of MTCC5279 in inducing plant growth promotion has been discussed in following heads.
Auxin responsive growth stimulus
Indole-3-acetic acid (IAA), cytokinins and gibberellins are important determinants of PGPRs26–28 of which IAA is most frequently reported.29–31 Overexpression of At4g36110 has been correlated with IAA production during plant-microbe interaction in Arabidopsis in earlier reports.20 Accordingly it is speculated that Pseudomonas putida MTCC5279 induces At4g36110 which results in increased IAA production. Topoisomerases, which catalyzes a transient breakage and reunion of double-stranded DNA was reported to be overexpressed in hormone treated pea plants supports the overexpression of At3g23890 (TopII) in MTCC5279 treated plants and speculate the view that MTCC 5279 treatment augment the hormonal level of the plant and enhance the catalysis step of transient breakage and reunion.32,33
Induction of amino acid biosynthetic pathway and nutrient uptake
Histidine is essential for plant growth and development, especially in root development in higher plants.34 Overexpression of histidinol dehydrogenases (At5g63890, HDH), responsible for the conversion of histidinol to histidine in histidine biosynthetic pathway is correlated with more greenish plants and better root development in MTCC5279 treated Arabidopsis thaliana plants is being reported for the first time in this study to the best of our knowledge. Higher expression of HDH in presence of MTCC5279 suggests that the plant is using histidine as a ET receptor and play role in transmembrane signaling through His-Asp phosphorelay.35 Overexpression of At2g16660, similar to nodulin-like protein is in accordance with the prior result suggesting that nodulin-like gene could be involved in signal perception or enhancement of plant mineral and nutrient uptake induced by PGPR colonization as evident by phosphate transportation by a PGPR Piriformospora indica.20,36
Signal transduction through calcium signaling components
The Ca2+ is one of a second messenger and is crucial for signal transduction processes during many metabolic interactions.24 Expression of some of calcium signaling components e.g., calcium-dependent protein kinase (At3g57530, CPK32) were found to be increased in Arabidopsis thaliana colonized with Pseudomonas putida MTCC 5279 (Table 3) which is in accordance with earlier report by Verhagen et al.37 The regulation of gene expression by elevations of cellular Ca2+ is crucial for plant defense against various stresses and interaction between Piriformospora indica and Arabidopsis thaliana.38,39 Upregulation of CPK32, an ABA signaling component is also in accordance with the prior result suggesting that MTCC5279 mediated regulation of ABA responsive genes via ABF4 in a ABA signaling pathway through Ca+2 elevations.40 ABA manipulation by endogenous signal is a promising means to improve productivity, performance and plant architecture.41–43 The overexpression of At5g17850 in MTCC5279 treated plants is involved in upregulated level of ABA (has regulatory roles in stress responses). Interestingly MTCC5279 overexpresses At2g29090 (CYP707A2) which is responsible for catabolic repression of ABA,45–47 hence suggesting a balanced level of ABA. Besides overexpression of At2g29090 (CYP707A2) is associated with upregulated level of nitrate as reported by Matakiadis et al.44 which probably increases nitrate uptake in plants which results in the enhanced vegetative growth.
Table 3. List of primers used for the validation of the microarray data by semiquantitative RT-PCR analysis.
| Gene(s)/locus tag | Up or downregulated | Accession number | Primer sequence | Annealing temperatures (⁰C) |
|---|---|---|---|---|
|
At5g20850 |
Upregulated (This study) |
NM_122092 |
Left Primer: 5′-ggccatgtacattgatgctg-3′ Right Primer: 5′-ttgcgcaactacttggtttg-3′ |
55 |
|
At5g63890 |
-do- |
NM_125784 |
Left Primer: 5′-aatcgatatgcctgctggtc-3′ Right Primer: 5′-agcctgcgttctcaatcagt-3′ |
55 |
|
At2g29090 |
-do- |
NM_128466 |
Left Primer: 5′-taaatggtggttgcactgga-3′ Right Primer: 5′-atggtatggaccttggtgga-3′ |
63 |
|
At2g46370 |
-do- |
NM_180122 |
Left Primer: 5′-gtcgtggagaattcggtgtt-3′ Right Primer: 5′-gtttcaggtccctgtgcatt-3′ |
55 |
|
At3g23890 |
-do- |
NM_113294 |
Left Primer: 5′-caacggcttgctcaatacaa-3′ Right Primer: 5′-ttctcgctgctctcctcttc-3′ |
52 |
|
