Abstract
Transcriptional co-activators of the multiprotein bridging factor 1 (MBF1) control gene expression by connecting transcription factors and the basal transcription machinery. In Arabidopsis thaliana functions of MBF1 genes have been related to stress tolerance and developmental alterations. Endogenous ABA plays a major role in the regulation of Arabidopsis seed dormancy and germination. Seed dormancy and ABA sensitivity are enhanced in ethylene insensitive mutants suggesting that ethylene signal transduction pathway is necessary to fully develop ABA-dependent germination. In this report we showed that a triple knock-down mutant for Arabidopsis MBF1 genes (abc-) has enhanced seed dormancy and displays hypersensitivity to exogenous ABA. In addition, higher ABA contents were detected in abc- seeds after imbibition. These evidences suggest a negative role of MBF1s genes in ABA-dependent inhibition of germination. The participation of MBF1s in ethylene signal transduction pathway is also discussed.
Keywords: ABA, ABA-ethylene cross talk, Arabidopsis thaliana, germination, transcriptional co-activators
Introduction
The multiprotein bridging factor 1 (MBF1) type controls gene expression by connecting transcription factors and the basal transcription machinery.1,2 In Arabidopsis thaliana there are three genes: AtMBF1a (At2g42680), AtMBF1b (At3g58680) and AtMBF1c (At3g24500), encoding MBF1 proteins.3 Several evidences relate AtMBF1 functions with tolerance to stress conditions.4-7 Analysis of AtMBF1c overexpressing plants and null mutant unraveled the role of MBF1c during osmotic stress and thermotolerance.5-7 Constitutive overexpression of AtMBF1a leads to elevated salt tolerance, insensitivity to glucose and resistance to fungal disease.4 Other reports link MBF1 functions with developmental alterations, such as plant size, leaf cell expansion and ploidy levels.8,9 In addition, a loss-of-function AtMBF1c mutant, showed a reduction on seed germination.8
Initially, MBF1 genes were related to ethylene signal transduction pathway. The first MBF1 gene was identified in tomato fruits and it was named ER24 by ethylene-responsive transcriptional co-activator.10 AtMBF1c overexpressing plants accumulate transcripts associated with ethylene signaling and exhibit a stronger triple-response phenotype.5 These results suggest that MBF1 genes are positive regulators of ethylene signaling.
We reported the analysis of an Arabidopsis thaliana triple knock-down mutant for MBF1 genes (abc-) under oxidative and osmotic stress conditions.11 We showed that abc- mutant seedlings were more sensitive than wild type (WT) to hydrogen peroxide (H2O2) and methyl viologen (MV). Inhibition of seed germination by oxidative treatments and osmotic stress was enhanced by the abc- mutation. In addition, we showed that AtMBF1s regulate the expression of the Abscisic Acid Repressor (ABR1) gene. ABR1 transcript levels were strongly reduced in the abc- mutant under normal conditions. WT seedlings treated with MV showed a reduction in ABR1 transcript levels meanwhile, abc- seedlings were unable to regulate ABR1 expression. ABR1 is a transcription factor of the Ethelyne Responsive Factor family (ERF) with an APETALA2 domain and it was described as an ABA response repressor during germination and root growth.12 In Arabidopsis, disruption of ABR1 gene leads to hypersensitivity to osmotic stress and to ABA application during seed germination and root growth assays.12 In our previous report we suggested that the reduced tolerance to oxidative stress in the abc- may be due to a perturbed regulation of the ABA signaling pathway.11 In addition, there are evidences connecting ABA and ethylene signaling cascades. The insensitive ethylene mutants etr1 and ein2 show phenotypes with enhanced dormancy, ABA hypersensitivity during germination and augmented levels of ABA, resembling ABA-signaling mutants.13,14 Ein 2 null mutant is also supersensitive to both salt and osmotic stress conditions.15 All these data suggest that a functional ethylene signal transduction pathway is necessary to fully develop several ABA responses. As we commented before there are evidences that MBF1 genes positively regulate ethylene signaling pathway. Thus, in this report we explored the impact of AtMBF1 genes on inhibition of germination, a typical ABA-dependent response.
