Abstract
HAIRY MERISTEM (HAM) proteins, members of the GRAS family of transcriptional regulators, are essential for maintenance of indeterminate growth in flowering plant shoots, loss-of-function ham mutants exhibiting a strikingly novel phenotype of shoot meristem arrest and differentiation. Specific cellular/molecular functions of HAM proteins underlying meristem maintenance are unknown. In this review, I highlight findings from recent analyses of Arabidopsis ham (Atham) loss-of-function phenotypes, including that HAM function limits the generation of clonally-derived meristem layers and that HAM function regulates CLAVATA3 expression. I consider how this new information both refines our understanding of the role of HAM proteins in regulating meristem structure and function, and may also suggest possible downstream HAM protein transcriptional targets. Finally, I note the significant phenotypic overlap between Atham phenotypes, and aintegumenta/anintegumenta-like6 double mutant phenotypes, suggesting meristem regulatory functions common to, and possible genetic interactions between, HAM and AINTEGUMENTA.
Keywords: AINTEGUMENTA, CLAVATA3, corpus, GRAS, HAM, indeterminacy, meristem, stem cell, tunica
Introduction
Vascular plants grow discontinuously throughout their life-spans, repeatedly initiating new shoot and root systems. This capacity for continuing organogenesis and growth throughout their life-spans, termed indeterminate growth or simply indeterminacy, permits plants to adaptively regulate their development in response to dynamic environments, which, as sessile organisms, they cannot relocate away from in response to adverse conditions. Indeterminate growth is also a fundamental aspect of the “life-strategy” of vascular plants, endowing woody perennials with the capacity for individuals to persist for thousands of years. A comprehensive understanding of the genetic basis of indeterminacy therefore ranks among the most fundamental goals of contemporary plant biology.
In 2002, Jeroen Stuurman, Fabienne Jäggi and Cris Kuhlemeier reported the characterization of a novel Petunia mutant defective in maintenance of shoot indeterminacy, which they named hairy meristem (ham).1 HAM function is required for maintenance of shoot indeterminacy in Petunia. Whereas wild-type Petunia plants produce as many as 19 leaves before transitioning to flowering, ham shoots typically exhibit cessation of both lateral organ and stem production (meristem arrest) following production of six to 14 leaves. Uniquely among genotypes exhibiting meristem arrest phenotypes,2-12 meristem arrest in ham mutants is accompanied by differentiation of the meristem, convincingly demonstrated by: progressive reduction in PhSHOOTMERISTEMLESS expression, a marker of meristem identity, within arrested ham apices; enlargement and vacuolization of internal cells of arrested ham shoot apices; and the appearance of typical stem trichomes on ham shoot apices following meristem arrest, the phenotype “hairy meristem” describes.
Exploiting the occasional reversion to wild type of transposon-tagged ham alleles, Stuurman and colleagues identified the HAM gene, which encodes a member of the GRAS family of transcriptional regulators.13-16 Despite ham loss-of-function alleles exhibiting differentiation at the meristem apex, HAM mRNA is confined to basal and peripheral regions of the meristem, most prominently in the provasculature, extending into initiating lateral organs. The absence of detectable HAM mRNA in the meristem apex suggests that HAM function entails non-cell-autonomous signaling from cells in the HAM expression domain of the basal meristem and/or lateral organ primordia, to more apical meristematic regions. Supporting this model, Stuurman and colleagues elegantly demonstrated, through analysis of HAM revertant sectors on ham mutant plants, that HAM expression in the basal meristem is likely sufficient to fully restore a wild-type phenotype.
HAM is therefore an essential component of a non-cell-autonomous signaling pathway required for maintenance of shoot meristem identity, and consequently for shoot indeterminacy. Yet despite the evident significance of HAM function to a central process of plant development, additional studies focusing upon HAM function in organ indeterminacy were not to appear for almost a decade following initial characterization of the ham mutant. How does HAM promote shoot meristem maintenance? What is the relationship of HAM function to other pathways regulating shoot meristem maintenance, such as the WUSCHEL(WUS)/CLAVATA(CLV) signaling pathway? Assuming (reasonably) that HAM is a transcription factor, what are HAM’s transcriptional targets? How is HAM function itself regulated at transcriptional, post-transcriptional and post-translational levels? Addressing these and additional questions would be greatly facilitated by translating HAM analysis from Petunia into Arabidopsis. Relative to Petunia, Arabidopsis offers a wider range of mutant genotypes for molecular genetic analysis of HAM function, along with visual reporters of gene expression and hormone signaling. Independent phylogenetic analyses of GRAS family proteins consistently identify three Arabidopsis genes, At2g45160, At3g60630 and At4g00150, as orthologs of Petunia HAM.14,15,17
In 2010, three studies independently reported phenotypes resulting from loss-of-function allele combinations of the three Arabidopsis HAM orthologs17-19 (AtHAMs, alternatively denoted SCARECROW-LIKE6 or LOST MERISTEMS). These studies collectively demonstrate that HAM protein function is required for maintenance of shoot indeterminacy in Arabidopsis, as well as Petunia, and therefore that HAM is likely to be a key component of meristem regulation across flowering plant diversity. Findings from these recent analyses of AtHAM function are significant not only in that they provide a foundation from which to launch molecular genetic analyses of HAM function, but also in that they provide new insights into the full range of HAM function. Rather than reviewing the full suite of known HAM functions, which encompass root as well as shoot development,17,18 this review will focus on, and attempt to integrate, aspects of the recent genetic analyses of AtHAM function that inform our understanding of how HAM proteins regulate shoot meristem maintenance, and thus shoot indeterminacy.
