Summary
It is now common to survey microbial communities by sequencing nucleic acid material extracted in bulk from a given environment. Comparative methods are needed that indicate the extent to which two communities differ given data sets of this type. UniFrac, which gives a somewhat ad hoc phylogenetics-based distance between two communities, is one of the most commonly used tools for these analyses. We provide a foundation for such methods by establishing that, if we equate a metagenomic sample with its empirical distribution on a reference phylogenetic tree, then the weighted UniFrac distance between two samples is just the classical Kantorovich–Rubinstein, or earth mover’s, distance between the corresponding empirical distributions. We demonstrate that this Kantorovich–Rubinstein distance and extensions incorporating uncertainty in the sample locations can be written as a readily computable integral over the tree, we develop Lp Zolotarev-type generalizations of the metric, and we show how the p-value of the resulting natural permutation test of the null hypothesis ‘no difference between two communities’ can be approximated by using a Gaussian process functional. We relate the L2-case to an analysis-of-variance type of decomposition, finding that the distribution of its associated Gaussian functional is that of a computable linear combination of independent random variables.
Keywords: Barycentre, CAT(κ)space, Gaussian process, Hadamard space, Metagenomics, Monte Carlo methods, Negative curvature, Optimal transport, Permutation test, Phylogenetics, Randomization test, Reproducing kernel Hilbert space, Wasserstein metric
1. Introduction
Next-generation sequencing technology enables sequencing from hundreds of thousands to millions of short deoxyribonucleic acid DNA sequences in a single experiment. This has led to a new methodology for characterizing the collection of microbes in a sample: rather than using observed morphology or the results of culturing experiments, it is possible to sequence directly genetic material extracted in bulk from the sample. This technology has revolutionized the possibilities for unbiased surveys of environmental microbial diversity, ranging from the human gut (Gill et al., 2006) to acid mine drainages (Baker and Banfield, 2003). We consider statistical comparison procedures for such DNA samples.
1.1. Introduction to UniFrac and its variants
In 2005, Lozupone and Knight introduced the UniFrac comparison measure to quantify the phylogenetic difference between microbial communities (Lozupone and Knight, 2005), and in 2007 they and others proposed a corresponding weighted version (Lozupone et al., 2007). These two references already have hundreds of citations in total, attesting to their centrality in microbial community analysis. Researchers have used UniFrac to analyse microbial communities on the human hand (Fierer et al., 2008), to establish the existence of a distinct gut microbial community associated with inflammatory bowel disease (Frank et al., 2007) and to demonstrate that host genetics play a major part in determining intestinal microbiota (Rawls et al., 2006). The distance matrices that are derived from the UniFrac method are also commonly employed as input to clustering algorithms, including hierarchical clustering and UPGMA (Lozupone et al., 2007). Furthermore, the distances are widely used in conjunction with ordination methods such as principal components analysis (Rintala et al., 2008) or to discover microbial community gradients with respect to another factor, such as ocean depth (Desnues et al., 2008). Two of the major metagenomic analysis ‘pipelines’ that were developed in 2010 had a UniFrac analysis as one of their end points (Caporaso et al., 2010; Hartman et al., 2010). Recently, the software that was used to compute the two UniFrac distances has been re-optimized for speed (Hamady et al., 2009) and it has been reimplemented in the heavily used mothur (Schloss et al., 2009) microbial analysis software package.
The unweighted UniFrac distance uses only presence–absence data and is defined as follows. Imagine that we have two samples A and B of sequences. Call each such sequence a read. Build a phylogenetic tree on the total collection of reads. Colour the tree according to the samples—if a given branch sits on a path between two reads from sample A, then it is coloured red, if it sits on a path between two reads from sample B, then it is coloured blue, and, if both, then it is coloured grey. Unweighted UniFrac is then the fraction of the total branch length that is ‘unique’ to one of the samples, i.e. it is the fraction of the total branch length that is either red or blue.