At5g62190 |
-do |
NM_125613 |
Left Primer: 5′-ggctcggttgattcctgata-3′ Right Primer: 5′-gttgccagctcttcctgttc-3′ |
52 |
|
At3g57530 |
-do- |
NM_115613 |
Left Primer: 5′-gtcgtggagaattcggtgtt-3′ Right Primer: 5′-gtttcaggtccctgtgcatt-3′ |
63 |
|
At5g17850 |
-do- |
NM_121791 |
Left Primer: 5′-tgggctatcactctgcctct-3′ Right Primer: 5′-cgcaactagctcttgtgctg-3′ |
52 |
|
At2g15890 |
Upregulated (Wang et al.20) |
NM_127149 |
Left Primer: 5′-gttcgacgcttgaaggtgat-3′ Right Primer: 5′-ggtcaggagcgtgagagaac-3′ |
55 |
|
At4g36110 |
-do- |
NM_119778 |
Left Primer: 5′-tcaacaccgaagtcgctatg-3′ Right Primer: 5′-catttgcttccgtcttctt-3′ |
55 |
|
At1g69490 |
-do- |
NM_105616 |
Left Primer: 5′-aattcgacccatggcaatta-3′ Right Primer: 5′-tccatgaaaccctcttgctc-3′ |
55 |
|
At5g17220 |
-do- |
NM_121728 |
Left Primer: 5′-ctctagagcaccgagccatc-3′ Right Primer: 5′-ctaccccgagccttaaccat-3′ |
55 |
|
At2g16660 |
-do- |
NM_099989 |
Left Primer: 5′-gtgagatcccaccagcatct-3′ Right Primer: 5′-ttcggagagagtaccggag-3′ |
52 |
|
At3g32920 |
Downregulated (This study) |
NM_114061 |
Left Primer: 5′-atcgggaaagacagcacttg-3′ Right Primer: 5′-ctgttggacctccaaatcgt-3′ |
60 |
|
At1g74930 |
Downregulated (Wang et al.20) |
NM_106151 |
Left Primer: 5′-ttacgacactcccgagaagg-3′ Right primer: 5′-tcctccgtaatcttcgatgg-3′ |
55 |
|
At2g44840 |
-do- |
NM_130048 |
Left Primer: 5′-gttcctcccgttacctctcc-3′ Right Primer: 5′-tctgctccgacgttaactga-3′ |
55 |
| At3g46520 | Wang et al.20 | - | Left primer: 5′-gatatggaaaagatctggcatcac-3′ Right primer: 5′-tcatactcggccttggagatccac-3′ |
57 |
Similarity in terms of usage of the oligo for PGPR
Induced systemic resistance (ISR)
Overexpression of jasmonate responsive gene (At2g46370, JAR1) in the present study demonstrate that MTCC5279 mediates ISR. The mechanisms leading to ISR seem to be highly dependent on the bacterial partners and the majority of genes were predicted to be regulated by either JA or ET, or both.6,48 Pseudomonas fluorescens WCS417r, elicited ISR via a salicylic acid (SA) independent pathway.49,50 MTCC5279 lead to ISR in a SA independent but JA dependent manner like Pseudomonas fluorescens strain, CHA0.15,16 Suppression of At2g44840, an ET-responsive transcription factor 13, which binds to the GCC-box of pathogenesis-related promoter in a stress dependent manner is in accordance with previous report37 and also supports ISR induction in a SA independent pathway without activating the PR proteins.51 Systemic induced resistance in Arabidopsis conferred by Piriformospora indica require JA signaling has also been reported by Stein et al.52
Induction of mitotic and meiotic division
Overexpression of a RNA helicase (PRH75, At5g62190) essential for cellular processes, regulate plant growth and development and expressed mainly in young and rapidly developing tissues is in accordance with the prior reports.37,53–55 Wang et al.20 has reported overexpression of At2g15890 (responsible for specifying petal and stamen identity in Arabidopsis thaliana) in presence of PGPR. On the contrary repression of At2g15890 was observed in the present study which may be due to sampling after maturity.56,57 RAD51 (At5g20850), a RecA functional homolog plays important role in mitotic and meiotic recombination and its mutant produce aborted siliques and anthers having immature pollens, it’s overexpression in MTCC5279 treated plants resulted in more silique formation as evident by physiological data (Table 2).58,59 Repression of AT3g32920, a putative DNA repair protein involved in recombination and DNA strand transfer in a process of rapid meiotic and mitotic division has been found. Overexpression of At1g69490, a gene responsible for leaf senescence in the present study is in accordance with the prior report, suggest the early maturation of the plant on MTCC5279 treatment.20,60,61 The overexpression of glutathione S-transferase (At5g17220) is in accordance with the prior result and supports prior reports of defense enzyme induction by the colonization of rhizobacteria. Overexpression of H+ cation exchanger (At5g17850) and the repression of At1g74930, which codes for ET suppression suggested the reduced level of ethylene resulting in better root elongation and plant architecture as evident by Camehl et al.62
Table 2. Genes showing significant increased /decreased expression in Arabidopsis plants colonized by Pseudomonas putida MTCC5279.
