Abstract
Plants have evolved resistance (R) proteins to detect pathogen effectors and trigger plant defense responses in the so named effector-triggered immunity (ETI). R proteins are under negative regulation in plants as upregulated activation of R protein is detrimental to plant growth. Autoimmune mutants have been instrumental in understanding the fine tuning of plant defense responses. Recently, a number of such mutants have been molecularly characterized, and some of them result from over-activation of SNC1, a TIR-NBS-LRR type of R protein. Studies of these mutants revealed a complex negative regulation of SNC1 activity from transcriptional to post-translational regulation. Here, we summarize studies on these SNC1-dependent autoimmune mutants and discuss the fine regulation of R proteins in plant immunity.
Keywords: autoimmune mutants, disease resistance, plant immunity, R protein, SNC1
To ward off pathogen invasion, plants have evolved at least two layers of defense responses. The first layer is triggered through the perception of pathogen associated molecular patterns (PAMP) by pattern recognition receptors (PRR), namely ‘PAMP triggered immunity’ (PTI). The second layer of defense is triggered through the recognition of pathogen-secreted effectors by resistance (R) proteins, namely ‘effector triggered immunity’ (ETI).1 Specific recognition of effectors by R proteins leads to a rapid programmed cell death called hypersensitive response (HR) which restricts the growth and spread of pathogens.2 Although R activation is essential for effective plant defense, R protein activity needs to be fine controlled to avoid extensive cell death by R protein over-activation. R proteins are normally expressed at a low level and assume an inactive state in the absence of pathogen triggers. The mechanisms underlying the negative regulation of R proteins are far from well understood. Here we will review the study of autoimmune mutants that have revealed negative regulation at multiple levels on SNC1, a TIR-NBS-LRR R protein.
snc1-1 and Other Auto-immune Mutants
In the last decade, a number of mutants were isolated in Arabidopsis showing dwarf morphology, constitutive expression of pathogen-related (PR) genes, and enhanced disease resistance. As they display immune responses without pathogen triggers, these mutants are named as auto-immune mutants. Since many such mutants also display microscopic cell deaths at least under some environmental conditions, they are called lesion mimic mutants as well.3,4 snc1 (referred to as snc1-1), isolated as a suppressor of npr1-15, has a missense mutation in a TIR-NBS-LRR R protein SNC1, leading to constitutive activation of SNC1 and thus autoimmune responses.5,6 The snc1-1 mutant can be suppressed to different extent by a set of modifer of snc1 (mos) mutants. Characterizations of these MOS genes indicate that SNC1-mediated resistance is regulated by many processes and pathways.7 The bon1/cpn1 mutants also exhibit dwarfism and auto-immune responses.8,9 Interestingly, this autoimmune phenotype is present in the Col-0 accession but not the Ws accession, and the study of this natural modification of bon1 reveals that the autoimmune response is dependent on the Col-0 haplotype-specific SNC1.8,10 In the last few years, many more autoimmune mutants were found to be SNC1 dependent, similarly to bon1. They include: bap1, bir1, srfr1, cpr1, and mkp1 (Table 1). Therefore, SNC1 seems to be under complex regulation by molecules with diverse functions and subcellular localizations (Table 1).
Table 1. SNC1 dependent auto-immune mutants.
| Gene name | Mutant name (in Col-0) |
Severity of dwarfness | suppression by snc1–11 | Subcellular localization | Biochemical function | References | |
|---|---|---|---|---|---|---|---|
|
BON1 |
cpn1-1 bon1-1 |
++ |
Fully |
Plasma membrane |
Phospholipid-binding Copine protein |
8–10 |
|
|
BAP1 |
bap1-1 |
+ |
Fully |
Membrane |
Phospholipid-binding protein |
8,12 |
|
|
BIR1 |
bir1-1 bir1-2 |
++++ |
Partially |
Plasma membrane |
Receptor like kinase |
13,17 |
|
|
SRFR1 |
srfr1-3 srfr1-4 srfr1-5 |
++++ |
Partially |
Cytoplasm and nucleus |
TPR1 domain protein |
24,26 |
|
|
CPR1 |
cpr-1 cpr1-2 (cpr30-1) cpr1-3 (cpr30-2) |
++ |
Fully |
Cytoplasm and nucleus |
F-box Protein |
20–23 |
|
| MKP1 | mkp1 | + | Partially | Cytoplasm | MAPK phosphatase | 27 | |
Note: more “+” means smaller size.