Flowering Plant Meristem Structure and Function
The shoot meristem is a population of undifferentiated cells located at shoot apices.20 In Arabidopsis, undifferentiated shoot meristem cells are marked by expression of the class I knox gene SHOOTMERISTEMLESS (STM),21 whose expression pattern in the shoot meristem is typical of class I knox gene expression across a wide spectrum of flowering plant diversity.1,22 Levels of STM expression in shoot meristems are non-uniform, exhibiting a gradient or bi-partite pattern of expression level, with the strongest expression at the meristem apex and a region of reduced expression in the more basal meristem, extending into the provasculature (Fig. 1A). STM expression is absent in lateral organ anlagen, thereby marking meristem cells fated to differentiate into stem tissue. Subtending the meristem, cells become enlarged and vacuolated relative to their meristematic precursors, and no longer express STM, consistent with differentiation into stem tissue. The boundary between meristem and subtending stem is typically quite distinct, the transition in cellular characteristics occurring across a span of one or a few cells (Fig. 1A). Factors regulating the transition from undifferentiated but differentiation-competent meristem cell, to differentiating stem tissue, are entirely obscure.
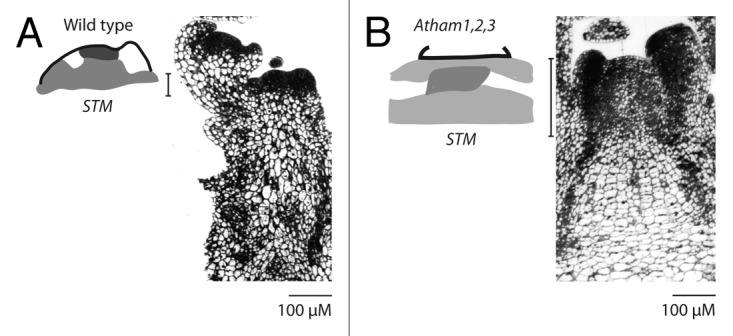
Figure 1.Atham1,2,3 inflorescence apices exhibit altered SHOOTMERISTEMLESS expression and a mixture of meristem and stem characters. (A) STM expression in a wild type vegetative primary shoot meristem, and median longitudinal section through a wild type primary inflorescence meristem. (B) STM expression in an Atham1,2,3 vegetative primary shoot meristem, and median longitudinal section through an Atham1,2,3 primary inflorescence meristem. Cartoons of STM expression are derived from the work of Schulze and colleagues.19 Darker gray levels reflect increased relative STM expression levels. Brackets to the left of longitudinal sections indicate the approximate depth of the zone in which cells are of typical meristematic dimensions. Cells of the Atham1,2,3 apex exhibit a mixture of characters typical of meristem cells (cell size), and of differentiating stem cells (vacuolization). Note that despite these abnormalities, the Atham1,2,3 meristem recently produced floral meristems.
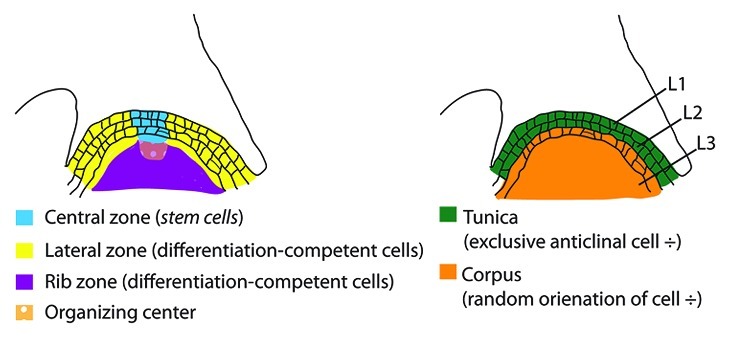
To achieve indeterminate growth, meristems must balance two competing functions: (1) maintenance of their population of totipotent, undifferentiated stem cells and (2) generation of differentiation-competent cells from stem cell precursors.23,24 [To avoid confusion, I refer to totipotent, undifferentiated cells as “stem cells”, in italics, while cells belonging to stem tissue are referred to as “stem cells,” without italics]. Both differentiation-competent cells and stem cells occupy well defined positions relative to one another along the apical-basal and central-peripheral axes of the stem (Fig. 2). Stem cells occupy the apical most several cell layers at the center of the stem apex, a region histologically defined by slow rates of cell division relative to surrounding meristem cells, and designated the central zone. As cells are displaced laterally or basally from the central zone, into the lateral and rib zones respectively, their rates of cell division increase and they acquire competence to undergo differentiation into stem tissue, or, in the lateral zone, to be co-opted for lateral organ development. The spatial relationship between indeterminate central zone and determinate lateral and rib zones, demonstrates that the developmental decision regulating the transition from stem cell identity to differentiation-competent cell is positionally determined, but significant gaps remain in our understanding of the inter-cellular signaling pathways utilized by meristem cells to assess their relative positions and to transduce positional information into appropriate cellular identities.