|
Upregulated pathways |
Probe set |
Putative function |
Fold change |
|
Carbohydrate metabolism 2.185% | |||
| Glyoxylate and dicarboxylate metabolism |
At4g17360 |
Formyl tetrahydrofolate deformylase |
4.65 |
| Fructose and mannose metabolism |
At1g73250 |
3, 5-epimerase-4-reductase |
3.94 |
|
Energy metabolism 3.32% | |||
| Oxidative phosphorylation |
At1g15690 |
Inorganic pyrophosphatase (h(+)-Ppase |
4.80 |
| Carbon fixation |
At2g45290 |
Transketolase |
4.087 |
| |
At5g08530 |
NADH dehydrogenase |
3.460 |
| |
At3g53620 |
Pyrophosphatase |
3.461 |
| |
At1g78900 |
Vacuolar ATP synthase |
3.147 |
| Nitrogen metabolism |
At3g47340 |
Glutamine-dependent asparagine synthase 1 |
3.080 |
| |
At3g10340 |
Ammonia ligase |
3.041 |
|
Lipid metabolism 4.83% | |||
| Glycerophospholipid metabolism |
At3g05630 |
Phospholipase D activity |
3.339 |
| Glycerolipid metabolism |
At4g31780 |
Udp-glycosyltransferase activity |
3.048 |
|
Amino acid metabolism 4.34% | |||
| Glycine, serine and threonine metabolism |
At4g13890 |
Glycine hydroxymethyl transferase |
3.836 |
| Lysine biosynthesis |
At3g11710 |
Aaspartate-t-RNA ligase |
3.765 |
| Alanine and aspartate metabolism |
At3g47340 |
Glutamine-dependent asparagine synthase 1 |
3.080 |
|
Glycan biosynthesis and metabolism 18.18% | |||
| N-glycan degradation |
At5g66150 |
Alpha-mannosidase |
3.559 |
|
Metabolism of cofactors and vitamins 16.6% | |||
| One carbon pool by folate |
At4g17360 |
Formyl tetrahydrofolate deformylase |
4.647 |
|
Biosynthesis of secondary metabolites 2.469% | |||
| |
At5g19880 |
Peroxidase |
4.663 |
| Phenylpropanoid biosynthesis |
At3g53280 |
Cytochrome p450 monooxygenase |
3.802 |
| |
At2g41480 |
Peroxidase |
3.193 |
| Benzoate degradation via CoA ligation |
At1g01140 |
CBl-interacting protein kinase |
4.410 |
|
Translation 1.33% | |||
| Ribosome |
At5g02960 |
Structural constituent of ribosome |
6.082 |
| |
At4g30800 |
Structural constituent of ribosome |
4.389 |
| |
At3g62250 |
Protein binding |
3.411 |
| Aminoacyl-tRNA biosynthesis |
At3g11710 |
Aspartate-tRNA ligase/ lysine-tRNA ligase |
3.765 |
|
Sorting and degradation 10.4% | |||
| Snare interactions in vesicular transport |
At1g15880 |
Intra-Golgi vesicle-mediated transport |
4.469 |
| |
At3g61450 |
Protein transporter activity |
4.105 |
| |
At3g58170 |
SNAP-receptor activity |
3.481 |
| |
At3g52400 |
Negative regulation of defense pathways |
3.137 |
| |
At1g32270 |
intra-Golgi vesicle-mediated transport |
3.011 |
|
Signal transduction 0.91% | |||
| Phosphatidylinositol signaling system |
At3g57530 |
Calcium-dependent protein kinaseC activity |
3.607 |
|
Behavior 5.71% | |||
| Circadian rhythm |
At1g01060 |
Myb-related putative transcription factor |
4.214 |
| |
At1g56650 |
Anthocyanin metabolism and radical scavenging |
3.040 |
|
Downregulated pathways |
Custom id |
Gene description |
Fold change |
|
Carbohydrate metabolism 5.88% | |||
| Butanoate metabolism |
At5g06580 |
Electron transport |
-3.621 |
| Galactose metabolism |
At5g17310 |
UTP-glucose-1-phosphate uridylyl transferase |
-3.236 |
| Starch and sucrose metabolism |
At1g02790 |
Polygalacturonase activity |
-3.0245 |
| |
At1g02640 |
Beta-xylosidase |
-3.672 |
| |
At3g43190 |
UDP-glycosyl transferase |
-3.183 |
| Pentose phosphate pathway |
At1g09420 |
Glucose-6-phosphate dehydrogenase activity |
-3.674 |
| Pentose and glucuronate interconversions |
At1g02790 |
Polygalacturonase activity |
-3.025 |
|
Energy metabolism 1.626% | |||
| Oxidative phosphorylation |
|
|
|
| |
At3g60330 |
Hydrogen-exporting ATPase activity |
-3.247 |
| |
At4g09650 |
Hydrogen-transporting ATP synthase |
-3.685 |
|
Nucleotide metabolism 2.65% | |||
| Purine and pyrimidine metabolism |
At1g08260 |
DNA polymerase epsilon catalytic subunit. |
-3.775 |
| |
At4g12440 |
Adenine phosphoribosyl transferase |
-4.103 |
|
Amino acid metabolism 2.83% | |||
| Histidine metabolism |
At3g46100 |
Histidyl-tRNA aminoacylation |
-3.998 |
| Phenylalanine metabolism |
At2g39040 |
Peroxidase |
-3.312 |
| Alanine and aspartate metabolism |
At5g14760 |
L-Aspartate oxidase |
-3.052 |
| Tryptophan metabolism |
At5g22300 |
3-Cyanoalanine hydratase activity |
-4.382 |
|
Metabolism of other amino acids 5.55% | |||
| Glutathione metabolism |