Regulation of SNC1 by BON1 Associated Proteins on the Plasma Membrane
BON1/CPN1 is a member of evolutionarily conserved copine proteins characterized by two C2 domains at the N-terminus and a VWA domain at the C-terminus.8 Though the C2 domain could confer a calcium-dependent phospholipid binding to the membranes, the localization of BON1 to the plasma membrane is mediated by the N-terminal myristoylation of BON1.11 Several BON1 interacting proteins have been isolated through the yeast two-hybrid screens. The first one identified is BAP1 (BON1-ASSOCIATED PROTEIN 1) which contains one C2 domain and a short C-terminal tail.8,12 Similar to bon1-1, the loss-of-function mutant bap1-1 shows auto-immune phenotype that is dependent on SNC1.12 Overexpression of BAP1 partially suppressed the bon1-1 phenotype, indicating that BON1 and BAP1 work together to negatively regulate SNC1.12 The BAP1 protein is membrane associated, however its exact location has not been determined. The second BON1 interacting protein identified is BIR1 (BAK1-INTERACTING RECEPTOR-LIKE KINASE 1) which encodes a receptor like kinase interacting with BAK1 (BRI1-ASSOCIATED RECEPTOR KINASE 1),13 a co-receptor of BRI1 (BRASSINOSTEROID INSENSITIVE 1) and FLS2 (FLAGELLIN SENSITIVE 2).14-16 BAK1 was also found to interact with BON1.17 The bir1 mutant shows a more severe growth defect than bon1-1 and can be partially rescued by BON1 overexpression. In addition, the bir1 mutant phenotype can be suppressed by a SNC1 loss-of-function mutant snc1-11, indicating that BIR1 and BON1 can function together to regulate SNC1.17 BAK1 is involved in PTI as well as in brassinosteroid signaling in development. The BAK1 loss-of-function mutant does not exhibit a severe growth defect but the double mutant of BAK1 and its homolog BKK1 has a very severe dwarf phenotype and cell death.18 It will be interesting to test if SNC1 contributes to this phenotype.
Although BIR1, BAK1 and BON1 could interact with each other in vitro and are all plasma membrane associated, the in vivo interaction is yet to be verified and the biological relevance is yet to be revealed. BIR1 has extracellular leucine-rich-repeats, and is presumably a receptor for an unknown ligand. As overexpression of BON1 and BIR1 can mutually partially suppress the loss-of-function mutant phenotypes of each other,17 they two probably do not work in a linear signaling pathway but rather in a protein complex or in parallel. BAK1 can interact with multiple RLKs including BRI1 and FLS2,14-16,19 and therefore could transduce multiple signals such as the pathogen signature flagellin and the plant hormone brassinolides. As BAK1 is shown to phosphorylate BON1 in vitro, BON1 could potentially be modified by multiple signals through BAK1 if indeed this phosphorylation has biological relevance. BON1 does not appear to have a kinase activity11 and therefore unlikely will serve as a phosphorylation transducer. Two other activities are shown to be critical for BON1 activity: its calcium binding and its interaction with BAP1.11 Therefore its modification (such as phosphorylation) by its interacting proteins could potentially regulate its calcium binding activities and/or BAP1 interacting activities. The output of BON1 activity could be a calcium signal and/or the BAP1 activity.
It is not entirely clear how the plasma-membrane localized proteins BON1 and BIR1 and the membrane associated BAP1 regulate SNC1 activity. In the bon1-1 and bap1-1 mutants, the transcript of SNC1 is upregulated, which is sufficient to cause an autoimmune phenotype. The upregulation is largely due to a feed-forward regulation by salicylic acid (SA) and PAD4/EDS1 as the SNC1 transcript level is greatly reduced in the NahG, pad4, or eds1 background.10,12 It therefore remains unclear if an initial small change of transcript level or protein activity leads to the final increase of SNC1 transcript. In either case, the regulation of these proteins on SNC1 is likely to be indirect as no physical interaction between BON1 and SNC1 has been detected (unpublished results).