Results
Seed dormancy is enhanced in ABA-hypersensitive mutants, such as era 1 as well as in the ethylene insensitive mutants (ctr1, ein2 ein3).13-17 We analyzed the effect of AtMBF1 mutations on seed dormancy of abc- seeds and compared with ein3–1 mutant and abc- complemented with AtMBF1c gene overexpression (abc- +c).11,16 Since stratification of Arabidopsis seeds breaks dormancy,18 we determine seed germination of WT or mutant seeds previously incubated or not for 4 d at 4°C (Fig. 1A). Germination rate did not differ among the lines upon 24 h stratification. However, the non-stratified abc- seeds showed a lower germination rate (10%) compared with WT (20%). Germination of ein3–1 was similar to abc-. However, germination of abc- +c was similar to WT indicating that AtMBF1c overexpression rescued the mutant phenotype. When developmental stages were analyzed at 48 h, according to Boyes et al.,19 only 40% of non-stratified abc- seeds reached stage 0.7 compared with 90% of the WT (Fig. 1B). Eventually all the non-stratified seeds germinated and reached stage 0.7 (data not shown).
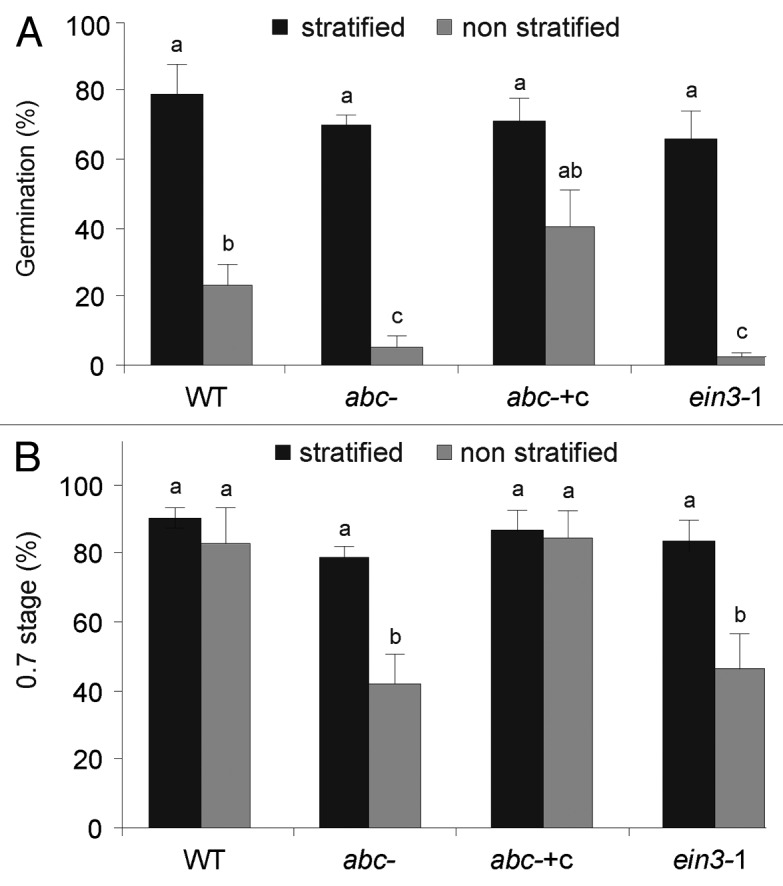
Figure 1. Mutations in AtMBF1 genes enhance seed dormancy. Seeds from WT, abc-, and abc- +c plants were surface-sterilized and stratified or not at 4°C for 2 d in the dark to break dormancy. Seeds were plated on ATS medium with 0.8% agar and placed on a growth chamber at 23°C with a 16 h-light photoperiod. (A) The percentage of germination was scored after 24 h. (B) The percentage of 0.7 stage according to Boyes et al.19 was evaluated after 48 h. Approximately, 100 seeds were processed per line in each experiment. Data are mean values (± SE) of five independent experiments. Different letters indicate a significant difference at p < 0.05 (Tukey's test).
