Abstract
The development of the angiosperm seed includes the accumulation of storage products, the loss of most of its water and the establishment of dormancy. While much is known about the pathways that initiate maturation during mid-embryogenesis or repress it after germination, only recently has it been shown that other mechanisms repress the program during early embryogenesis. Two recent reports have shown that microRNAs are critical regulators of maturation in Arabidopsis early embryogenesis. Two closely related trihelix transcription factors, ASIL1 and ASIL2, were identified as probable partially redundant repressors of early maturation downstream of the microRNA-synthesizing enzyme DICER-LIKE1. An overlap between the genes upregulated in asil1-1 and dcl1-15 mutants support this conclusion. ASIL2 orthologs are found across seed plants, indicating that their role in maturation might be conserved. ASIL1 arose from the ancestral ASIL2 clade by a gene duplication event in the Brassicaceae, although it is not clear whether its function has diverged.
Keywords: Arabidopsis, embryogenesis, maturation, microRNA, trihelix transcription factor
Seed maturation encompasses a set of processes that lead to the accumulation of storage products, desiccation tolerance and dormancy.1 The onset of the seed maturation program in Arabidopsis occurs around the heart stage of embryogenesis. A number of transcriptional regulators have been identified that promote maturation. Central to the process appear to be the B3 domain transcription factors ABA INSENSITIVE3 (ABI3), LEAFY-COTYLEDON2 (LEC2) and FUSCA3 (FUS3), as well as two proteins that are B subunits of the NF-Y family of trimeric transcription factors [LEC1 (also called NF-YB9)] and LEC1-LIKE (L1L, NF-YB6). Other transcriptions factors from the bZIP, MYB and AGL families, and the hormone ABA have also been implicated, directly or indirectly, in positively regulating the process.2
It is known that the expression and accumulation of storage products and other embryonic traits are actively repressed after germination. Most of the repressors that have been described so far involve proteins that downregulate transcription by acting at the chromatin level: histone deacetylases (HDA6/SIL1 and HDA19)3, SWI/SNF2 chromatin remodelers (BRAHMA, BUSHY GROWTH and ATSWI3c)4, Polycomb group proteins (SWINGER, CURLY LEAF, EMBRYONIC FLOWER2 and FERTILIZATION INDEPENDENT ENDOSPERM)5,6 and Trithorax group proteins (PICKLE and PICKLE-RELATED)7. Only a handful of transcription factors that repress the expression of maturation genes have been described so far: the B3 domain proteins VP1/ABI3-LIKE1 (VAL1, also called HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENE2 or HSI2) and VAL2/HSL1, and the trihelix protein ARABIDOPSIS 6B-INTERACTING PROTEIN1-LIKE1 (ASIL1).8-10
Because maturation does not start until mid-embryogenesis, it was natural to assume that there were also unknown factors preventing its premature onset. We and David Bartel’s lab recently showed that microRNAs (miRNAs) are key repressors of maturation during early embryogenesis.11,12 As early as the early globular stage, embryos homozygous for loss-of-function mutations in DICER-LIKE1 (DCL1) that significantly reduce or eliminate the synthesis of miRNAs also show the development of chloroplasts, accumulation of chlorophyll, and the synthesis of storage products. Nodine and Bartel11 showed that the upregulation of the miR156 targets SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE10 (SPL10) and SPL11 in dcl1 is partly responsible for this effect. Willmann et al.12 demonstrated that ASIL1, its closest homolog ASIL2, and HDA6/SIL1 redundantly repress maturation during early embryonic development. Early accumulation of chlorophyll is seen in asil1 and asil2 single mutants and is further enhanced in all three double mutant combinations of asil1, asil2, and sil1. The normal timing of maturation might be explained by the ability of these genes to repress maturation combined with the significantly decreasing expression of ASIL2 and SIL1 during embryonic development, as shown in a developmental time series microarray experiment from the Goldberg and Harada labs.12 The expression levels of these genes are likely indirectly controlled at least in part by miRNAs, because their expression is decreased in dcl1 embryos. Thus, it is likely that their expression is repressed directly or indirectly by a miRNA target; whether SPL10 and/or SPL11 could be this miRNA target remains unknown. The phenotypes of the double mutants of asil1, asil2 and sil1 are still weaker than that of dcl1, however, indicating that there are other factors downstream of miRNAs involved in this process, as well. Interestingly, the involvement of ASIL1, ASIL2 and SIL1 in repressing early maturation shows that at least some of the pathways involved in the post-embryonic repression of maturation function similarly during early embryogenesis.