Figure 2. Structure of the flowering plant shoot meristem.
Does HAM Function Set the Meristem-Stem Boundary?
Both Petunia and Arabidopsis ham mutants exhibit differentiation of shoot meristems, with differentiating cells most likely acquiring stem identity.1,17 In Arabidopsis, meristem arrest and differentiation occur robustly in secondary meristems, but with incomplete penetrance in primary meristems, and, under long-day growth conditions, exclusively in the inflorescence.17-19 Arrested meristems of Petunia ham mutants exhibit greatly reduced STM expression levels,1 consistent with meristem differentiation. Therefore, HAM function is required to repress cellular differentiation within the meristem itself.
A complete loss of stem cells to differentiation must necessarily result in meristem arrest and loss of organ indeterminacy. But is meristem differentiation the primary or sole cause of meristem arrest? Or is generation of lateral organ and stem tissue compromised in Atham1,2,3 mutants independently of meristem differentiation?
Atham1,2,3 primary shoots exhibit abnormalities preceding full meristem arrest, including aberrant phyllotaxis, broadening and flattening of the shoot apex, and altered STM expression.17,19 Schulze and colleagues report STM expression in an Atham1,2,3 shoot apex, which is likely not to have undergone meristem arrest, judging from a recently initiated lateral organ.19STM expression in this Atham1,2,3 shoot apex is both reduced in expression level relative to wild type, and expanded spatially (Fig. 1B). The Atham1,2,3 STM expression maxima no longer occurs at the meristem apex, but is internalized, displaced basally relative to wild type by four or five cell layers, and STM expression extends deeper into the subtending shoot. This pattern of STM expression in Atham1,2,3 apices suggests repression of stem tissue differentiation and extended retention of an undifferentiated cell state. HAM function may be required for lateral and rib zone cells to transition from undifferentiated meristem cells to differentiating stem cells.
Consistent with a failure of lateral and rib zone cells to transition to stem differentiation in the Atham1,2,3 genotype, Atham1,2,3 shoot apices do not exhibit the distinct boundary between meristem and subtending, differentiating stem tissue (Fig. 1B). Instead, Atham1,2,3 shoot apices exhibit an expanded region in which cells exhibit meristematic cellular dimensions17,19 and STM expression.19 The possibility that Atham1,2,3 meristems retain a capacity for lateral organ initiation at a stage where production of stem tissue is impaired, could account for the deviations from regular phyllotactic patterning characteristic of Atham1,2,3 mutants (Fig. 1B).
ham phenotypes therefore suggest what may appear to be a paradoxical set of HAM functions; promoting differentiation, in the context of the meristem to stem transition, and inhibiting differentiation, in the context of the meristem. This paradox is largely resolved if HAM is modeled to function in the generation and/or maintenance of the boundary between the meristem and subtending stem, restricting both the acropetal transmission of hypothetical factors promoting differentiation, and the basipetal transmission of hypothetical factors promoting an undifferentiated cell state. Such a model of HAM protein function was earlier proposed by Goldschmidt and colleagues,25 and is similar to the proposed function for the closely related GRAS protein LATERAL SUPPRESSOR, which is expressed at meristem/lateral organ boundaries and mediates non-cell-autonomous signaling from differentiating lateral organs to shoot meristems. Consistent with a hybrid meristem/stem identity of Atham1,2,3 shoot apices, cell vacuolization in Atham1,2,3 apices occurs throughout the entire domain of meristematic cell dimensions at a frequency intermediate between the low levels of vacuolization observed in wild type meristems and the high levels of vacuolization observed in wild type stem cortex (Fig. 1B).17
If this model, by which HAM establishes or promotes the meristem-stem boundary, is correct, then meristem arrest is not necessarily a consequence of meristem differentiation in Atham1,2,3 mutants. Rather, meristem arrest and meristem differentiation can be uncoupled as causally distinct Atham1,2,3 phenotypes. Designing experiments to effectively further test this model may prove a major challenge of future investigations into HAM function.
Do Atham Mutants Provide A Window into the “Why” of Tunica-Corpus Organization?