Weighted UniFrac incorporates information about the frequencies of reads from the two samples by assigning weights to branch lengths that are not just 0 or 1. Assume there are m reads from sample A and n reads in sample B, and that we build a phylogenetic tree T from all m + n reads. For a given branch i of the tree T, let li be the length of branch i and define fi to be the branch length fraction of branch i, i.e. li divided by the total branch length of T. The formula for the (raw) weighted UniFrac distance between the two samples is
| (1) |
where Ai and Bi are the respective number of descendants of branch i from communities A and B (Lozupone et al. 2007). To determine whether or not a read is a descendant of a branch, we need to prescribe a vertex of the tree as being the root, but it turns out that different choices of the root lead to the same value of the distance because
| (2) |
and the quantity on the right-hand side depends only on the proportions of reads in each sample that are in the two disjoint subtrees obtained by deleting the branch i. Also, similar reasoning shows that the (unweighted) UniFrac distance is, up to a factor of , given by a formula that is similar to expression (1) in which Ai/m and Bi/n respectively are replaced by a quantity that is either 1 or 0 depending on whether there are any descendants of branch i in the A or B sample and the branch length li is replaced by the branch length fraction fi. Using the quantities li rather than the fi simply changes the resulting distance by a multiplicative constant, the total branch length of the tree T.
The UniFrac distances can also be calculated by using a pre-existing tree (rather than a tree built from samples) by performing a sequence comparison such as BLAST to associate a read with a previously identified sequence and attaching the read to that sequence’s leaf in the pre-existing tree with an intervening branch of zero length. Using this mapping strategy, the tree that is used for comparison can be adjusted depending on the purpose of the analysis. For example, the user may prefer an ‘ultrametric’ tree (a tree with the same total branch length from the root to each tip) instead of a tree made with branch lengths that reflect amounts of molecular evolution.
With the goal of making reported UniFrac values comparable across different trees, it is common to divide by a suitable scalar to fit them into the unit interval. Given a rooted tree T and counts Ai and Bi as above, the raw weighted UniFrac value is bounded above by
| (3) |
where di is the distance from the root to the leaf side of edge i (Lozupone et al., 2007). When divided by this factor, the resulting scaled UniFrac values sit in the unit interval; a scaled Uni-Frac value of 1 means that there is a branch adjacent to the root which can be cut to separate the two samples. Note that the factor D, and consequently the ‘normalized’ weighted UniFrac value, does depend on the position of the root.
A statistical significance for the observed UniFrac distance is typically assigned by a permutation procedure that we review here for completeness. The idea of a permutation test (which is also known as a randomization test) goes back to Fisher (1935) and Pitman (1937a, b, 1938) (see Good (2005) and Edgington and Onghena (2007) for guides to the more recent literature). Suppose that our data are a pair of samples with counts m and n, and that we have computed the UniFrac distance between the samples. Imagine creating a new pair of ‘samples’ by taking some other subset of size m and its complement from the set of all m + n reads and then computing the distance between the two new samples. The proportion of the choices of such pairs of samples that result in a distance that is larger than that observed in the data is an indication of the significance of the observed distance. Of course, we can rephrase this procedure as taking a uniform random subset of reads of size m and its complement (call such an object a random pair of pseudosamples) and asking for the probability that the distance between these is greater than the observed distance. Consequently, it is possible (and computationally necessary for large values of m and n) to approximate the proportion or probability in question by taking repeated independent choices of the random subset and recording the proportion of choices for which there is a distance between the pair of pseudosamples that is greater than the observed distance. We call the distribution of the distance between a random pair of pseudosamples produced from a uniform random subset of reads of size m and its complement of size n the distribution of the distance under the null hypothesis of no clustering.
1.2. Phylogenetic placement and probability distributions on a phylogenetic tree
We now describe how it is natural to begin with a fixed reference phylogenetic tree constructed from previously characterized DNA sequences and then use likelihood-based phylogenetic methods to map a DNA sample from some environment to a collection of phylogenetic placements on the reference tree. This collection of placements can then be thought of as a probability distribution on the reference tree.
In classical likelihood-based phylogenetics (see, for example, Felsenstein (2004)), one has data consisting of DNA sequences from a collection of taxa (e.g. species) and a probability model for those data. The probability model is composed of two ingredients. The first ingredient is a tree with branch lengths that has its leaves labelled by the taxa and describes their evolutionary relationship. The second ingredient is a Markovian stochastic mechanism for the evolution of DNA along the branches of the tree. The parameters of the model are the tree (its topology and branch lengths) and the rate parameters in the DNA evolution model. The likelihood of the data is, as usual, the function on the parameter space that gives the probability of the observed data. The tree and rate parameters can be estimated by using standard approaches such as maximum likelihood or Bayesian methods.