CPR1/CPR30 and Possibly SRFR1 Regulate the SNC1 Protein Stability
cpr1 is one of the first auto-immune Arabidopsis mutants isolated in a screen of constitutive expressor of PR genes (cpr) mutants.20cpr30 was more recently identified as a cpr like mutant from a background mutation in a T-DNA line, and recently CPR1 and CPR30 were found to be the same gene.21CPR1/CPR30 encodes a functional F-box protein that interacts with multiple SKP1 homologs,22 and the cpr1 phenotype is SNC1-dependent. The SNC1 protein is upregulated in both mutants and downregulated in CPR1 overexpression lines. CPR1 can physically interact with SNC1 and affect SNC1 protein accumulation through the 26S proteasome mediated protein degradation.21,23 Therefore, CPR1 regulates the protein stability of SNC1. The SUPPRESSOR OF rps4-RLD (SRFR1) likely regulates SNC1 at the protein level as well. The srfr1 mutants in Col-0 background show a SNC1-dependent auto-immune phenotype similar to that of bon1-1.24,25SRFR1 encodes a conserved TPR domain-containing protein, and it can physically interacts with SGT1 (SUPPRESSOR OF THE G2 ALLELE OF SKP1) proteins that are involved in the control of stability of multiple R proteins.25 The SNC1 protein level is elevated in both srfr1 and sgt1b mutants, suggesting a regulation of SNC1 protein stability by SRFR1 and SGT1 proteins.26
Negative Regulation of SNC1 by MKP1
MKP1 (MAP KINASE PHOSPHATASE 1) and PTP1 (PROTEIN TYROSINE PHOSPHATASE 1) both dephosphorylate MPK6 (MAP KINASE6). The mkp1 null mutant in Col-0 but not in Ws exhibits auto-immune mutant phenotype that is enhanced by ptp1 and dependent on MPK3 and MPK6, key signaling components in PTI.27,28 The mkp1 and the mkp1ptp1 phenotypes could be partially suppressed by snc1-11, suggesting that MKP1 is another negative regulator of SNC1. Because the SNC1 transcription level is not significantly changed in the mkp1 mutant,27 MKP1 might regulate SNC1 at posttranscriptional level. It will be of interest to determine the relevance between MPK3 and MPK6 dephosphorylation and SNC1 negative regulation, which might shed light on the link between PTI and ETI.
Model for the Complex Regulation of SNC1 by Multiple Proteins
The identification of multiple regulators of SNC1 indicates a complex regulation of SNC1 gene function at different levels as summarized in the working model (Fig. 1). SNC1 is repressed in the wild type in the absence of pathogen invasion. Perturbation in various host genes induces activation of SNC1 and this could be at both the transcriptional and posttranscriptional levels. Some of them directly regulate the stability of SNC1 protein. SNC1 is targeted by the F-box protein CPR1, ubiquitinated, and degraded by the 26S proteasome. SGT1 proteins, which interact directly with the SKP1 proteins, might affect SNC1 activity through its regulation of the SCF complex stability. SRFR1, which interacts directly with SGT1, might also regulate SNC1 by affecting the stability of the SCF complex. Other host genes might modulate SNC1 expression or activity indirectly. Signals generated from the loss of BON1 and BIR1 at the plasma membrane could be transduced through BAP1 or a secondary messenger like Ca2+ to regulate the SNC1 transcription in the nucleus. Indeed, MOS1, identified as a suppressor of snc1, regulates the transcript level of SNC1 possibly through chromatin remodeling and DNA methylation.29 However, it cannot be excluded at this point that BON1 and its interacting proteins could affect the stability of SCF complex that regulates SNC1 in a yet unkown mechanism. MKP1 is a negative regulator of MPK3 and MPK6 as well as SNC1, suggesting a cross regulation of both PTI and ETI. It will be interesting to reveal whether the regulation of SNC1 by MKP1 is dependent on its regulation on MPK3 and MPK6.
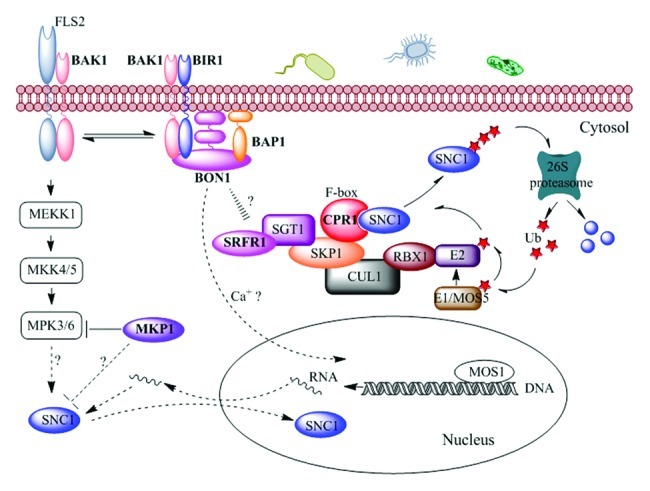
Figure 1. Working model for the regulation of SNC1.
The pressing question is to further understand the roles, if any, of these regulators of SNC1 in plant immunity. Presumably, they are involved in the perception and signaling of pathogen invasions if they do not function only in general production, processing and degradation of transcripts and proteins. One interesting observation is that some of the negative regulators of SNC1 including BON1, CPR1, and BAP1 are induced upon pathogen invasion.12,23,30 Presumably, SNC1 activity will be consequently reduced during pathogen invasion. It will be interesting to determine if SNC1 is initially activated and then repressed during pathogen invasion, which would indicate a fine tuning of defense responses by modulating R activities. In sum, SNC1 activities are under tight controls by different signaling components. Further study of these components and their pathways will enhance our understanding of complex regulation of plant immunity.