At1g09420 |
Glucose-6-phosphate dehydrogenase activity |
-3.673 |
| Cyanoamino acid metabolism |
At1g02640 |
Beta-xylosidase |
-3.672 |
|
Biosynthesis of secondary metabolites 1.92% | |||
| Phenylpropanoid biosynthesis |
|
|
|
| |
At1g51680 |
4-Coumarate-coA ligase activity |
-3.028 |
| |
At2g39040 |
Peroxidase activity |
-3.312 |
|
Xenobiotics biodegradation and metabolism 4.05% | |||
| Styrene degradation |
At5g22300 |
|
-4.382 |
| Caprolactam degradation |
At5g56650 |
IAA conjugate hydrolases |
-4.598 |
|
Translation 0.77% | |||
| Ribosome |
At5g41520 |
Structural constituent of ribosome |
-3.081 |
|
Sorting and degradation 2.27% | |||
| Proteasome |
At2g27020 |
Endopeptidase activity |
-4.884 |
|
Signal transduction 0.91% | |||
| Phosphatidyl inositol signaling system | At1g68000 | CDP-diacylglycerol-inositol 3-phosphatidyl transferase activity | -4.694 |
In conclusion we can say that MTCC5279 mediated PGP in Arabidopsis thaliana is governed by many reported and new up and downregulated genes identified in this study. Further experimentation and manipulation of the identified candidate genes and processes could lead to generate important information on plant-microbial interactions, especially on plant growth promotion and their role in the interaction of bacteria with plants.
Materials and Methods
Plant growth promotion using Arabidopsis thaliana as a host plant
Bacterial culture Pseudomonas putida MTCC5279 was isolated from hot deserts of Rajasthan, India. The strain was characterized for plant growth promotional trait following standard protocols like P-solubilization, auxin production, abiotic stress tolerance.63–65 Bacterial culture was grown in Nutrient broth at 30°C for 48 h in a rotary shaker at 180 rpm. Bacterial cells were pelleted after 48 h at 6000 rpm for 5 min and washed pellets was used for inoculating the Arabidopsis thaliana plants after resuspending in sterile 0.85% saline, as described earlier.8
Plant growth conditions
The plant growth promotary effect of MTCC5279 on Arabidopsis thaliana was assessed in sterile soilrite. Seeds of Arabidopsis thaliana ecotype Columbia (Col-0) were surface-sterilized by 0.4% NaClO and 70% ethanol followed by repeated washing with distilled water. Surface sterilized seeds of Arabidopsis thaliana were sown (~10 seeds) on sterile soilrite and pots were transferred to temperature controlled culture room (set at 22°C) in continuous light conditions after 3 d of cold treatment. After 7 d of germination, a 48 h grown culture of Pseudomonas putida MTCC 5279 resuspended in 0.85% saline (final density of 109 CFU ml−1), was used for inoculating the plants to maintain the moisture up to 40%. The control set were inoculated with same amount of sterile saline water. Plants were irrigated weekly with the nutrient solution and the plants were grown for 6 weeks. Plant growth promotional effect of the Arabidopsis thaliana plants grown as control and treated sets were evaluated by taking the root length, shoot length, number of branches, number of siliques and dry weight of the four plants from each replicates and each treatment has six replicates. Rhizosphere and phyllosphere colonization of MTCC5279 was evaluated as described earlier.65
RNA extraction and microarray analysis
Rosette leaves of 6 week old Arabidopsis thaliana plants were stored in RNA laterTM (Qiagen) and stored at -80°C. Preserved leaf material of control and treated samples was used for the isolation of total RNA using RNeasy mini plant kit (Qiagen) as per manufacturer’s instructions. Quality of total RNA was assessed spectrophotometrically (NanoDrop) and integrity was checked using agilent bioanalyzer (Agilent Technologies).
Ten micrograms of the total RNA was converted to cRNA using ExpressArt aminoallyl mRNA amplification Kit (AmpTec GmbH) as per the manufacturer’s instruction. Amplified aminoallyl cRNA was purified and 20 µg was coupled with fluorescent Cy3 and Cy5 dye for a dual channel experiment with dye swap. Fifteen µg of labeled control and treated samples were mixed in 100 µl of the hybridization buffer and was hybridized to Arabidopsis thaliana 25 K OciChip™ (Ocimum Biosolutions Ltd.) on the automated Hyb station (Tecan HS4800 Pro; Tecan Groups Ltd).
