Abstract
Network analysis is now a common statistical tool for molecular biologists. Network algorithms are readily used to model gene, protein and metabolic correlations providing insight into pathways driving biological phenomenon. One output from such an analysis is a candidate gene list that can be responsible, in part, for the biological process of interest. The question remains, however, as to whether molecular network analysis can be used to inform process models at higher levels of biological organization. In our previous work, transcriptional networks derived from three plant species were constructed, interrogated for orthology and then correlated with photosynthetic inhibition at elevated temperature. One unique aspect of that study was the link from co-expression networks to net photosynthesis. In this addendum, we propose a conceptual model where traditional network analysis can be linked to whole-plant models thereby informing predictions on key processes such as photosynthesis, nutrient uptake and assimilation, and C partitioning.
Keywords: ecology, modeling, molecular biology, networks, photosynthesis, transcriptomics
Recent interest in modeling molecular network interrelationships can be attributed, in part, to the technical advances in high-throughput technologies (e.g., cDNA sequencing, metabolomics) and the need to reduce the dimensionality of those data to better understand the plant as a system. This is commonly termed ‘systems biology,’ and although numerous definitions exist most agree that systems biology entails the study of biological processes in terms of dynamic interactions among the components that identify emergent properties of the respective system.1 Numerous studies have leveraged publicly available microarray data and subjected them to network algorithms in which individual gene transcripts are clustered into subnetworks called modules based on expression pattern.2,3 Such clusters exploit the “guilt by association paradigm,” whereby genes that are highly correlated in expression pattern are likely to be involved in the same biological process. Lists of candidate genes and potential pathways can be created and further interrogated using molecular biology techniques (e.g., transgenesis, immunolocalization) to decipher gene function underlying pathways of interest.
Grouping genes and metabolites into like functions with network analysis is becoming a useful way to describe the biological system, especially if the system represents a relatively simple bacterium.4 Plants, however, encompass additional layers of biological complexity including numerous organelles, tissues, organs and genome complexity (e.g., ploidy levels, alternative splicing). Therefore, for network models to begin to accurately describe plant processes, they must first come to terms with this added complexity. Recently, we characterized a network for the heat stress transcriptome of three species (Populus, Arabidopsis and soybean) and discovered gene orthologs representing functional equivalents based on gene expression and enzyme activity assays.5 An interesting outcome from this study was that the expression of one module was negatively correlated with leaf-level photosynthesis. Although this represents an initial step in scaling molecular networks to higher levels of biological organization, a common framework is still needed to test, improve and define these relationships mathematically.
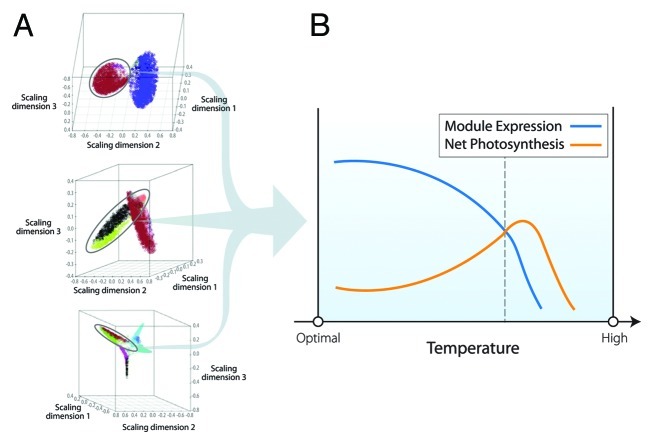
We propose a conceptual model that uses our recent work5 as an initial starting point and is expanded to whole-plant scales using examples from the literature. Tissue samples for biochemistry and gene expression were taken at defined states of photosynthetic thermoinhibition for Arabidopsis, soybean and Populus. Network algorithms clustered genes into respective modules, one of which was enriched for genes encoding heat shock proteins and antioxidant enzymes. This heat responsive module were then evaluated for orthologous gene members among species to reveal a conserved representation of the response of the underlying network to warming. A fundamental property of functional gene networks is the coalescence of many genes into simplified modules and the ability to correlate module information (e.g., eigenvalue) to external information, photosynthesis in our case.6,7 Figure 1 shows a schematic representation of the relationship among net photosynthesis and the heat responsive module to temperature. Expression of this module is negatively correlated with net photosynthesis until a critical temperature where cellular function is disrupted.