To further explore the connections between DCL1 and these genes, we compared microarray data sets generated for dcl1-15 (a strong allele) and asil1-1 (probably a null allele).9,12 If miRNAs and ASIL1 regulate the same processes, we expect to see a significant overlap between them, particularly between the sets of upregulated genes. We expected the comparisons to be noisy, though, as we are comparing microarrays generated from torpedo stage embryos (dcl1-15) and two-week-old seedlings treated with ABA (asil1-1) (Fig. 1). In spite of that, we saw a highly significant overlap between the sets of genes that are upregulated in both mutants. This analysis strengthens the argument that miRNAs and ASIL1 both repress maturation.
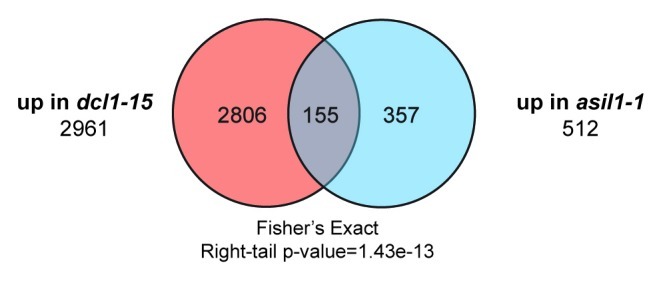
Figure 1. Comparison of microarray data for dcl1-15 and asil1-1. The overlaps were generated using Partek Genomics Suite 6.5. The statistical test used to show significance of the overlap was a Fisher’s exact test.
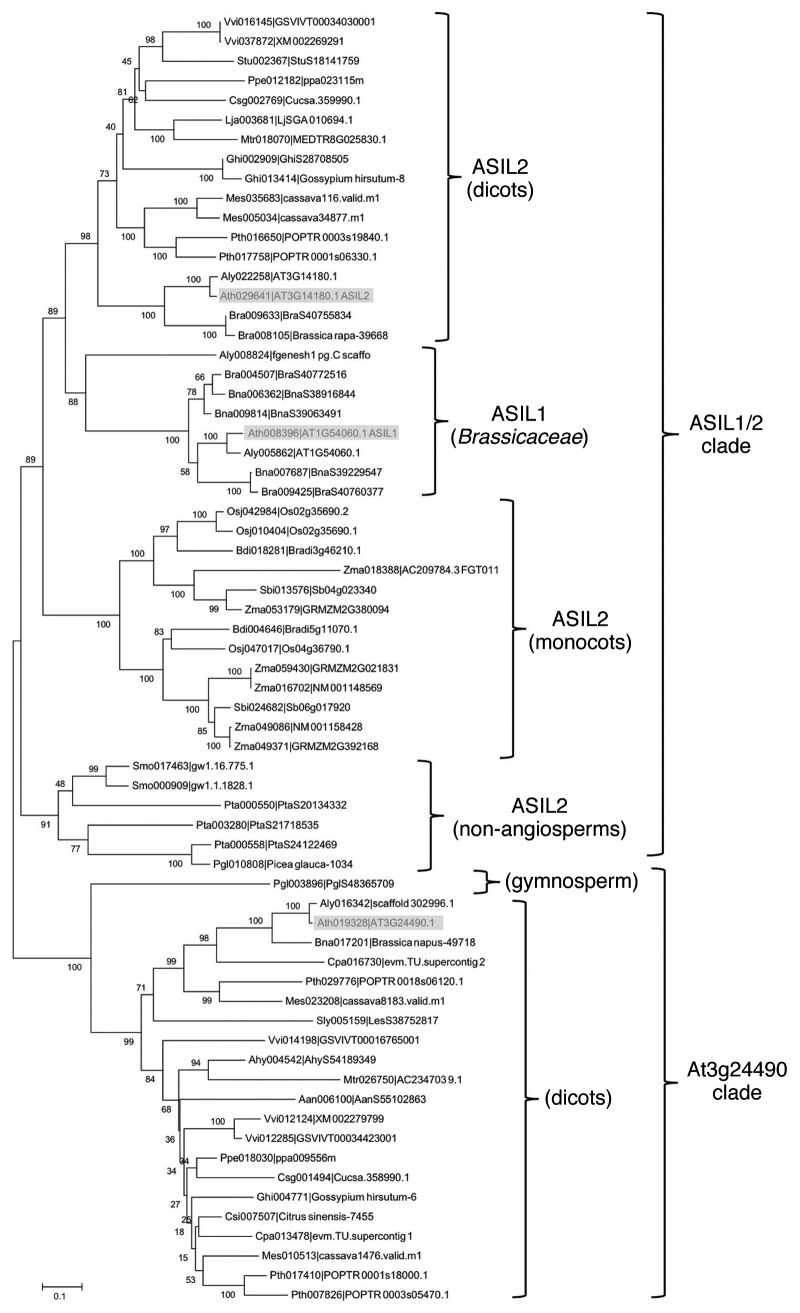
The trihelix family of transcription factors, to which ASIL1 and ASIL2 belong, is specific to the land plant lineage. There are 33 members in Arabidopsis.9 To better understand the relationships of ASIL1 and ASIL2 with the rest of the trihelix family, we performed a BLAST search within the Plant Transcription Factors Database (PlantTFDB) (http://planttfdb.cbi.pku.edu.cn/)13 using their protein sequences. We picked all the sequences matching the full-length protein with high scores (> 200, e-value < 10-15). A phylogenetic tree derived from the alignment of the full-length proteins shows four distinct, well-supported clades: one each for the orthologs of the Arabidopsis genes At3g24490, At3g58630 and At3g11100/At5g05550 and one that includes both ASIL1 and ASIL2 (ASIL1/2) (not shown). These four clades were also found, with various levels of support, by previous analyses of Arabidopsis and rice trihelix family transcription factors.9,14 To increase the support of the nodes, we re-built our tree using just the sister clades