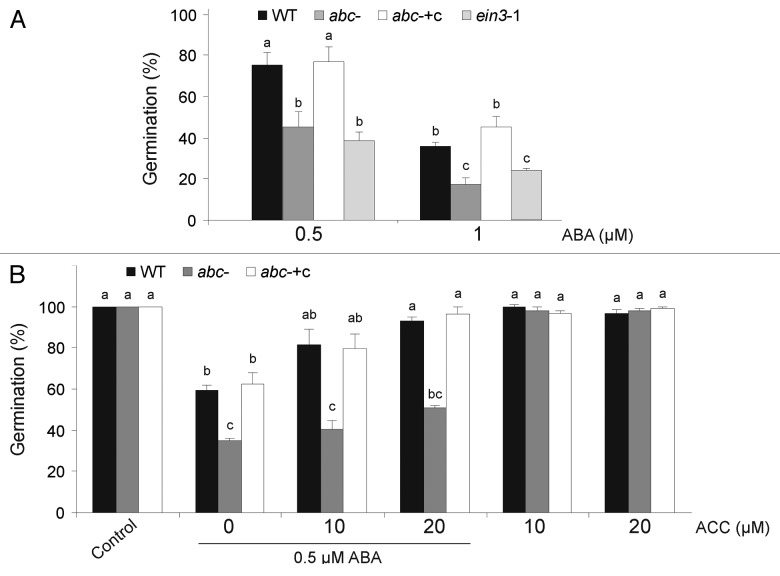
In Arabidopsis, mutants with enhanced seed dormancy also showed increased sensitivity to exogenous ABA during germination.13,14,17 Therefore, to clarify ABA sensitivity of the abc- mutant, ABA dose-response experiments were performed during germination of stratified WT and mutant seeds (Fig. 2A). Germination rates of abc- and ein3–1 seeds were 20% lower than WT for the tested concentrations. abc- +c showed germination rates similar to WT.
Figure 2.abc- mutant is hypersensitive to ABA during germination. (A) Sterilized and stratified WT, abc-, abc- +c and ein3-1 seeds were plated on ATS agar medium supplemented with the indicated concentrations of ABA and incubated in the growth chamber. (B) Sterilized and stratified WT, abc- and abc- +c seeds were plated on ATS agar supplemented with ABA, ACC or the combination of both as indicated. In (A) and (B) the percentage of germination was scored after 24 h. Approximately, 100 seeds were processed per line in each experiment. Data are mean values (± SE) of five independent experiments. Different letters indicate a significant difference at p < 0.05 (Tukey's test).
It is well known that 1-aminocyclopropane-1-carboxylic acid (ACC) is an intermediate in the conversion of methionine to ethylene and that ACC synthesis, mediated by the enzyme ACC synthase, determines the rate of ethylene production. Exogenous ACC induces ACC oxidase leading to ethylene biosynthesis.20 Moreover, ABA regulates ACC synthase and ACC oxidase genes in mung bean and during tomato fruit ripening.21,22 Ghassemian et al.14 described that ABA-dependent inhibition of WT germination is partially rescued in a dose dependent manner by ACC, while ACC alone does not enhance germination in Arabidopsis. Effects of ACC on enhancement of germination in abc- were weaker than those in WT and the complemented line (Fig. 2B). Taken together these results suggest that MBF1 negatively regulates ABA-dependent inhibition of germination by positively regulating ethylene signaling.
Next, we analyzed ABA content in non-stratified mature seeds of WT, abc- and abc- +c lines imbibed for different times (Fig. 3). At 8 h after imbibition all seeds showed a similar increase in ABA content. A similar increase on ABA levels has been reported for non dormant Arabidopsis seeds during the first hours after imbibition.23 At 16 h, ABA content decreased at a slower rate in abc- compared with abc- +c and WT seeds.
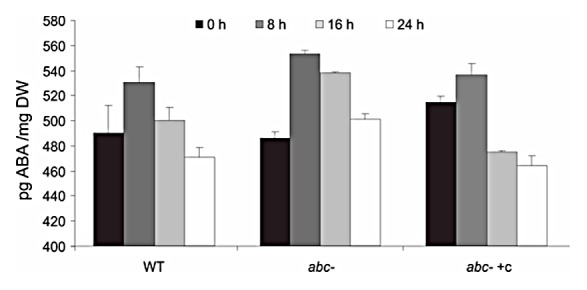
Figure 3.abc- mutant accumulates ABA in seeds. Seeds from WT, abc-, and abc- +c plants were imbibed in sterile water for the designated times and assayed for ABA content by radioimmunoassay. Each sample was assessed twice. The results presented are the mean value of two biological replicates ± SE.