Acknowledgment
Research in J.H.'s lab is supported by grants from National Science Foundation IOS-0642289 and IOS-0919914.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/18884
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–9. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Hammond-Kosack KE, Jones JD. Resistance gene-dependent plant defense responses. Plant Cell. 1996;8:1773–91. doi: 10.1105/tpc.8.10.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorrain S, Vailleau F, Balague´ C, Roby D. Lesion mimic mutants: keys for deciphering cell death and defense pathways in plants? Trends Plant Sci. 2003;8:263–71. doi: 10.1016/S1360-1385(03)00108-0. [DOI] [PubMed] [Google Scholar]
- 4.Moeder W, Yoshioka K. Lesion mimic mutants: A classical, yet still fundamental approach to study programmed cell death. Plant Signal Behav. 2008;3:764–7. doi: 10.4161/psb.3.10.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X, Clarke JD, Zhang Y, Dong X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact. 2001;14:1131–9. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–46. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monaghan J, Germain H, Weihmann T, Li X. Dissecting plant defence signal transduction: modifiers of snc1 in Arabidopsis. Can J Plant Pathol. 2010;32:35–42. doi: 10.1080/07060661003621001. [DOI] [Google Scholar]
- 8.Hua J, Grisafi P, Cheng SH, Fink GR. Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev. 2001;15:2263–72. doi: 10.1101/gad.918101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jambunathan N, Siani JM, McNellis TW. A humidity-sensitive Arabidopsis copine mutant exhibits precocious cell death and increased disease resistance. Plant Cell. 2001;13:2225–40. doi: 10.1105/tpc.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang S, Hua J. A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell. 2004;16:1060–71. doi: 10.1105/tpc.020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Gou M, Sun Q, Hua J. Requirement of calcium binding, myristoylation, and protein-protein interaction for the Copine BON1 function in Arabidopsis. J Biol Chem. 2010;285:29884–91. doi: 10.1074/jbc.M109.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Li Y, Hua J. The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J. 2006;48:238–48. doi: 10.1111/j.1365-313X.2006.02869.x. [DOI] [PubMed] [Google Scholar]
- 13.Gao M, Wang X, Wang D, Xu F, Ding X, Zhang Z, et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe. 2009;6:34–44. doi: 10.1016/j.chom.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 14.Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 15.Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–22. doi: 10.1016/S0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 16.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–12. doi: 10.1016/S0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, Meng P, Zhang X, Ren D, Yang S. BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J. 2011;67:1081–93. doi: 10.1111/j.1365-313X.2011.04659.x. [DOI] [PubMed] [Google Scholar]
- 18.He K, Gou X, Yuan T, Lin H, Asami T, Yoshida S, et al. BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr Biol. 2007;17:1109–15. doi: 10.1016/j.cub.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 19.Postel S, Küfner I, Beuter C, Mazzotta S, Schwedt A, Borlotti A, et al. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol. 2010;89:169–74. doi: 10.1016/j.ejcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Bowling SA, Guo A, Cao H, Gordon AS, Klessig DF, Dong X. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–57. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, et al. Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci U S A. 2011;108:14694–9. doi: 10.1073/pnas.1105685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gou M, Su N, Zheng J, Huai J, Wu G, Zhao J, et al. An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 2009;60:757–70. doi: 10.1111/j.1365-313X.2009.03995.x. [DOI] [PubMed] [Google Scholar]
- 23.Gou M, Shi Z, Zhu Y, Bao Z, Wang G, Hua J. The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J. 2011 doi: 10.1111/j.1365-313X.2011.04799.x. In press. [DOI] [PubMed] [Google Scholar]
- 24.Kim SH, Gao F, Bhattacharjee S, Adiasor JA, Nam JC, Gassmann W. The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 2010;6:e1001172. doi: 10.1371/journal.ppat.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kadota Y, Shirasu K, Guerois R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends Biochem Sci. 2010;35:199–207. doi: 10.1016/j.tibs.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y. SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 2010;6:e1001111. doi: 10.1371/journal.ppat.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartels S, Anderson JC, Gonz´lez Besteiro MA, Carreri A, Hirt H, Buchala A, et al. MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell. 2009;21:2884–97. doi: 10.1105/tpc.109.067678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–83. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Tessaro MJ, Li X, Zhang Y. Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol. 2010;153:1425–34. doi: 10.1104/pp.110.156240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee TF, McNellis TW. Evidence that the BONZAI1/COPINE1 protein is a calcium- and pathogen-responsive defense suppressor. Plant Mol Biol. 2009;69:155–66. doi: 10.1007/s11103-008-9413-6. [DOI] [PubMed] [Google Scholar]