Data analyses
The 24652 probes on Arabidopsis thaliana 25 K OciChip™ were used to compare across samples and MAD scaling was performed on each sample.66 Differentially expressed genes were identified by outlier detection of a contaminated bivariate distribution.67 To determine the differentially expressed candidate genes from these outliers, the fold-change value were calculated providing up and downregulated genes across the samples in respect to control. The biological significance and functional classification of differentially expressed genes was performed using Gene Ontology.
Semi-quantitative RT-PCR analysis
Validation of the transcriptome profiling experiments was performed by semiquantitative RT-PCR on selected candidate genes identified from the microarray experiments and some of the selected genes from others report.20 Total RNA for RT-PCR verification was extracted from the liquid nitrogen frozen plant materials (control and treated) from the same set of experimental plant leaves. Total RNA was extracted with the RNeasy plant mini kit (Qiagen Sciences), according to the manufacturer’s instructions using on-column DNaseI digestion. RNA preparations were first reverse transcribed for first-strand cDNA synthesis using oligo-dT primer using revertaid Hminus first strand cDNA synthesis kit (Fermentas). Subsequently, cDNA made were used for PCRs with the gene of interest by adding the corresponding gene specific primer combinations. The genes were amplified for 28 cycles in a PCR program, 95°C for 3 min followed by 94°C for 30 sec, 30 sec at the annealing temps (52°C-63°C), and 30 sec for 72°C followed by the final extension of 72°C for 10 min (Table 3).
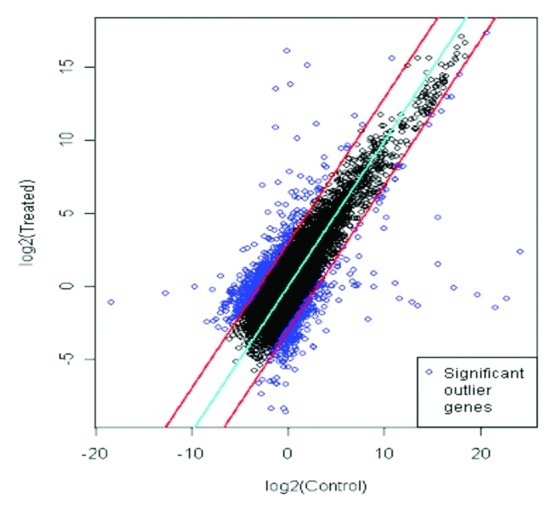
Figure 2. Scatter plot of differentially expressed genes in Arabidopsis plants with and without inoculation with MTCC5279. The upper and lower lines demarcate the limits of 3-fold differences in expression levels, dots outside the lines identify 882 genes differentially expressed between with and without inoculation of Pseudomonas putida MTCC5279 in Arabidopsis thaliana plants.
Figure 3. Functional classification of the upregulated and downregulated Arabidopsis thaliana genes in terms of their geneontology (GO terms), relative to their representation in the genome involved in plant growth promotion of Arabidopsis thaliana by Pseudomonas putida MTCC5279.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are indebted to Prof S.K. Sopory, Vice Chancellor, Jawahar al Nehru University, New Delhi, India for his valuable suggestions and critical editing of the manuscript. The study was partially supported by TATA Innovation Fellowship, Department of Biotechnology, Government of India, New Delhi and Task Force grant NWP-006 from Council of Scientific and Industrial Research (CSIR), New Delhi awarded to C.S.N.
Glossary
Abbreviations:
- MTCC5279
Pseudomonas putida MTCC5279
- PGPR
plant-growth-promoting rhizobacteria
- IAA
indole-3-acetic acid
- ACCS
1-aminocyclopropane-1-carboxylase synthase
- TopII
topoisomerase II
- HDH
histidinol dehydrogenases
- ABA
abcissic acid
- ET
ethylene
- JAR1
jasmonate responsive gene
- PRH75
plant RNA helicase 75
- MAD
median absolute deviation
- ISR
induced systemic resistance
- JA
jasmonic acid
- SA
salicylic acid
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18957
References
- 1.Barea JM, Pozo MJ, Azco´n R, Azco´n-Aguilar C. Microbial co-operation in the rhizosphere. J Exp Bot. 2005;56:1761–78. doi: 10.1093/jxb/eri197. [DOI] [PubMed] [Google Scholar]
- 2.Loper JE. Influence of bacterial sources of indole-3-acetic acid on root elongation of sugar beet. Phytopathology. 1986;76:76–386. doi: 10.1094/Phyto-76-386. [DOI] [Google Scholar]
- 3.Idris EE, Iglesias DJ, Talon M, Borriss R. Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol Plant Microbe Interact. 2007;20:619–26. doi: 10.1094/MPMI-20-6-0619. [DOI] [PubMed] [Google Scholar]
- 4.Joo G-J, Kim Y-M, Kim J-T, Rhee I-K, Kim J-H, Lee I-J. Gibberellins-producing rhizobacteria increase endogenous gibberellins content and promote growth of red peppers. J Microbiol. 2005;43:510–5. [PubMed] [Google Scholar]
- 5.Orti´z-Castro R, Valencia-Cantero E, Lo´pez-Bucio J. Plant growth promotion by Bacillus megaterium involves cytokinin signaling. Plant Signal Behav. 2008;3:263–5. doi: 10.4161/psb.3.4.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pieterse CMJ, Van Pelt JA, Ton J, Parchmann S, Mueller MJ, Buchala AJ, et al. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol Mol Plant Pathol. 2000;57:123–34. doi: 10.1006/pmpp.2000.0291. [DOI] [Google Scholar]