Suppose that we already have, from whatever source, DNA sequences for each of a number of taxa along with a corresponding phylogenetic tree and rate parameters, and that a new sequence, the query sequence, arrives. Rather than estimate a new tree and rate parameters ab initio, we can take the rate parameters as given and consider only trees that consist of the existing tree, the reference tree, augmented by a branch of some length leading from an attachment point on the reference tree to a leaf labelled by the new taxon. The relevant likelihood is now the conditional probability of the query sequence as a function of the attachment point and the pendant branch length, and we can input this likelihood into maximum likelihood or Bayesian methods to estimate these two parameters. For example, a maximum likelihood point phylogenetic placement for a given query sequence is the maximum likelihood estimate of the attachment point of the sequence to the tree and the pendant branch length leading to the sequence. Such estimates are produced by various algorithms (Von Mering et al., 2007; Monier et al., 2008; Berger and Stamatakis, 2010; Matsen et al., 2010). Typically, if there is more than one query sequence, then this procedure is applied in isolation to each one using the same reference tree, i.e. the taxa corresponding to the successive query sequences are not used to enlarge the reference tree. By fixing a reference tree rather than attempting to build a phylogenetic tree for the sample de novo, recent algorithms of this type can place tens of thousands of query sequences per hour per processor on a reference tree of 1000 taxa, with linear performance scaling in the number of reference taxa.
For the purposes of this paper, the data that we retain from a collection of point phylogenetic placements will simply be the attachment locations of those placements on the reference phylogenetic tree. We shall call these positions placement locations. We can identify such a set of placement locations with its empirical distribution, i.e. with the probability distribution that places an equal mass at each placement. In this way, starting with a reference tree and an aligned collection of reads, we arrive at a probability distribution on the reference tree representing the distribution of those reads across the tree.
We can also adopt a Bayesian perspective and assume a prior probability on the branch to which the attachment is made, the attachment location within that branch and the pendant branch length, to calculate a posterior probability distribution for a placement. For example, we might take a prior for the attachment location and pendant branch length that assumes that these quantities are independent, with the prior distribution for the attachment location being uniform over branches and uniform within each branch and with the prior distribution of the pendant branch length being exponential or uniform over some range. By integrating out the pendant branch length, we obtain a posterior probability distribution μi on the tree for query sequence i. We call such a probability distribution a spread placement: with priors such as those above, μi will have a density with respect to the natural length measure on the tree. It is natural to associate this collection of probability distributions with the single distribution Σi μi/n, where n is the number of query sequences.
For large data sets, it is not practical to record detailed information about the posterior probability distribution. Thus, in the implementation of Matsen et al. (2010), the posterior probability is computed branch by branch for a given query sequence by integrating out the attachment location and the pendant branch length, resulting in a probability for each branch. The mass is then assigned to the attachment location of the maximum likelihood phylogenetic placement. With this simplification, we are back in the point placement situation in which each query sequence is assigned to a single point on the reference tree and the collection of assignments is described by the empirical distribution of this set of points. However, since it is possible in principle to work with a representation of a sample that is not just a discrete distribution with equal masses on each point, we develop the theory in this greater level of generality.
1.3. Comparing probability distributions on a phylogenetic tree
If we wished to use the standard Neyman–Pearson framework for statistical inference to determine whether two metagenomic samples came from communities with the ‘same’ or ‘different’ constituents, we would first have to propose a family of probability distributions that described the outcomes of sampling from a range of communities and then construct a test of the hypothesis that the two samples were realizations from the same distribution in the family. However, there does not appear to be such a family of distributions that is appropriate in this setting.
We are thus led to the idea of representing the two samples as probability distributions on the reference tree in the manner that was described in Section 1.2, calculating a suitable distance between these two probability distributions, and using the general permutation or randomization test approach that was reviewed in Section 1.1 to assign a statistical significance to the observed distance.
The key element in implementing this proposal is the choice of a suitable metric on the space of probability distributions on the reference tree. There are, of course, a multitude of choices: chapter 6 of Villani (2009) notes that there are ‘dozens and dozens’ of them and provides a discussion of their similarities, differences and various virtues.
Perhaps the most familiar metric is the total variation distance, which is just the supremum over all (Borel) sets of the difference between the masses assigned to the set by the two distributions. The total variation distance is clearly inappropriate for our purposes, however, because it pays no attention to the evolutionary distance structure on the tree: if we took k point placements and constructed another set of placements by perturbing each of the original placements by a tiny amount so that the two sets of placements were disjoint, then the total variation distance between the corresponding probability distributions would be 1, the largest it can be for any pair of probability distributions, even though we would regard the two sets of placements as being very close. Note that even genetic material from organisms of the same species can result in slightly different placements due to genetic variation within species and experimental error.