Flowering plant shoot meristems maintain a structure of clonally distinct layers, resulting from the apical-most meristem cells undergoing cytokinesis in a strictly or predominantly anticlinal division plane26-28 (Fig. 2). Both Petunia and Arabidopsis shoot meristems are organized into three layers, Layer 1 (L1) designating the apical-most layer generated by anticlinal cell divisions, L2 designating an additional layer generated by anticlinal cell divisions immediately subtending the L1, and L3 designating the remainder of the meristem subtending the L2, in which planes of cell division become variable. Meristem layers generated by anticlinal cell divisions are collectively termed tunica, while the meristem region in which cell division planes are variable is termed corpus. Stratification of the shoot meristem into layers thereby generates lineage relationships in which specific tissues are derived from descendants of a specific meristem layer: for example shoot epidermis is derived exclusively from meristem L1 and gametophytes are derived exclusively from meristem L2.27 Tunica-corpus meristem organization is likely an ancestral trait of extant flowering plants.20
Although the tunica-corpus concept was first articulated by Schmidt in 1924,26 the functional significance of tunica-corpus organization remains poorly understood to this day. Why the meristem should be structured to generate lineage relationships among differentiating tissues is particularly puzzling in light of the fact that cell position, rather than cell lineage, is the principle determinant of cell differentiation until late in organogenesis.29,30
Atham1,2,3 mutants exhibit an expansion in the number of meristem cell layers generated by anticlinal cell divisions in the inflorescence apex under both long- and short-day growth conditions, and in the vegetative apex under short-day conditions, from the two tunica layers observed in wild type, to as many as seven readily distinguishable cell layers in Atham1,2,3 apices17,19 (Fig. 3). Expression of the L1 marker gene ATML1 remains confined to the outer-most layer in Atham1,2,3 apices, demonstrating that supernumerary layering of Atham1,2,3 shoot apices is a consequence of either expanded L2 identity, or of increased levels of anticlinal division in the apical corpus.19HAM function is therefore required to restrict exclusively anticlinal cell division to the apical-most two cell layers.
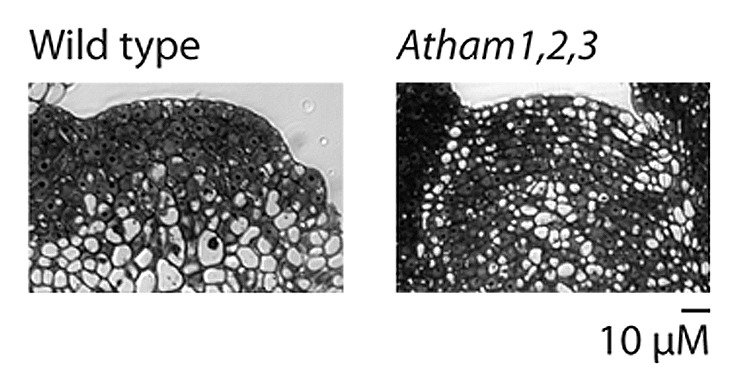
Figure 3. Wild type and Atham1,2,3 inflorescence apices. Supernumerary cell layering is evident in the Atham1,2,3 apex. Scale bar conforms to both wild type and Atham1,2,3 panels.
If organ and tissue abnormalities of Atham1,2,3 mutants are shown to be attributable to supernumerary layering in Atham1,2,3 apices, the functional correlation would suggest a developmental function for tunica-corpus organization. Atham1,2,3 cauline leaves are thicker than wild type, resulting from mesophyll expansion along the adaxial-abaxial axis.17 Might expansion of the Atham1,2,3 cauline leaf mesophyll result from supernumerary cell layering of the Atham1,2,3 inflorescence apex, leading to recruitment of what would amount to altered proportions of tunica/corpus into initiating cauline leaf primordia? Or does expansion of cauline leaf mesophyll result from increased anticlinal cell divisions within cauline leaf primordia following leaf initiation? It remains unclear if there are causal relationships between ectopic anticlinal cell divisions of Atham1,2,3 apices and other Atham1,2,3 phenotypes, including leaf thickness, meristem arrest and loss-of-indeterminacy, but future investigations of Atham1,2,3 mutants may provide significant insights into an enduring enigma, the function (or absence of function?) of tunica-corpus meristem organization in plant development.
HAM Proteins Regulate CLAVATA3 expression
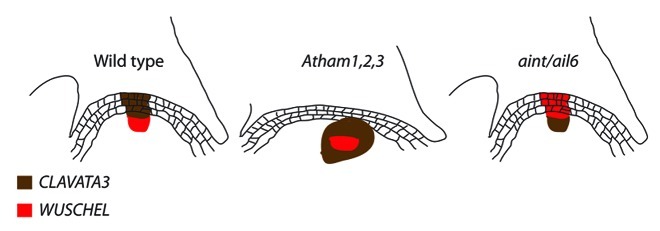
Maintenance of stem cell identity, the crux of organ indeterminacy, depends upon non-cell-autonomous signaling from meristem organizing centers.23,24 In the shoot apex, the organizing center is comprised of a small population of cells located within the rib meristem, adjacent and basal to the central zone and delineated by expression of the homeodomain transcription factor WUSCHEL (WUS)2 (Fig. 2). Recent evidence indicates that the non-cell-autonomous signal emanating from the organizing center and promoting stem cell identity in the central zone is the WUS protein itself, which translocates acropetally from the organizing center in a size-restricted manner consistent with trafficking through plasmodesmata.31 The region of WUS movement encompasses the entirety of the central zone, in which WUS directly activates transcription of CLAVATA3 (CLV3). CLV3 encodes the precursor of a mobile, 12-amino acid peptide signaling ligand that diffuses basally and laterally away from the central zone,32-34 binding to CLV1, an LRR-receptor kinase, and to the CLV2/CORYNE (CRN) LRR-receptor kinase complex,35,36 both localized to a region encompassing the basal central zone and rib meristem. Binding of CLV3 to CLV1 and CLV2/CRN both limits CLV3 diffusion, and represses WUS expression, thereby completing a feedback signaling loop from organizing center to central zone and back to the organizing center: WUS promotes CLV3 expression while CLV3 restricts WUS expression.31,33 Analysis of wus and clv loss-of-function phenotypes demonstrates that the WUS-CLV signaling pathway functions in both meristem maintenance and in regulation of meristem size.33,37
Specification and maintenance of WUS expression and organizing-center identity occurs in response to interpretation of cellular location along the apical-basal and central-peripheral axes of the plant body. WUS expression is promoted by cytokinins, likely synthesized within the meristem, when cytokinin levels exceed a critical concentration threshold.38-40 WUS in turn negatively regulates cytokinin signal transduction, generating a second feedback loop regulating WUS expression.41 Is superimposition of the WUS/CLV and WUS/cytokinin signaling loops sufficient to generate wild-type WUS expression at the apex of the rib zone? Modeling suggests that this may indeed be the case,38 but major questions remain with regard to the intercellular signaling pathways regulating CLV3 expression. WUS is observed to traffic into cells of the lateral zone adjoining the central zone, indicating that WUS activity alone is not sufficient to specify CLV3 expression. What factors in addition to WUS are necessary to specify CLV3 expression? What is the relationship between central zone stem cell identity and CLV3 expression? Finally, what restricts WUS to moving acropetally, rather than uniformly in all directions?