ASIL1/2 and At3g24490 (Fig. 2). The tree shows that there are ASIL2 orthologs in dicots, monocots, gymnosperms, and lycophytes. The PlantTFDB contains 41 trihelix transcription factors from the bryophyte Physcomitrella patens, but none of them fall into these clades. ASIL1 orthologs, however, are only present in the Brassicaceae, suggesting a gene duplication event in the earliest ancestor of this family. Although nothing is known yet about the function of the ASIL1/2 orthologs in other plants, it is possible that they are involved in maturation as well. Interestingly, there are orthologs of the other regulators of maturation (VAL genes, FUS3/LEC2 and LEC1) in all lineages of land plants, including seedless plants.15 One wonders what their ancestral role was, before they were co-opted for the regulation of seed maturation. Protection against abiotic stress is an intriguing possibility because ABA and ABI3, which are important for seed maturation in Arabidopsis, are involved in desiccation tolerance in the moss Physcomitrella patens.16
Figure 2. Neighbor-joining phylogenetic tree for the ASIL1/2 and At3g24490 clades. Full-length protein sequences were aligned using ClustalW, and the tree was built using MEGA4. For each protein, the first number is the one assigned by the PlantTFDB, while the second number is the one assigned by the corresponding sequencing project. Arabidopsis genes are highlighted in gray boxes. Species are identified as follow: Aan, Artemisia annua; Ahy, Arachis hypogaea; Ath, Arabidopsis thaliana; Aly, Arabidopsis lyrata; Bdi, Brachypodium distachyon; Bna, Brassica napus; Bra, Brassica rapa; Cpa, Carica papaya; Csg, Cucumis sativus; Csi, Citrus sinensis; Ghi, Gossypium hirsutum; Ppe, Prunus persica; Lja, Lotus japonicas; Mes, Manihot esculenta; Mtr, Medicago truncatula; Osj, Oryza sativa subsp Japonica; Pta, Pinus taeda; Pgl, Picea glauca; Pth, Populus trichocarpa; Sbi, Sorghum bicolor; Sly, Solanum lycopersicon; Smo, Selaginella moellendorffii; Stu, Solanum tuberosum; Vvi, Vitis vinifera; Zma, Zea mays.