Figure 1. Visualization and characterization of network architecture in relation to net photosynthesis. (A) Arabidopsis, soybean and Populus multi-dimensional scaling plots of co-expression network. The distance between circles is a function of topological overlap and provides a visual representation of gene and module relationships within network. Circled modules are enriched for genes participating in warming (e.g., heat shock protein and antioxidant enzymes). (B) The relationship between the expression of the warming module and net photosynthesis. Data from reference 5 produced a significant negative correlation between warming module expression and net photosynthesis (Pearson r = -0.82, p = 0.02). Note that data was not available at high temperatures compromising membrane integrity (dotted vertical line), so relationships beyond this point are entirely conceptual.
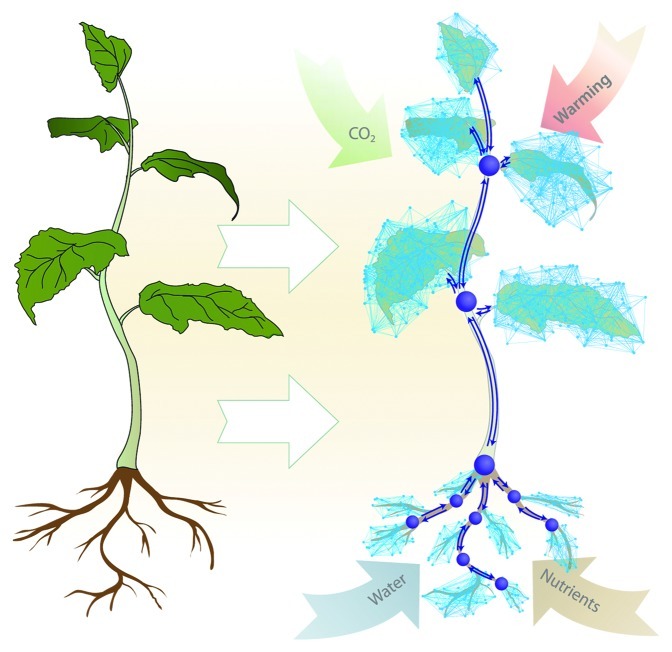
The final step, and by far the most challenging aspect to this conceptual framework is the integration of molecular networks with whole-plant processes (Fig. 2). Although a single representative module is useful in predicting our response variable (net photosynthesis) from our independent variable (temperature), we still need to consider the complicating factors associated with biological complexity and environment. For example, how do we account for C partitioning into different processes, such as defense and stress pathways or root growth? These processes can modify sink strength and carbon skeleton supply, thereby influencing photosynthesis.
Figure 2. Hypothetical representation of a whole-plant network interrelationship model. The plant system can be modeled as suggested by Lucas and colleagues,8 whereby a series of nodes are connected to neighboring nodes and the surrounding environment. Functions association with edge nodes could include water and nutrients fluxes. Hierarchical functions could be assigned to hub nodes (large purple nodes) allowing the determination of which signals to pass to corresponding networks at higher-levels of biological organization.
Network and systems biology approaches are being applied independently at tissue and plant scales,8 suggesting that we may be able to link these levels to molecular networks. One commonality between many tissue and whole-plant models is that both relate spatial development of the plant (or tissue) to its physiology.8 This relationship allows the plant to be spatially represented by network nodes that are connected to neighboring nodes (Fig. 2). Node edges are associated with mathematical functions that can describe various biological processes and hierarchical concepts can be included to determine which signals are passed to higher levels of biological organization (purple hub nodes; Fig. 2). For example, ion concentrations within the rhizosphere initiates plant signaling and hormonal networks driving root growth that ultimately influence whole-plant development.9 The sink strength provided by this signaling cascade would ultimately determine the availability of C skeletons for proteins mitigating leaf stress and defense responses, both of which are metabolically costly.10,11 Models based on source-sink relationships assume that C partitioning depends on the relative ability of different sinks to import available assimilates from sources. They can provide a basis for changing partitioning in relation to environmental heterogeneity.12 The rules for determining sink strength can be difficult to determine, and this level of mechanistic detail is currently missing from whole-plant models; molecular networks could be used to parameterize such functions.