PhWUS expression in recently arrested meristems of Petunia ham mutants is comparable in location and expression level to wild type, suggesting that HAM function is not required to define wild-type WUS expression.1 Excellent in situ hybridization analyses by Schulze and colleagues show that WUS expression in the vegetative meristem of an Atham1,2,3 mutant grown in short-day conditions is comparable to wild-type WUS expression, though shifted slightly basipetally relative to wild type19 (although comparison of WUS expression in wild type and Atham1,2,3 mutants is complicated by the alterations to meristem structure in Atham1,2,3 mutants). However, in contrast to WUS, CLV3 expression in Atham1,2,3 meristems is both significantly enlarged and shifted basipitally relative to wild type, resulting in internalization of CLV3 expression, and in the superimposition of WUS and CLV3 expression (Fig. 4). HAM function is required to generate wild-type CLV3 expression.
Figure 4. Expression of WUSCHEL and CLAVATA3 in wild type, Atham1,2,3 and aintegumenta/aintegumentalike6 genotypes.
As the level of WUS expression appears to be minimally disrupted in Atham1,2,3 mutants, it is plausible to speculate that altered patterning of CLV3 expression in Atham1,2,3 mutants is not significantly a consequence of the amount of WUS expression. Alternative mechanisms by which HAM proteins might regulate CLV3 expression include regulating acropetal intercellular trafficking of WUS from the organizing center to the central zone, and/or regulating CLV3 expression via a WUS-independent pathway.
How is WUS movement away from the organizing center directed acropetally rather than uniformly in all directions? One possibility, following from the observation that WUS movement appears to occur via symplastic transport, is that plasmodesmata location itself channels WUS movement, that is, that there are no plasmodesmata located at organizing center cell surfaces through which WUS could move laterally or basally. As noted by Yadav and colleagues, defining meristem symplastic domains, which would address this model, is an important priority for future research.31
Alternatively, plasmodesmata may be available to facilitate WUS transport in multiple directions, but specific protein-protein interactions constrain WUS from moving other than acropetally. The HAM gene expression domain, roughly the inverse of the combined CLV3 and WUS expression domains, suggests that HAM proteins are localized appropriately to function in limiting WUS movement, a model that could be readily tested by comparison of GFP-WUS trafficking in wild type and Atham1,2,3 meristems. SCARECROW (SCR), a GRAS protein related to HAMs,42 regulates movement of the GRAS protein SHORT-ROOT (SHR), restricting SHR movement from the stele to the adjoining endodermis by sequestering SHR to the nucleus, thereby preventing transport through additional root cell layers.43 The SCR/SHR interaction is essential for wild-type root patterning, and demonstrates a precedent for GRAS protein function in limiting transcription factor transport through plasmodesmata. To my knowledge there are no known examples of GRAS proteins physically interacting with homeodomain transcription factors.
What of HAMs regulating CLV3 expression independently of WUS? Modeling of CLV3 expression suggests that an L1-derived intercellular signal acts in concert with WUS to generate wild-type CLV3 expression in the central zone.44 While this model currently has limited molecular genetic support, it is interesting to consider in view of Atham1,2,3 shoot phenotypes. As noted above, the generation of supernumerary cell layers in Atham1,2,3 apices does not coincide with an expansion of L1 identity, as expression of the L1 marker ATML1 remains restricted to the outermost cell layer in Atham1,2,3 shoot apices, as in wild type.19 However, the distance separating the L1 layer from the WUS expression domain is increased in Atham1,2,3 meristems relative to wild type,19 possibly as a result of the expansion in anticlinal cell divisions. If both basipetal transport of an L1-derived signal and acropetal transport of an organizing center-derived signal (WUS) are required for wild-type CLV3 expression, an increase in the distance separating L1 and the organizing center could result in altered expression of CLV3 similar to that observed in Atham1,2,3 meristems, particularly if WUS translocation is effected to a greater extent than transmission of the L1-derived signal.