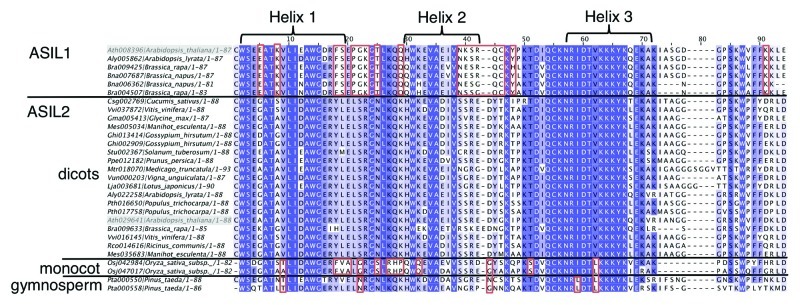
We selected representatives of the ASIL1/2 clade and aligned their full-length protein sequences to see whether there are lineage-specific differences. We found that there are several positions in the N-terminal proline-rich domain (not shown) and in the trihelix domain (specifically in helices 1 and 2) (Fig. 3) that consistently differ between the dicot representatives of ASIL1 and ASIL2. This suggests that the ASIL1 orthologs have significantly diverged from their ASIL2 counterparts since their duplication. There are also some positions in the trihelix domain that appear to be different in rice. Overall, the trihelix domains of the dicot ASIL2 proteins are more similar to those of rice and pine than to ASIL1 proteins. Because the trihelix domain is a DNA binding domain, these changes may result in changes in the DNA sequence recognized. However, the only known structure of a trihelix domain, that of the GT-1 protein of Arabidopsis, suggests that helix 3 interacts with the major groove of DNA and determines binding specificity.17 None of the observed ASIL1 and dicot ASIL2 lineage differences affect this particular helix. Further work will be needed to see whether these differences affect DNA binding properties or function.
Figure 3. Alignment of the trihelix domains of selected proteins of the ASIL1/2 clade. The trihelix domains were aligned using ClustalW. The position of the helices is based on.9 The numbers identifying the proteins correspond to those on Figure 2. Differences present in only one lineage (dicot, rice or pine) are boxed in red. Arabidopsis genes are highlighted in gray boxes.
Additional evidence of functional divergence of ASIL1 and ASIL2 comes from the Arabidopsis protein interactome recently built using a yeast two-hybrid system, which shows that they have largely different potential interactors.18 Although the biological significance of all the resulting interactions needs to be validated in planta, it is interesting to note that out of 63 putative interactors (48 for ASIL1 and 15 for ASIL2), only three proteins interacted with both of them: the protein kinase AKIN11/SnRK1.2 (At3g29160), the 60S ribosomal subunit RPL4A (At3g09630) and a transposase (At3g19120). There is also a provocative connection to maturation: an antisense line for an AKIN11/SnRK1.2 ortholog in pea showed defects in seed maturation.19 While we have yet no data to support a connection, it is tempting to speculate that SnRK1 kinase might act in part by negatively regulating ASIL1 or ASIL2 activity.