Discussion
The enhanced seed dormancy of abc- mutant seeds and their hypersensitivity to exogenous ABA (Figs. 1 and 2A) suggest a negative role of MBF1s genes in ABA-dependent inhibition of germination. Unlike ethylene insensitive mutants (ein2, ein3 or etr1)14,15 abc- root growth was not altered in the presence of exogenous ABA (data not shown), suggesting that MBF1s may modulate specific ABA-dependent responses.
Our results unravel new evidence that connects AtMBF1 genes to the ethylene signal transduction pathway. First, abc- mutant resembled ein3 responses in all the assays. Second, ABA inhibition of germination could not be fully rescue by exogenous ACC (Fig. 1B), suggesting that abc- ability to sense ACC is compromised.
An Oryza sativa MBF1 was reported to interact “in vitro” with ERF2 and ERF4 transcription factors.24 Furthermore, Arabidopsis thaliana plants overexpressing ERF4 have less sensitivity to ABA and are hypersensitive to osmotic stress.25 Thus, we speculate that AtMBF1s might be interacting with specific ERFs such as ABR1 and ERF4 to modulate ABA-dependent germination in Arabidopsis seeds.
Moreover, higher ABA contents were detected in abc- seeds at 16 h after imbibition, suggesting that AtMBF1 genes positively regulate ABA degradation during early hours after imbibition (Fig. 3). Supporting our data, Arabidopsis mutants insensitive to ethylene also show increased endogenous ABA concentrations. The ethylene insensitive mutant ein2 accumulates ABA in green tissue and this accumulation is related to an increased of ABA biosynthesis.14 ABA levels in mature dry seeds of the etr1–2 mutant were 10-fold higher than in WT seeds.26
The expression pattern of some key genes regulated by ABA or ethylene in the mutants could provide additional evidences on MBF1-mediated interaction between these two hormones. Since it has been suggested that ABA and ethylene may control the hormonal biosynthesis, catabolism, or signaling of each other to enhance their antagonistic effects upon seed germination,27 it would be interesting to further explore the influence of MBF1 genes on these hormone cross-talks.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana (L.) Heynh. wild type, abc- and ein3–1 mutants used in this study were of ecotype Columbia. The abc- mutant line is a T-DNA insertion mutant for AtMBF1a (At2g42680), AtMBF1b (At3g58680) and AtMBF1c (At3g24500) genes. Their genetic and phenotypic characteristics have been described by Arce et al.11 The ein3–1 is an ethylene insensitive mutant with a loss-of-function mutation for EIN3 gene (At3g20770).17 Plants were grown at 22–24°C under fluorescent light 120 µmol photons m−2 s−1 with a16-h-photoperiod. Seeds were sown on organic substrate placed for 2 d at 4°C in the dark to break residual dormancy and then transferred to normal growth conditions. Plants were watered twice a week until senescence.
Germination Assays
To quantify dormancy, seeds 1 mo-old after harvest were surface-sterilized in 30% commercial bleach and 0.02% Triton X-100 for 15 min, rinsed four times with sterile water and stratified or not at 4°C for 2 d in the dark to break dormancy. Then, seeds were plated on ATS medium with 0.8% agar and placed on a growth chamber at 23°C with a 16 h-light photoperiod. The percentage of germination (fully emerged radicle tip) was evaluated after 24 h of incubation. The percentage of 0.7 stage according to Boyes et al.19 was determined after 48 h of incubation. Measurements of ABA sensitivity were conducted with 1 to 3 mo-old seeds. Seeds were surface-sterilized, stratified at 4°C for 2 d in the dark. Seeds were plated on ATS medium with 0.8% agar containing various concentrations of ABA in combination or not with various concentrations of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid and placed on a growth chamber. The percentage of germination was scored after 24 h.