We therefore need a metric that is compatible with the evolutionary distance on the reference tree and measures two distributions as being close if one is obtained from the other by short-range redistributions of mass. The Kantorovich–Rubinstein (KR) metric, which can be defined for probability distributions on an arbitrary metric space, is a classical and widely used distance that meets this requirement and, as we shall see, has other desirable properties such as being easily computable on a tree. It is defined rigorously in Section 2 below, but it can be described intuitively in physical terms as follows. Picture each of two probability distributions on a metric space as a collection of piles of sand with unit total mass: the mass of sand in the pile at a given point is equal to the probability mass at that point. Suppose that the amount of ‘work’ that is required to transport an amount of sand from one place to another is proportional to the mass of the sand moved times the distance that it has to travel. Then, the KR distance between two probability distributions P and Q is simply the minimum amount of work required to move sand in the configuration corresponding to P into the configuration corresponding to Q. It will require little effort to move sand between the configurations corresponding to two similar probability distributions, whereas more will be needed for two distributions that place most of their respective masses on disjoint regions of the metric space. As noted by Villani (2009), the KR metric is also called the Wasserstein(1) metric or, in the engineering literature, the earth mover’s distance. We note that mass transport ideas have already been used in evolutionary bioinformatics for the comparison and clustering of ‘evolutionary fingerprints’—such a fingerprint being defined by Kosakovsky Pond et al. (2010) as a discrete bivariate distribution on synonymous and non-synonymous mutation rates for a given gene.
1.4. Overview of results
Our first result is that, in the phylogenetic case, the optimization that is implicit in the definition of the KR metric can be done analytically, resulting in a closed form expression that can be evaluated in linear time, thereby enabling analysis of the volume of data produced by large-scale sequencing studies. Indeed, as shown in Section 2, the metric can be represented as a single integral over the tree, and for point placements the integral reduces to a summation with a number of terms of the order of the number of placements. In contrast, computing the KR metric in Euclidean spaces of dimension greater than 1 requires a linear programming optimization step. It is remarkable that the point version of this closed form expression for the phylogenetic KR distance (although apparently not the optimal mass transport justification for the distance) was intuited by microbial ecologists and is nothing other than the weighted UniFrac distance that was recalled in Section 1.1 above.
We introduce Lp-generalizations of the KR metric that are analogous to metrics on the real line due to Zolotarev (Rachev, 1991; Rachev and Rüschendorf, 1998)—the KR metric corresponds to the case p = 1. Small p emphasizes primarily differences due to separation of samples across the tree, whereas large p emphasizes large mass differences. The generalizations do not arise from optimal mass transport considerations, but we remark in Section 5.3 that the square of the p = 2 version does have an appealing analysis of variance like interpretation as the amount of variability in a pooling of the two samples that is not accounted for by the variability in each of them.
We show in Section 3 that the distribution of the distance under the null hypothesis of no clustering is approximately that of a readily computable functional of a Gaussian process indexed by the tree and that this Gaussian process is relatively simple to simulate. Moreover, we observe that when p = 2 this approximate distribution is that of the square root of a weighted sum of random variables. We also discuss the interpretation of the resulting p-value when the data exhibit local ‘clumps’ that might be viewed as being the objects of fundamental biological interest rather than the individual reads.
In Section 5, we discuss alternative approaches to sample comparison. In particular, we remark that any probability distribution on a tree has a well-defined barycentre (i.e. centre of mass) that can be computed effectively. Thus, we can obtain a one-point summary of the location of a sample by considering the barycentre of the associated probability distribution and measure the similarity of two samples by computing the distance between the corresponding barycentres.
2. Phylogenetic Kantorovich–Rubinstein metric
In this section we more formally describe the phylogenetic KR metric, which is a particular case of the family of Wasserstein metrics. We then use a dual formulation of the KR metric to show that it can be calculated in linear time via a simple integral over the tree. We also introduce a Zolotarev-type Lp-generalization.
Let T be a tree with branch lengths. Write d for the path distance on T. We assume that probability distributions have been given on the tree via collections of either ‘point’ or ‘spread’ placements as described in Section 1.