- 7.Glick BR, Todorovic B, Czarny J, Cheng Z, Duan J, McConkey B. Promotion of plant growth by bacterial ACC deaminase. Crit Rev Plant Sci. 2007;26:227–42. doi: 10.1080/07352680701572966. [DOI] [Google Scholar]
- 8.Nautiyal CS. Selection of Chickpea-Rhizosphere-Competent Pseudomonas fluorescens NBRI1303 Antagonistic to Fusarium oxysporum f. sp. ciceri, Rhizoctonia bataticola and Pythium sp. Curr Microbiol. 1997;35:52–8. doi: 10.1007/s002849900211. [DOI] [Google Scholar]
- 9.de Freitas JR, Banerjee MR, Germida JJ. Phosphate-solubilizing rhizobacteria enhance the growth and yield but not phosphorus uptake of canola (Brassica napus L.) Biol Fertil Soils. 1997;24:358–64. doi: 10.1007/s003740050258. [DOI] [Google Scholar]
- 10.Dimkpa C, Weinand T, Asch F. Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 2009;32:1682–94. doi: 10.1111/j.1365-3040.2009.02028.x. [DOI] [PubMed] [Google Scholar]
- 11.Saleem M, Arshad M, Hussain S, Bhatti AS. Perspective of plant growth promoting rhizobacteria (PGPR) containing ACC deaminase in stress agriculture. J Ind Microbiol Biotechnol. 2007;34:635–48. doi: 10.1007/s10295-007-0240-6. [DOI] [PubMed] [Google Scholar]
- 12.van Loon LC, Bakker PA, Pieterse CM. Systemic resistance induced by rhizosphere bacteria. Annu Rev Phytopathol. 1998;36:453–83. doi: 10.1146/annurev.phyto.36.1.453. [DOI] [PubMed] [Google Scholar]
- 13.Heil M, Bostock RM. Induced systemic resistance (ISR) against pathogens in the context of induced plant defences. Ann Bot. 2002;89:503–12. doi: 10.1093/aob/mcf076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berg G. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol. 2009;84:11–8. doi: 10.1007/s00253-009-2092-7. [DOI] [PubMed] [Google Scholar]
- 15.Pieterse CM, van Wees SC, Hoffland E, van Pelt JA, van Loon LC. Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell. 1996;8:1225–37. doi: 10.1105/tpc.8.8.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Wees SC, Pieterse CM, Trijssenaar A, Van ’t Westende YA, Hartog F, Van Loon LC. Differential induction of systemic resistance in Arabidopsis by biocontrol bacteria. Mol Plant Microbe Interact. 1997;10:716–24. doi: 10.1094/MPMI.1997.10.6.716. [DOI] [PubMed] [Google Scholar]
- 17.Knoester M, Pieterse CM, Bol JF, Van Loon LC. Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol Plant Microbe Interact. 1999;12:720–7. doi: 10.1094/MPMI.1999.12.8.720. [DOI] [PubMed] [Google Scholar]
- 18.Li W, Estrada-de los Santos P, Matthijs S, Xie GL, Busson R, Cornelis P, et al. Promysalin, a salicylate-containing Pseudomonas putida antibiotic, promotes surface colonization and selectively targets other Pseudomonas. Chem Biol. 2011;18:1320–30. doi: 10.1016/j.chembiol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Wu X, Monchy S, Taghavi S, Zhu W, Ramos J, van der Lelie D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol Rev. 2011;35:299–323. doi: 10.1111/j.1574-6976.2010.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Ohara Y, Nakayashiki H, Tosa Y, Mayama S. Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol Plant Microbe Interact. 2005;18:385–96. doi: 10.1094/MPMI-18-0385. [DOI] [PubMed] [Google Scholar]
- 21.Paszkowski U. A journey through signaling in arbuscular mycorrhizal symbioses 2006. New Phytol. 2006;172:35–46. doi: 10.1111/j.1469-8137.2006.01840.x. [DOI] [PubMed] [Google Scholar]
- 22.Johnson JM, Oelmüller R. Mutualism or parasitism: life in an unstable continuum. Endocytobiosis Cell Res. 2009;19:81–111. [Google Scholar]
- 23.Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;444:139–58. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 24.Tuteja N. Integrated calcium signaling in plants, In Signaling in plants, Baluska F and Mancuso S, eds. (Springer-Verloag Press, Germany) 2009; 29–49. [Google Scholar]
- 25.Newman EI, Reddel P. The distribution of mycorrhizas among families of vascular plants. New Phytol. 1987;106:745–51. doi: 10.1111/j.1469-8137.1987.tb00175.x. [DOI] [PubMed] [Google Scholar]
- 26.Prikryl Z, Vancura V, Wurst M. Auxin formation by rhizosphere bacteria as a factor of root growth. Biol Plant. 1985;27:159–63. doi: 10.1007/BF02902155. [DOI] [Google Scholar]
- 27.Bottini R, Cass´n F, Piccoli P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl Microbiol Biotechnol. 2004;65:497–503. doi: 10.1007/s00253-004-1696-1. [DOI] [PubMed] [Google Scholar]