ABA determination
One-mo-old seeds were imbibed in sterile water for different times at 22°C, lyophilized, powdered, weighed and stored at –20°C. ABA content was determined by radioimmunoassay as described in Steinbach et al.28 This method uses the monoclonal antibody AFRC MAC 25229 and tritiated-ABA (Amersham-Pharmacia). Each sample was assessed twice.
Statistical analysis
The values shown in each figure are mean values ± SE. The data were subjected to analysis of variance (one-way ANOVA) and post hoc comparisons were done with Tukey’s multiple range test at p < 0.05 level. The statistical software program used was SigmaStat 3.1.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This research was supported by grants from UNMDP, CONICET, ANPCyT, Argentina. E.M.V., R.B.A. and C.A.C. are members of CONICET. MJM is a fellow of the same institution. D.P.A. and M.F.M. were fellows of CONICET and UNMDP, respectively, at the time of this work. The authors wish to thank Dr Claudia Tono´n for her helpful assistance.
Glossary
Abbreviations:
- ABA
abscisic acid
- abc-
Arabidopsis thaliana triple knock-down mutant for MBF1 genes
- abc- +c
abc- mutant complemented with AtMBF1c gene over-expression
- ABR1
absisic acid repressor
- ACC
1-aminocyclopropane-1-carboxylic acid
- ERF
ethelyne responsive factor family
- H2O2, hydrogen peroxide
- MBF1
multiprotein bridging factor 1
- MV
methyl viologen
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18843
References
- 1.Takemaru K, Li FQ, Ueda H, Hirose S. Multiprotein bridging factor 1 (MBF1) is an evolutionarily conserved transcriptional coactivator that connects a regulatory factor and TATA element-binding protein. Proc Natl Acad Sci U S A. 1997;94:7251–6. doi: 10.1073/pnas.94.14.7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsuda K, Tsuji T, Hirose S, Yamazaki K. Three Arabidopsis MBF1 homologs with distinct expression profiles play roles as transcriptional co-activators. Plant Cell Physiol. 2004;45:225–31. doi: 10.1093/pcp/pch017. [DOI] [PubMed] [Google Scholar]
- 3.Tsuda K, Yamazaki K. Structure and expression analysis of three subtypes of Arabidopsis MBF1 genes. Biochim Biophys Acta 2004; 1680:1-10. [DOI] [PubMed]
- 4.Kim MJ, Lim GH, Kim ES, Ko CB, Yang KY, Jeong JA, et al. Abiotic and biotic stress tolerance in Arabidopsis overexpressing the multiprotein bridging factor 1a (MBF1a) transcriptional coactivator gene. Biochem Biophys Res Commun. 2007;354:440–6. doi: 10.1016/j.bbrc.2006.12.212. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki N, Rizhsky L, Liang H, Shuman J, Shulaev V, Mittler R. Enhanced tolerance to environmental stress in transgenic plants expressing the transcriptional coactivator multiprotein bridging factor 1c. Plant Physiol. 2005;139:1313–22. doi: 10.1104/pp.105.070110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki N, Bajad S, Shuman J, Shulaev V, Mittler R. The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J Biol Chem. 2008;283:9269–75. doi: 10.1074/jbc.M709187200. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki N, Sejima H, Tam R, Schlauch K, Mittler R. Identification of the MBF1 heat-response regulon of Arabidopsis thaliana. Plant J. 2011;66:844–51. doi: 10.1111/j.1365-313X.2011.04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hommel M, Khalil-Ahmad Q, Jaimes-Miranda F, Mila I, Pouzet C, Latche´a A, et al. Over-expression of a chimeric gene of the transcriptional co-activator MBF1 fused to the EAR repressor motif causes developmental alteration in Arabidopsis and tomato. Plant Sci. 2008;175:168–77. doi: 10.1016/j.plantsci.2008.01.019. [DOI] [Google Scholar]
- 9.Tojo T, Tsuda K, Yoshizumi T, Ikeda A, Yamaguchi J, Matsui M, et al. Arabidopsis MBF1s control leaf cell cycle and its expansion. Plant Cell Physiol. 2009;50:254–64. doi: 10.1093/pcp/pcn187. [DOI] [PubMed] [Google Scholar]