For a metric space (S, r), the KR distance Z(P, Q) between two Borel probability distributions P and Q on S is defined as follows. Let
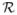 (P, Q) denote the set of probability distributions R on the product space S × S with the property R(A × S) = P(A) and R(S × B) = Q(B) for all Borel sets A and B (i.e. the two marginal distributions of R are P and Q). Then,
(P, Q) denote the set of probability distributions R on the product space S × S with the property R(A × S) = P(A) and R(S × B) = Q(B) for all Borel sets A and B (i.e. the two marginal distributions of R are P and Q). Then,
| (4) |
see, for example, Rachev (1991), Rachev and Rüschendorf (1998), Villani (2003, 2009) and Ambrosio et al. (2008).
There is an alternative formula for Z(P, Q) that comes from convex duality. Write
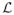 for the set of functions f: S → ℝ with the Lipschitz property |f(x) − f(y)| ≤ r(x, y) for all x, y ∈ S. Then,
for the set of functions f: S → ℝ with the Lipschitz property |f(x) − f(y)| ≤ r(x, y) for all x, y ∈ S. Then,
We can use this expression to obtain a simple explicit formula for Z(P, Q) when (S, r) = (T, d).
Given any two points x, y ∈ T, let [x, y] be the arc between them. There is a unique Borel measure λ on T such that λ([x, y]) = d(x, y) for all x, y ∈ T. We call λ the length measure; it is analogous to Lebesgue measure on the real line. Fix a distinguished point ρ ∈ T, which we call the ‘root’ of the tree. For any f ∈
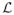 with f(ρ) = 0, there is a λ almost everywhere unique Borel function g:T → [−1, 1] such that f(x) = ∫[ρ,
x]
g(y)λ(dy) (this follows easily from the analogous fact for the real line).
with f(ρ) = 0, there is a λ almost everywhere unique Borel function g:T → [−1, 1] such that f(x) = ∫[ρ,
x]
g(y)λ(dy) (this follows easily from the analogous fact for the real line).
Given x ∈ T, put τ(x):= {y ∈ T: x ∈ [ρ, y]}; i.e., if we draw the tree with the root ρ at the top of the page, then τ (x) is the subtree below x. Observe that, if h: T → ℝ is a bounded Borel function and μ is a Borel probability distribution on T, then we have the integration-by-parts formula
Thus, if P and Q are two Borel probability distributions on T and f: T → ℝ is given by f(x) = ∫[ρ, x] g(y) λ(dy), then we have
and an analogous formula holds for Q. Hence,
It is clear that the integral is maximized by taking g(y) = 1 and g(y) = −1 when respectively P{τ(y)} > Q{τ(y)} and P{τ(y)} < Q{τ(y)}, so that
| (5) |
Note that equation (1) is the special case of equation (5) that arises when P assigns point mass 1/m to each of the leaves in community A, and Q assigns point mass 1/n to each of the leaves in community B.
We can generalize the definition of the KR distance by taking any pseudometric f on [0, 1] and setting
This object will be a pseudometric on the space of probability distributions on the tree T. All the distances considered so far are of the form Ẑf for an appropriate choice of the pseudometric f: (unweighted) UniFrac results from taking f(x, y) equal to 1 when exactly one of x or y is greater than 0, and Z arises when f(x, y) = |x − y|.
Furthermore, if f(x, y) = f(1 − x, 1 − y), then Ẑf is invariant with respect to the position of the root. Indeed, for λ almost everywhere y ∈ T we have that y is in the interior of a branch and P({y}) = Q({y}) = 0 so that, for such y, P{τ (y)} and P{T \ t (y)}, and Q{t (y)} and Q{T \ t (y)} are respectively the P-masses and Q-masses of the two disjoint connected components of T produced by removing y (see equation (2)), and hence these quantities do not depend on the choice of the root. Because
for any y ∈ T, the claimed invariance follows on integrating with respect to λ. In particular, we see that the distance Z is invariant to the position of the root: a fact that is already apparent from the original definition (4).
In a similar spirit, the KR distance as defined by the integral (5) can be generalized to an Lp Zolotarev-type version by setting
for 0 < p < ∞—see Rachev (1991) and Rachev and Rüschendorf (1998) for a discussion of analogous metrics for probability distributions on the real line. Intuitively, large p gives more weight in the distance to parts of the tree which are maximally different in terms of P and Q, whereas small p gives more weight to differences which require a large amount of transport. The position of the root ρ also does not matter for this generalization of Z by the argument above.