Better representation of C partitioning at the whole-plant level could have implications at still higher levels of biological organization. Ecosystem and terrestrial C cycling models would be improved with a more dynamic representation of partitioning of photosynthate into various metabolites and tissues. Partitioning within the plant alters the subsequent fate of C in ecosystems,13 but incomplete understanding of carbon partitioning currently limits the capacity to >model forest ecosystem metabolism and accurately predict the effects of global change on carbon cycling.14 We have described a framework for the potential integration of metabolic processes indicated by molecular networks into whole-plant models for application to higher-level processes (e.g., net photosynthesis, nutrient uptake and assimilation, antioxidant production). In this framework, molecular networks provide a means by which mechanism can be linked to key parameters used in estimating metabolic processes governing C partitioning. As the links between molecular biology, physiology and ecology develop, we will have the opportunity to consider ways in which inference from one discipline can inform another. Here, we suggest a start to this exciting endeavor, and the maturation of this collaboration will improve our ability to not only to understand, but to predict whole-plant performance.
Acknowledgments
Research was conducted at Oak Ridge National Laboratory, and supported by the US. Department of Energy (DOE), Office of Science, Biological and Environmental Research; and the Laboratory Directed Research and Development Program of Oak Ridge National Laboratory, managed by UT-Battelle, L.L.C., for the US Department of Energy under contract DE-AC05–000R22725.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18802
References
- 1.Bennett M, Monk N. The flowering of systems approaches in plant and crop biology. New Phytol. 2008;179:567–8. doi: 10.1111/j.1469-8137.2008.02565.x. [DOI] [PubMed] [Google Scholar]
- 2.Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, et al. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 2007;50:347–63. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- 3.Weston DJ, Gunter LE, Rogers A, Wullschleger SD. Connecting genes, coexpression modules, and molecular signatures to environmental stress phenotypes in plants. BMC Syst Biol. 2008;2:16. doi: 10.1186/1752-0509-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teusink B, Westerhoff HV, Bruggeman FJ. Comparative systems biology: from bacteria to man. Wiley Interdiscip Rev Syst Biol Med. 2010;2:518–32. doi: 10.1002/wsbm.74. [DOI] [PubMed] [Google Scholar]
- 5.Weston DJ, Karve AA, Gunter LE, Jawdy SS, Yang X, Allen SM, et al. Comparative physiology and transcriptional networks underlying the heat shock response in Populus trichocarpa, Arabidopsis thaliana and Glycine max. Plant Cell Environ. 2011;34:1488–506. doi: 10.1111/j.1365-3040.2011.02347.x. [DOI] [PubMed] [Google Scholar]
- 6.Dong J, Horvath S. Understanding network concepts in modules. BMC Syst Biol. 2007;1:24. doi: 10.1186/1752-0509-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:e17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 8.Lucas M, Laplaze L, Bennett MJ. Plant systems biology: network matters. Plant Cell Environ. 2011;34:535–53. doi: 10.1111/j.1365-3040.2010.02273.x. [DOI] [PubMed] [Google Scholar]
- 9.Gutie´rrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007;8:R7. doi: 10.1186/gb-2007-8-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohnert HJ, Gong Q, Li P, Ma S. Unraveling abiotic stress tolerance mechanisms--getting genomics going. Curr Opin Plant Biol. 2006;9:180–8. doi: 10.1016/j.pbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 11.van Hulten M, Pelser M, van Loon LC, Pieterse CML, Ton J. Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103:5602–7. doi: 10.1073/pnas.0510213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang ZJ, Midmore DJ. Modelling plant resource allocation and growth partitioning in response to environmental heterogeneity. Ecol Modell. 2005;181:59–77. doi: 10.1016/j.ecolmodel.2004.06.023. [DOI] [Google Scholar]
- 13.Norby RJ, Zak DR. Ecological lessons from free-air CO2 enrichment (FACE) experiments. Annu Rev Ecol Evol Syst. 2011;42:181–203. doi: 10.1146/annurev-ecolsys-102209-144647. [DOI] [Google Scholar]
- 14.Litton CM, Raich JW, Ryan MG. Carbon allocation in forest ecosystems. Glob Change Biol. 2007;13:2089–109. doi: 10.1111/j.1365-2486.2007.01420.x. [DOI] [Google Scholar]