The available data so far indicate that the trihelix transcription factors ASIL1 and ASIL2 repress at least certain aspects of the maturation program: the accumulation of storage products (in embryos and seedlings), the development of chloroplasts (embryos), and the expression of regulators of the pathway (seedlings). Our data also support the idea that they mediate some of the effects of miRNAs in the embryos. The conservation of ASIL2 hints at the conservation of this repressive pathway across seed plants. Further experiments will help clarify their respective contributions and any divergences in roles between ASIL1 and ASIL2.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This project was funded by Franklin and Marshall College. M.L.B. was supported by an award to Franklin and Marshall College from the Howard Hughes Medical Institute's Undergraduate Science Education Program. M.R.W. was supported by an NIH NRSA postdoctoral fellowship. We would like to thank Jaime Blair (Franklin and Marshall College) for help with phylogenetic analyses and Jinpu Jin (PlantTFDB) for collating the trihelix protein sequences.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18893
References
- 1.Holdsworth MJ, Bentsink L, Soppe WJ. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 2.Vicente-Carbajosa J, Carbonero P. Seed maturation: developing an intrusive phase to accomplish a quiescent state. Int J Dev Biol. 2005;49:645–51. doi: 10.1387/ijdb.052046jc. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka M, Kikuchi A, Kamada H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008;146:149–61. doi: 10.1104/pp.107.111674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang X, Hou A, Babu M, Nguyen V, Hurtado L, Lu Q, et al. The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 2008;147:1143–57. doi: 10.1104/pp.108.121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouyer D, Roudier F, Heese M, Andersen ED, Gey D, Nowack MK, et al. Polycomb repressive complex 2 controls the embryo-to-seedling phase transition. PLoS Genet. 2011;7:e1002014. doi: 10.1371/journal.pgen.1002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, et al. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep. 2006;7:947–52. doi: 10.1038/sj.embor.7400760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aichinger E, Villar CBR, Farrona S, Reyes JC, Hennig L, Köhler C. CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet. 2009;5:e1000605. doi: 10.1371/journal.pgen.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsukagoshi H, Morikami A, Nakamura K. Two B3 domain transcriptional repressors prevent sugar-inducible expression of seed maturation genes in Arabidopsis seedlings. Proc Natl Acad Sci U S A. 2007;104:2543–7. doi: 10.1073/pnas.0607940104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao M-J, Lydiate DJ, Li X, Lui H, Gjetvaj B, Hegedus DD, et al. Repression of seed maturation genes by a trihelix transcriptional repressor in Arabidopsis seedlings. Plant Cell. 2009;21:54–71. doi: 10.1105/tpc.108.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki M, Wang HHY, McCarty DR. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 2007;143:902–11. doi: 10.1104/pp.106.092320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nodine MD, Bartel DP. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010;24:2678–92. doi: 10.1101/gad.1986710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willmann MR, Mehalick AJ, Packer RL, Jenik PD. MicroRNAs regulate the timing of embryo maturation in Arabidopsis. Plant Physiol. 2011;155:1871–84. doi: 10.1104/pp.110.171355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, et al. PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011;39(Database issue):D1114–7. doi: 10.1093/nar/gkq1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Y, Xie K, Hou X, Hu H, Xiong L. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol Genet Genomics. 2010;283:157–69. doi: 10.1007/s00438-009-0507-x. [DOI] [PubMed] [Google Scholar]
- 15.Banks JA, Nishiyama T, Hasebe M, Bowman JL, Gribskov M, dePamphilis C, et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science. 2011;332:960–3. doi: 10.1126/science.1203810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khandelwal A, Cho SH, Marella H, Sakata Y, Perroud P-F, Pan A, et al. Role of ABA and ABI3 in desiccation tolerance. Science. 2010;327:546. doi: 10.1126/science.1183672. [DOI] [PubMed] [Google Scholar]
- 17.Nagata T, Niyada E, Fujimoto N, Nagasaki Y, Noto K, Miyanoiri Y, et al. Solution structures of the trihelix DNA-binding domains of the wild-type and a phosphomimetic mutant of Arabidopsis GT-1: mechanism for an increase in DNA-binding affinity through phosphorylation. Proteins. 2010;78:3033–47. doi: 10.1002/prot.22827. [DOI] [PubMed] [Google Scholar]
- 18.Consortium A, Arabidopsis Interactome Mapping Consortium Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333:601–7. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Radchuk R, Radchuk V, Weschke W, Borisjuk L, Weber H. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 2006;140:263–78. doi: 10.1104/pp.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]