As the following computations show, the distance Z2 has a particularly appealing interpretation. First note that
Now, the product of indicator functions 1[ρ, ν] 1[ρ, w] is the indicator function of the set [ρ, ν] ∩ [ρ, w]. This set is an arc of the form [ρ, ν ∧ w], where ν ∧ w is the ‘most recent common ancestor’ of v and w relative to the root ρ. Hence, ∫T 1[ρ, ν](u)1[ρ, w](u)λ(du) = λ([ρ, ν ∧ w]) is . Therefore,
Thus, if X′, X″, Y′ and Y″ are independent T -valued random variables, where X′ and X″ both have distribution P and Y′ and Y″ both have distribution Q, then
| (6) |
Analogous to weighted UniFrac, we can ‘normalize’ the KR distance by dividing it by a scalar. The most direct analogue of the scaling factor D used for weighted UniFrac on a rooted tree (3) would be twice the KR distance between (P + Q)/2 and a point mass at the root. This is an upper bound by the triangle inequality. A root invariant version would be instead to place the point mass at the centre of mass (i.e. the barycentre; see Section 5.2) of (P + Q)/2, and twice the analogous distance is again an upper bound by the triangle inequality. It is clear from the original definition of the KR distance (4) that Z1(P, Q) is bounded above by the diameter of the tree (i.e. maxx, y{d(x, y)}) or by the possibly smaller similar quantity that arises by restricting x and y to the respective supports of P and Q. Any of these upper bounds can be used as a ‘normalization factor’.
The goal of introducing such normalizations would be to permit better comparisons between distances obtained for different pairs of samples. However, some care needs to be exercised here: it is not clear how to scale distances for pairs on two very different reference trees so that similar values of the scaled distances convey any readily interpretable indication of the extent to which the elements of the two pairs differ from each other in a ‘similar’ way. In short, when comparing results between trees, the KR distance and its generalizations are more useful as test statistics than as descriptive summary statistics.
3. Assessing significance
To assess the significance of the observed distance between the probability distributions that are associated with a pair of samples of placed reads of size m and n, we use the permutation strategy that was mentioned in Section 1 for assigning significance to observed UniFrac distances. In general, we have a pair of probability distributions representing the pair of samples that is of the form and , where πk is a probability distribution on the reference tree T representing the placement of the kth read in a pooling of the two samples (in the point placement case, each πk is just a unit point mass at some point wk ∈ T). We imagine creating all pairs of ‘samples’ that arise from placing m of the reads from the pool into one sample and the remaining n into the other, computing the distances between the two probability distributions on the reference tree that result from the placed reads and determining what proportion of these distances exceed the distance observed in the data. This proportion may be thought of as a p-value for a test of the null hypothesis of no clustering against an alternative of some degree of clustering.
Of course, for most values of m and n it is infeasible actually to perform this exhaustive listing of distances. We observe that, if I ⊆ {1, …, m + n} is a uniformly distributed random subset with cardinality m (i.e. all values are equally likely), J:= Ic is the complement of I, P̃ is the random probability distribution (1/m) Σi∈I πi and Q̃ is the random probability distribution (1/n)Σj∈J πj, then the proportion of interest is simply the probability that the distance between P̃ and Q̃ exceeds the distance between P and Q. We can approximate this probability in the obvious way by taking independent replicates of (I, J) and hence of (P̃, Q̃) and looking at the proportion of them that result in distances that are greater than the observed distance. We illustrate this Monte Carlo approximation procedure in Section 4.
3.1. Gaussian approximation
Although the above Monte Carlo approach to approximating a p-value is conceptually straightforward, it is tempting to explore whether there are further approximations to the outcome of this procedure that give satisfactory results but require less computation.
Recall that π1, …, πm+n is the pooled collection of placed reads and that P̃ = (1/m)Σi∈I πi and Q̃ = (1/n)Σj∈J πj, where I is a uniformly distributed random subset of {1, …, m + n} and J is its complement. Write
where we recall that τ(u) is the tree below u relative to the root ρ. Define a T -indexed stochastic process X = (X(u))u∈T by
Then,
If Hk, 1 ≤ k ≤ m + n, is the indicator random variable for the event {k ∈ I}, then
Writing
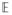 ,
,
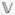 and ℂ for expectation, variance and covariance, we have
and ℂ for expectation, variance and covariance, we have
and
It follows that
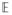 [X(u)] = 0 and
[X(u)] = 0 and
when m + n is large, where Ḡ(u):= {1/(m + n)}Σk Gk(u).