Schulze and colleagues also examined expression of AINTENGUMENTA (ANT) in Atham1,2,3 apicies.19 AINTEGUMENTA regulates cell division during lateral organ development,45 and upregulation of ANT occurs early in the definition of lateral organ anlagen.46 ANT expression is also observed within the shoot meristem, at lower levels relative to organ anlagen, in a band of expression extending across the meristem apex.19 Wild-type expression of ANT within the shoot meristem may reflect meristematic ANT function(s). In a non-arrested Atham1,2,3 vegetative meristem, ANT expression is elevated and expanded, with expression still observed in a band of cells across the meristem apex, comparable in position to wild-type ANT expression, but extending deeper into the provasculature, and at a more uniform level of expression.19
Krizek recently reported that Arabidopsis doubly homozygous for null loss-of-function alleles of ANT and the related gene AINTEGUMENTA-LIKE6 (AIL6) exhibit altered localization of both WUS and CLV3 expression47 (Fig. 4). In both inflorescence and flower meristems of ant/ail double mutants, WUS expression is shifted acropetally, in the inflorescence meristem encompassing the region normally associated with CLV3 expression, while in flower meristems, WUS expression also expands further outward peripherally along the apical meristem flanks. CLV3 expression in ant/ail double mutant inflorescence meristems is both shifted basipetally and expanded laterally, with the maxima of CLV3 expression overlapping with the region in which WUS is expressed in wild-type meristems, very similar to CLV3 expression in Atham1,2,3 apices. WUS and CLV3 expression domains thus are essentially reversed in ant/ail double mutants, demonstrating that ANT and AIL6 function is, like HAM proteins, required for wild-type patterning of CLV3 expression. Notably, ant/ail6 double mutants exhibit loss of indeterminacy in the both the inflorescence and in agamous flowers.
Although the full extent to which ANT/AIL6 function in promoting shoot indeterminacy parallels that of HAM function remains unclear, comparison of Atham1,2,3 and ant/ail6 phenotypes suggests the possibilities both of a genetic interaction between HAM and ANT/AIL6, and of a causal relationship between CLV3 internalization and loss of meristem maintenance. Does internalization of CLV3 expression in Atham1,2,3 and ant/ail6 correspond to internalization of the meristem stem cell population? Or does internalization of CLV3 expression induce loss of stem cell identity? These questions may hold the key to answering the fundamental question of how HAM proteins promote organ indeterminacy.
Conclusions and Perspective
While Atham1,2,3 phenotypes support the conclusions of Stuurman and colleagues, that HAM proteins are required for shoot meristem maintenance and thereby for shoot indeterminacy, the expanded suite of ham shoot meristem phenotypes revealed by histological and gene expression analyses of Atham1,2,3, may appear to complicate, rather than facilitate, our ability to infer the underlying cellular/molecular basis for meristem arrest and loss-of-indeterminacy in ham mutants. How might repression of meristem differentiation, promotion of stem differentiation, restriction of anticlinal cell division within the meristem, and regulation of CLV3 expression share a common underlying regulatory mechanism? GRAS proteins function as transcription factors and regulation of gene expression by promoter binding or interactions with promoter binding proteins is well characterized for several GRAS proteins.48-51 Does the spectrum of Atham1,2,3 phenotypes contain a common motif that may suggest specific transcriptional targets of HAMs?
The transition from meristem cell to differentiating stem cell entails a switch from growth by cell division to growth by cell enlargement. Similarly, the distinction between central zone stem cells, which in wild-type meristems express CLV3, and differentiation-competent cells of the meristem lateral and rib zones, is one of relative rate of cell division, stem cells entering the cell cycle at lower frequency. A model by which HAM proteins regulate transcription of genes encoding regulators of cell cycle progression, such as cyclins and cyclin-dependent kinases, may account for the differentiation phenotypes of ham mutants, and possibly for the CLV3 internalization phenotype as well should Atham1,2,3 CLV3 expression prove to reflect ectopic localization of stem cell identity. What of the expansion of clonally distinct meristem layers in Atham1,2,3 mutants? Expansion of anticlinal cell division resulting from a loss-of-function suggests that extensive meristem layering may be an evolutionary default in flowering plants, and that HAM function is needed to promote periclinal cell divisions generating the corpus. In the root, the periclinal cell division responsible for generating the cortex and endodermal layers is regulated by a Cyclin D whose transcription is promoted by the GRAS protein SHR, demonstrating both a possible functional link between cell cycle progression and the orientation of cell division, and a precedent for transcriptional regulation of cell cycle regulatory genes by a GRAS protein.51 Regulation of cell cycle regulatory gene transcription by HAM proteins may plausibly account for supernumerary cell layering in Atham1,2,3 apices. Identification of transcriptional targets of HAM proteins is clearly a priority for future investigations of HAM function, and cell cycle regulatory genes are excellent candidates for examination.