- 28.Lo´pez-Bucio J, Campos-Cuevas JC, Hern´ndez-Caldero´n E, Vel´squez-Becerra C, Fari´as-Rodri´guez R, Maci´as-Rodri´guez LI, et al. Bacillus megaterium rhizobacteria promote growth and alter root-system architecture through an auxin- and ethylene-independent signaling mechanism in Arabidopsis thaliana. Mol Plant Microbe Interact. 2007;20:207–17. doi: 10.1094/MPMI-20-2-0207. [DOI] [PubMed] [Google Scholar]
- 29.Bloemberg GV, Lugtenberg BJJ. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol. 2001;4:343–50. doi: 10.1016/S1369-5266(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 30.Espinosa-Urgel M. Plant-associated Pseudomonas populations: molecular biology, DNA dynamics, and gene transfer. Plasmid. 2004;52:139–50. doi: 10.1016/j.plasmid.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 31.Desbrosses G, Contesto C, Varoquaux F, Galland M, Touraine B. PGPR-Arabidopsis interactions is a useful system to study signaling pathways involved in plant developmental control. Plant Signal Behav. 2009;4:321–3. doi: 10.4161/psb.4.4.8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuvier O, Hirano T. A role of topoisomerase II in linking DNA replication to chromosome condensation. J Cell Biol. 2003;160:645–55. doi: 10.1083/jcb.200209023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh BN, Mudgil Y, Sopory SK, Reddy MK. Molecular characterization of a nuclear topoisomerase II from Nicotiana tabacum that functionally complements a temperature-sensitive topoisomerase II yeast mutant. Plant Mol Biol. 2003;52:1063–76. doi: 10.1023/A:1025427700337. [DOI] [PubMed] [Google Scholar]
- 34.Mo X, Zhu Q, Li X, Li J, Zeng Q, Rong H, et al. The hpa1 mutant of Arabidopsis reveals a crucial role of histidine homeostasis in root meristem maintenance. Plant Physiol. 2006;141:1425–35. doi: 10.1104/pp.106.084178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Z, Wen CK, Binder BM, Chen YF, Chang J, Chiang YH, et al. Heteromeric interactions among ethylene receptors mediate signaling in Arabidopsis. J Biol Chem. 2008;283:23801–10. doi: 10.1074/jbc.M800641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, et al. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem. 2010;285:26532–44. doi: 10.1074/jbc.M110.111021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Verhagen BWM, Glazebrook J, Zhu T, Chang H-S, van Loon LC, Pieterse CMJ. The transcriptome of rhizobacteria-induced systemic resistance in arabidopsis. Mol Plant Microbe Interact. 2004;17:895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- 38.Tuteja N, Mahajan S. Calcium signaling network in plants: an overview. Plant Signal Behav. 2007;2:79–85. doi: 10.4161/psb.2.2.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vadassery J, Oelmüller R. Calcium signaling in pathogenic and beneficial plant microbe interactions: what can we learn from the interaction between Piriformospora indica and Arabidopsis thaliana. Plant Signal Behav. 2009;4:215–6. doi: 10.4161/psb.4.11.9800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi HI, Park HJ, Park JH, Kim S, Im MY, Seo HH, et al. Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, a transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 2005;139:1750–61. doi: 10.1104/pp.105.069757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou R, Cutler AJ, Ambrose SJ, Galka MM, Nelson KM, Squires TM, et al. A new abscisic acid catabolic pathway. Plant Physiol. 2004;134:361–9. doi: 10.1104/pp.103.030734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuteja N. Abscisic Acid and abiotic stress signaling. Plant Signal Behav. 2007;2:135–8. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12:343–51. doi: 10.1016/j.tplants.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 44.Matakiadis T, Alboresi A, Jikumaru Y, Tatematsu K, Pichon O, Renou J-P, et al. The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 2009;149:949–60. doi: 10.1104/pp.108.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, et al. Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 2004;134:1439–49. doi: 10.1104/pp.103.037614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E. Integration of abscisic acid signalling into plant responses. Plant Biol (Stuttg) 2006;8:314–25. doi: 10.1055/s-2006-924120. [DOI] [PubMed] [Google Scholar]
- 47.Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, et al. CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cartieaux F, Contesto C, Gallou A, Desbrosses G, Kopka J, Taconnat L, et al. Simultaneous interaction of Arabidopsis thaliana with Bradyrhizobium Sp. strain ORS278 and Pseudomonas syringae pv. tomato DC3000 leads to complex transcriptome changes. Mol Plant Microbe Interact. 2008;21:244–59. doi: 10.1094/MPMI-21-2-0244. [DOI] [PubMed] [Google Scholar]