Remark 1
In the case of point placements, with the probability distribution πk being the point mass at wk ∈ T for 1 ≤ k ≤ m + n, then
By a standard central limit theorem for exchangeable random variables (see, for example, theorem 16.23 of Kallenberg (2001)), the process X is approximately Gaussian with covariance kernel Γ when m + n is large. A straightforward calculation shows that we may construct a Gaussian process ξ with covariance kernel Γ by taking independent standard Gaussian random variables η1, …, ηm+n and setting
It follows that the distribution of Zp(P̃, Q̃) is approximately that of the random variable
| (7) |
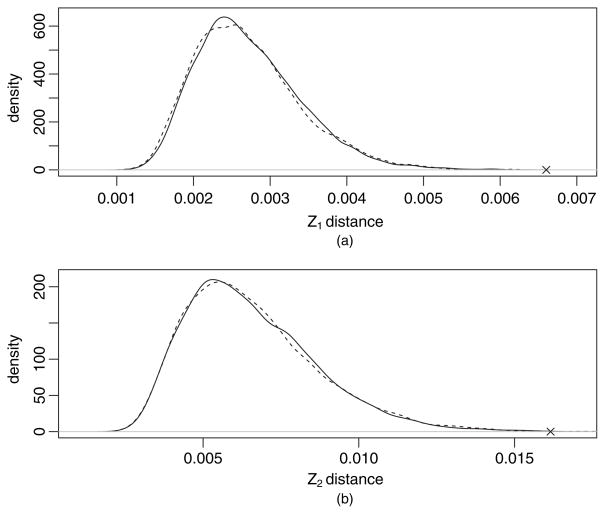
We can repeatedly sample the normal random variates ηi and numerically integrate expression (7) to approximate the distribution of this integral. In the example application of Section 4, this provides a reasonable though not perfect approximation (Fig. 3).
Fig. 3.
Comparison of the distribution of (a) Z1- and (b) Z2-distances obtained by shuffling (———), Gaussian approximation (– – –) and the observed value (×) for the example data set
There is an even simpler approach for the case p = 2. Let , k = 1, 2, …, and ψk, k = 1, 2, …, be the positive eigenvalues and corresponding normalized eigenfunctions of the non-negative definite, self-adjoint, compact operator on L2(λ) that maps the function f to the function ∫T Γ(·, ν) f(ν)λ(dν). The functions μkψk, k = 1, 2, …, form an orthonormal basis for the reproducing kernel Hilbert space associated with Γ and the Gaussian process ξ has the Karhunen–Loève expansion ξ(u) = Σk μk ψk(u)ηk, where ηk, k = 1, 2, …, are independent standard Gaussian random variables—see Jain and Marcus (1978) for a review of the theory of reproducing kernel Hilbert spaces and the Karhunen–Loève expansion.
Therefore, , and the distribution of is approximately that of a certain positive linear combination of independent random variables.
The eigenvalues of the operator that is associated with Γ can be found by calculating the eigenvalues of a related matrix as follows. Define an (m + n) × (m + n) non-negative definite, self-adjoint matrix M given by
If we have point placements at locations wk ∈ T for 1 ≤ k ≤ m + n as in remark 1, then
where I is the identity matrix, 1 is the vector which has 1 for every entry and the matrix N has (i, j) entry given by the distance from the root to the ‘most recent common ancestor’ of wi and wj.
Suppose that x is an eigenvector of M for the positive eigenvalue ν2. Set
| (8) |
Observe that
and so ψ is an (unnormalized) eigenfunction of the operator on L2(λ) defined by the covariance kernel Γ with eigenvalue ν2.
Conversely, suppose that μ2 is an eigenvalue of the operator with eigenfunction φ. Set xj:=∫T{Gj(ν) − Ḡ(ν)φ(ν) λ(dν). Then,
so that μ2 is an eigenvalue of M with (unnormalized) eigenvector of x.
It follows that the positive eigenvalues of the operator that is associated with Γ coincide with those of the matrix M and have the same multiplicities.
However, we do not actually need to compute the eigenvalues of M to implement this approximation. Because M is orthogonally equivalent to a diagonal matrix with the eigenvalues of M on the diagonal, we have from the invariance under orthogonal transformations of the distribution of the random vector η: (η1, …, ηm+n)T that has the same distribution as ηTMη. Thus, the distribution of the random variable is approximately that of Σij Mijηiηj.