The full power of Arabidospsis as a model system has yet to be brought to bear on the molecular genetic analysis of HAM function in plant development. Despite the considerable advantages of studying HAM function in Arabidopsis relative to Petunia, Arabidopsis confers the ironic disadvantage of functional redundancy among genetically unlinked AtHAMs, requiring double and triple mutant lines to generate loss-of-function phenotypes, a requirement that complicates both introgression of reporter fusion constructs, and the generation of multiple mutant lines for epistasis analysis. One potential means to sidestep the problem of AtHAM functional redundancy is provided by the fact that AtHAMs are targets of post-transcriptional regulation by microRNAs.52,53 MicroRNA regulation has been exploited for constructing microRNA-resistant HAM alleles,18 which may generate informative gain-of-function phenotypes, and for constructing genotypes constitutively overexpressing an HAM microRNA, which generates a ham loss-of-function phenotype with a much simpler heritability than the Atham1,2,3 mutant.54 With the HAM saga now securely established in Arabidopsis as well as Petunia, there is good reason to anticipate that the pace of discovery in our understanding of HAM function in plant development will increase dramatically relative to the past decade.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18958
References
- 1.Stuurman J, Jäggi F, Kuhlemeier C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002;16:2213–8. doi: 10.1101/gad.230702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laux T, Mayer KF, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 3.Waites R, Selvadurai HRN, Oliver IR, Hudson A. The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell. 1998;93:779–89. doi: 10.1016/S0092-8674(00)81439-7. [DOI] [PubMed] [Google Scholar]
- 4.Talbert PB, Adler HT, Parks DW, Comai L. The REVOLUTA gene is necessary for apical meristem development and for limiting cell divisions in the leaves and stems of Arabidopsis thaliana. Development. 1995;121:2723–35. doi: 10.1242/dev.121.9.2723. [DOI] [PubMed] [Google Scholar]
- 5.Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996;10:967–79. doi: 10.1046/j.1365-313X.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- 6.Smith HMS, Campbell BC, Hake S. Competence to respond to floral inductive signals requires the homeobox genes PENNYWISE and POUND-FOOLISH. Curr Biol. 2004;14:812–7. doi: 10.1016/j.cub.2004.04.032. [DOI] [PubMed] [Google Scholar]
- 7.Rutjens B, Bao D, van Eck-Stouten E, Brand M, Smeekens S, Proveniers M. Shoot apical meristem function in Arabidopsis requires the combined activities of three BEL1-like homeodomain proteins. Plant J. 2009;58:641–54. doi: 10.1111/j.1365-313X.2009.03809.x. [DOI] [PubMed] [Google Scholar]
- 8.Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–70. doi: 10.1016/S0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- 9.Weir I, Lu J, Cook H, Causier B, Schwarz-Sommer Z, Davies B. CUPULIFORMIS establishes lateral organ boundaries in Antirrhinum. Development. 2004;131:915–22. doi: 10.1242/dev.00993. [DOI] [PubMed] [Google Scholar]
- 10.Lee DK, Geisler M, Springer PS. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development. 2009;136:2423–32. doi: 10.1242/dev.031971. [DOI] [PubMed] [Google Scholar]
- 11.Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13:1768–74. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 12.Sarojam R, Sappl PG, Goldshmidt A, Efroni I, Floyd SK, Eshed Y, et al. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell. 2010;22:2113–30. doi: 10.1105/tpc.110.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN. The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999;18:111–9. doi: 10.1046/j.1365-313X.1999.00431.x. [DOI] [PubMed] [Google Scholar]
- 14.Bolle C. The role of GRAS proteins in plant signal transduction and development. Planta. 2004;218:683–92. doi: 10.1007/s00425-004-1203-z. [DOI] [PubMed] [Google Scholar]
- 15.Tian C, Wan P, Sun S, Li J, Chen M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol Biol. 2004;54:519–32. doi: 10.1023/B:PLAN.0000038256.89809.57. [DOI] [PubMed] [Google Scholar]
- 16.Lee MH, Kim B, Song SK, Heo JO, Yu NI, Lee SA, et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol Biol. 2008;67:659–70. doi: 10.1007/s11103-008-9345-1. [DOI] [PubMed] [Google Scholar]
- 17.Engstrom EM, Andersen CM, Gumulak-Smith J, Hu J, Orlova E, Sozzani R, et al. Arabidopsis homologs of the petunia hairy meristem gene are required for maintenance of shoot and root indeterminacy. Plant Physiol. 2011;155:735–50. doi: 10.1104/pp.110.168757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L, Mai YX, Zhang YC, Luo Q, Yang HQ. MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant. 2010;3:794–806. doi: 10.1093/mp/ssq042. [DOI] [PubMed] [Google Scholar]
- 19.Schulze S, Schäfer BN, Parizotto EA, Voinnet O, Theres K. LOST MERISTEMS genes regulate cell differentiation of central zone descendants in Arabidopsis shoot meristems. Plant J. 2010;64:668–78. doi: 10.1111/j.1365-313X.2010.04359.x. [DOI] [PubMed] [Google Scholar]
- 20.Steeves TA, Sussex IM. (1972) Patterns in Plant Development. Prentice Hall, Inc., Englewood Cliffs, New Jersey. [Google Scholar]
- 21.Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–9. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- 22.Jackson D, Veit B, Hake S. Expression of maize KNOTTED1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development. 1994;120:405–13. [Google Scholar]
- 23.Dinneny JR, Benfey PN. Plant stem cell niches: standing the test of time. Cell. 2008;132:553–7. doi: 10.1016/j.cell.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 24.Sablowski R. Plant stem cell niches: from signalling to execution. Curr Opin Plant Biol. 2011;14:4–9. doi: 10.1016/j.pbi.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Goldshmidt A, Alvarez JP, Bowman JL, Eshed Y. Signals derived from YABBY gene activities in organ primordia regulate growth and partitioning of Arabidopsis shoot apical meristems. Plant Cell. 2008;20:1217–30. doi: 10.1105/tpc.107.057877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt A. Histologische Studien an Phanerogamen Vegetationspunkten. Botany Archives. 1924;8:345–404. [Google Scholar]
- 27.Satina S, Blakeslee AF, Avery AG. Demonstration of the Three Germ Layers in the Shoot Apex of Datura by Means of Induced Polyploidy in Periclinal Chimeras. Am J Bot. 1940;27:895–905. doi: 10.2307/2436558. [DOI] [Google Scholar]
- 28.Foster AS, Gifford EM. Comparative Morphology of Vascular Plants, second edition. San Francisco: W.H. Freeman and Company, 1974.