- 10.Zegzouti H, Jones B, Frasse P, Marty C, Maitre B, Latch A, et al. Ethylene-regulated gene expression in tomato fruit: characterization of novel ethylene-responsive and ripening-related genes isolated by differential display. Plant J. 1999;18:589–600. doi: 10.1046/j.1365-313x.1999.00483.x. [DOI] [PubMed] [Google Scholar]
- 11.Arce DP, Godoy AV, Tsuda K, Yamazaki K, Valle EM, Iglesias MJ, et al. The analysis of an Arabidopsis triple knock-down mutant reveals functions for MBF1 genes under oxidative stress conditions. J Plant Physiol. 2010;167:194–200. doi: 10.1016/j.jplph.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S. ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 2005;139:1185–93. doi: 10.1104/pp.105.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beaudoin N, Serizet C, Gosti F, Giraudat J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell. 2000;12:1103–15. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P. Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell. 2000;12:1117–26. doi: 10.1105/tpc.12.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Liu C, Li K, Sun F, Hu H, Li X, et al. Arabidopsis EIN2 modulates stress response through abscisic acid response pathway. Plant Mol Biol. 2007;64:633–44. doi: 10.1007/s11103-007-9182-7. [DOI] [PubMed] [Google Scholar]
- 16.Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–44. doi: 10.1016/S0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- 17.Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273:1239–41. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- 18.Bethke PC, Gubler F, Jacobsen JV, Jones RL. Dormancy of Arabidopsis seeds and barley grains can be broken by nitric oxide. Planta. 2004;219:847–55. doi: 10.1007/s00425-004-1282-x. [DOI] [PubMed] [Google Scholar]
- 19.Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, et al. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Annu Rev Plant Physiol. 1984;35:155–89. doi: 10.1146/annurev.pp.35.060184.001103. [DOI] [Google Scholar]
- 21.Kim JH. Ethylene-Regulated Expression of ACC Oxidase and ACC Synthase Genes in Mung Bean Hypocotyls. J Plant Biol. 2006;49:291–7. doi: 10.1007/BF03031158. [DOI] [PubMed] [Google Scholar]
- 22.Zhang M, Yuan B, Leng P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J Exp Bot. 2009;60:1579–88. doi: 10.1093/jxb/erp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ali-Rachedi S, Bouinot D, Wagner MH, Bonnet M, Sotta B, Grappin P, et al. Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta. 2004;219:479–88. doi: 10.1007/s00425-004-1251-4. [DOI] [PubMed] [Google Scholar]
- 24.Yamazaki K, Hirose S. Ethylene-Responsive transcription coactivator in plant, Patent (US) 2007; No.: 7.238.857 B2.
- 25.Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol Biol. 2005;58:585–96. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
- 26.Chiwocha SD, Cutler AJ, Abrams SR, Ambrose SJ, Yang J, Ross AR, et al. The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 2005;42:35–48. doi: 10.1111/j.1365-313X.2005.02359.x. [DOI] [PubMed] [Google Scholar]
- 27.Cheng WH, Chiang MH, Hwang SG, Lin PC. Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol Biol. 2009;71:61–80. doi: 10.1007/s11103-009-9509-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinbach HS, Benech-Arnold RL, Kristof G, S´nchez RA, Marcucci-Poltri S. Physiological basis of pre-harvest sprouting resistance in Sorghum bicolor (L.) Moench. ABA levels and sensitivity in developing embryos of sprouting-resistant and sprouting-susceptible varieties. J Exp Bot. 1995;46:701–9. doi: 10.1093/jxb/46.6.701. [DOI] [Google Scholar]
- 29.Quarrie SA, Whithford PN, Appleford NE, Wang TL, Cook SK, Henson IE, et al. A monoclonal antibody to (S)-abscisic acid:its characterization and use in a radioinmunoassay for measuring abscisic acid in crude extracts of cereal an lupin leaves. Planta. 1988;163:330–9. doi: 10.1007/BF00401020. [DOI] [PubMed] [Google Scholar]