- 49.Ton J, Davison S, Van Wees SC, Van Loon L, Pieterse CM. The arabidopsis ISR1 locus controlling rhizobacteria-mediated induced systemic resistance is involved in ethylene signaling. Plant Physiol. 2001;125:652–61. doi: 10.1104/pp.125.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuominen H, Overmyer K, Keinänen M, Kollist H, Kangasjärvi J. Mutual antagonism of ethylene and jasmonic acid regulates ozone-induced spreading cell death in Arabidopsis. Plant J. 2004;39:59–69. doi: 10.1111/j.1365-313X.2004.02107.x. [DOI] [PubMed] [Google Scholar]
- 51.Verhagen BWM, Trotel-Aziz P, Couderchet M, Höfte M, Aziz A. Pseudomonas spp.-induced systemic resistance to Botrytis cinerea is associated with induction and priming of defence responses in grapevine. J Exp Bot. 2010;61:249–60. doi: 10.1093/jxb/erp295. [DOI] [PubMed] [Google Scholar]
- 52.Stein E, Molitor A, Kogel KH, Waller F. Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant Cell Physiol. 2008;49:1747–51. doi: 10.1093/pcp/pcn147. [DOI] [PubMed] [Google Scholar]
- 53.Lorkovic´ ZJ, Herrmann RG, Oelmüller R. PRH75, a new nucleus-localized member of the DEAD-box protein family from higher plants. Mol Cell Biol. 1997;17:2257–65. doi: 10.1128/mcb.17.4.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kushwaha HR, Singh AK, Sopory SK, Singla-Pareek SL, Pareek A. Genome wide expression analysis of CBS domain containing proteins in Arabidopsis thaliana (L.) Heynh and Oryza sativa L. reveals their developmental and stress regulation. BMC Genomics. 2009;10:200. doi: 10.1186/1471-2164-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanan-Mishra N, Pham XH, Sopory SK, Tuteja N. Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc Natl Acad Sci U S A. 2005;102:509–14. doi: 10.1073/pnas.0406485102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krizek BA, Meyerowitz EM. The Arabidopsis homeotic genes APETALA3 and PISTILLATA are sufficient to provide the B class organ identity function. Development. 1996;122:11–22. doi: 10.1242/dev.122.1.11. [DOI] [PubMed] [Google Scholar]
- 57.Zik M, Irish VF. Global identification of target genes regulated by APETALA3 and PISTILLATA floral homeotic gene action. Plant Cell. 2003;15:207–22. doi: 10.1105/tpc.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, et al. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci U S A. 2004;101:10596–601. doi: 10.1073/pnas.0404110101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minorsky PV. A gene involved in homologous recombination. Plant Physiol. 2005;139:561–2. doi: 10.1104/pp.104.900175. [DOI] [Google Scholar]
- 60.Guo Y, Gan S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006;46:601–12. doi: 10.1111/j.1365-313X.2006.02723.x. [DOI] [PubMed] [Google Scholar]
- 61.Kunieda T, Mitsuda N, Ohme-Takagi M, Takeda S, Aida M, Tasaka M, et al. NAC family proteins NARS1/NAC2 and NARS2/NAM in the outer integument regulate embryogenesis in Arabidopsis. Plant Cell. 2008;20:2631–42. doi: 10.1105/tpc.108.060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Camehl I, Sherameti I, Venus Y, Bethke G, Varma A, Lee J, et al. Ethylene signalling and ethylene-targeted transcription factors are required to balance beneficial and nonbeneficial traits in the symbiosis between the endophytic fungus Piriformospora indica and Arabidopsis thaliana. New Phytol. 2010;185:1062–73. doi: 10.1111/j.1469-8137.2009.03149.x. [DOI] [PubMed] [Google Scholar]
- 63.Bric JM, Bostock RM, Silverstone SE. Rapid in situ assay for indoleacetic Acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol. 1991;57:535–8. doi: 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mehta S, Nautiyal CS. An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr Microbiol. 2001;43:51–6. doi: 10.1007/s002840010259. [DOI] [PubMed] [Google Scholar]
- 65.Nautiyal CS, Rehman A, Chauhan PS. Environmental Escherichia coli occur as natural plant growth-promoting soil bacterium. Arch Microbiol. 2010;192:185–93. doi: 10.1007/s00203-010-0544-1. [DOI] [PubMed] [Google Scholar]
- 66.Bengtsson A, Bengtsson H. Microarray image analysis: background estimation using quantile and morphological filters. BMC Bioinformatics. 2006;7:96. doi: 10.1186/1471-2105-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loguinov AV, Mian IS, Vulpe CD. Exploratory differential gene expression analysis in microarray experiments with no or limited replication. Genome Biol. 2004;5:R18. doi: 10.1186/gb-2004-5-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.