- 29.Stewart RN, Burk LG. Independence of Tissues Derived from Apical Layers in Ontogeny of the Tobacco Leaf and Ovary. Am J Bot. 1970;57:1010–6. doi: 10.2307/2441000. [DOI] [Google Scholar]
- 30.Kessler S, Seiki S, Sinha N. Xcl1 causes delayed oblique periclinal cell divisions in developing maize leaves, leading to cellular differentiation by lineage instead of position. Development. 2002;129:1859–69. doi: 10.1242/dev.129.8.1859. [DOI] [PubMed] [Google Scholar]
- 31.Yadav RK, Perales M, Gruel J, Girke T, Jönsson H, Reddy GV. WUSCHEL protein movement mediates stem cell homeostasis in the Arabidopsis shoot apex. Genes Dev. 2011;25:2025–30. doi: 10.1101/gad.17258511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–4. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 33.Lenhard M, Laux T. Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development. 2003;130:3163–73. doi: 10.1242/dev.00525. [DOI] [PubMed] [Google Scholar]
- 34.Kondo T, Sawa S, Kinoshita A, Mizuno S, Kakimoto T, Fukuda H, et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science. 2006;313:845–8. doi: 10.1126/science.1128439. [DOI] [PubMed] [Google Scholar]
- 35.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–85. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 36.Bleckmann A, Weidtkamp-Peters S, Seidel CAM, Simon R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 2010;152:166–76. doi: 10.1104/pp.109.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoof H, Lenhard M, Haecker A, Mayer KFX, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–44. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 38.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci U S A. 2009;106:16529–34. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurakawa T, Ueda N, Maekawa M, Kobayashi K, Kojima M, Nagato Y, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–5. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 40.Veit B. Hormone mediated regulation of the shoot apical meristem. Plant Mol Biol. 2009;69:397–408. doi: 10.1007/s11103-008-9396-3. [DOI] [PubMed] [Google Scholar]
- 41.Leibfried A, To JPC, Busch W, Stehling S, Kehle A, Demar M, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–5. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- 42.Engstrom EM. Phylogenetic analysis of GRAS proteins from moss, lycophyte and vascular plant lineages reveals that GRAS genes arose and underwent substantial diversification in the ancestral lineage common to bryophytes and vascular plants. Plant Signal Behav. 2011;6:850–4. doi: 10.4161/psb.6.6.15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, et al. An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science. 2007;316:421–5. doi: 10.1126/science.1139531. [DOI] [PubMed] [Google Scholar]
- 44.Jonsson H, Shapiro BE, Meyerowitz EM, Mjolsness E. Signaling in multicellular models of plant development. In: S Kumar, PJ Bentley, eds. On Growth, Form and Computers. Great Britain: Elsevier Ltd. 2003. [Google Scholar]
- 45.Mizukami Y, Fischer RL. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc Natl Acad Sci U S A. 2000;97:942–7. doi: 10.1073/pnas.97.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–68. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krizek B. AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiol. 2009;150:1916–29. doi: 10.1104/pp.109.141119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fode B, Siemsen T, Thurow C, Weigel R, Gatz C. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell. 2008;20:3122–35. doi: 10.1105/tpc.108.058974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Lucas M, Davière JM, Rodri´guez-Falco´n M, Pontin M, Iglesias-Pedraz JM, Lorrain S, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–4. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 50.Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GED. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21:545–57. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sozzani R, Cui H, Moreno-Risueno MA, Busch W, Van Norman JM, Vernoux T, et al. Spatiotemporal regulation of cell-cycle genes by SHORTROOT links patterning and growth. Nature. 2010;466:128–32. doi: 10.1038/nature09143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–20. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 53.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–6. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 54.Song L, Axtell MJ, Fedoroff NV. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr Biol. 2010;20:37–41. doi: 10.1016/j.cub.2009.10.076. [DOI] [PubMed] [Google Scholar]