We might hope to go even further in the case p = 2 and obtain an analytic approximation for the distribution or a useful upper bound for its right-hand tail. It is shown in Hwang (1980) that, if we order the positive eigenvalues so that and assume that , then
in the sense that the ratio of the two sides converges to 1 as r → ∞. It is not clear what the rate of convergence is in this result and it appears to require a detailed knowledge of the spectrum of the matrix M to apply it.
Gaussian concentration inequalities such as Borell’s inequality (see, for example, section 4.3 of Bogachev (1998)) give bounds on the right-hand tail that only require knowledge of and , but these bounds are far too conservative for the example in Section 4.
There is a substantial literature on various series expansions of densities of positive linear combinations of independent random variables. Some representative references are Robbins and Pitman (1949), Gurland (1955), Pachares (1955), Ruben (1962), Kotz et al. (1967) and Gideon and Gurland (1976)). However, it seems that applying such results would also require detailed knowledge of the spectrum of the matrix M as well as a certain amount of additional computation to obtain the coefficients in the expansion and then to integrate the resulting densities, and this may not be warranted given the relative ease with which it is possible to simulate the random variable ηTMη repeatedly.
Even though these more sophisticated ways of using the Gaussian approximation may not provide tight bounds, the process of repeatedly sampling normal random variates ηi and numerically integrating the resulting Gaussian approximation (7) does provide a useful way of approximating the distribution that is obtained by shuffling. This approximation is significantly faster to compute for larger collections of placements. For example, we considered a reference tree with 652 leaves and five samples with sizes varying from 3372 to 15 633 placements. For each of the 10 pairs of samples, we approximated the distribution of the Z1-distance under the null hypothesis of no difference by both creating pseudosamples via random assignment of reads to each member of the pair (‘shuffling’) and by simulating the Gaussian process functional with a distribution that approximates that of the Z1-distance between two such random pseudosamples. We used 1000 Monte Carlo steps for both approaches. The (shuffle, Gaussian) run times in seconds ranged from (494.1, 36.8) to (36.1, 2.2); in general, the Gaussian procedure ran an order of magnitude faster than the shuffle procedure.
3.2. Interpretation of p-values
Although the above-described permutation procedure is commonly used to assess the statistical significance of an observed distance, we discuss in this section how its interpretation is not completely straightforward.
In terms of the classical Neyman–Pearson framework for hypothesis testing, we are computing a p-value for the null hypothesis that an observed subdivision of a set of m + n objects into two groups of size m and n looks like a uniformly distributed random subdivision against the complementary alternative hypothesis. For many purposes, this turns out to be a reasonable proxy for the imperfectly defined notion that the two groups are ‘the same’ rather than ‘different’.
However, a rejection of the null hypothesis may not have the interpretation that is often sought in the microbial context—namely that the two collections of reads represent communities that are different in biologically relevant ways. For example, assume that m = n = NK for integers N and K. Suppose that the placements in each sample are obtained by independently laying down N points uniformly (i.e. according to the normalized version of the measure λ) and then putting K placements at each of those points. The stochastic mechanism generating the two samples is identical and they are certainly not different in any interesting way, but if K is large relative to N the resulting collections of placements will exhibit a substantial ‘clustering’ that will be less pronounced in the random pseudosamples, and the randomization procedure will tend to produce a ‘significant’ p-value for the observed KR distance if the clustering is not taken into account.
These considerations motivate consideration of randomization tests performed on data which are ‘clustered’ on an organismal level. Clustering reads by organism is a difficult task and an active research topic (White et al., 2010). A thoroughgoing exploration of the effect of different clustering techniques is beyond the scope of this paper, but we examine the effect of some simple approaches in the next section.
4. Example application
To demonstrate the use of the Zp-metric in an example application, we investigated variation in expression levels for the psbA gene for an experiment in the Sargasso Sea (Vila-Costa et al., 2010). Metatranscriptomic data were downloaded from http://camera.calit2.net/, and a psbA alignment was supplied by Robin Kodner. Searching and alignment was performed by using HMMER (Eddy, 1998), a reference tree was inferred by using RAxML (Stamatakis, 2006) and phylogenetic placement was performed by using pplacer (Matsen et al., 2010). The calculations that are presented here were performed by using the ‘guppy’ binary that is available as part of the pplacer suite of programs (http://matsen.fhcrc.org/pplacer).
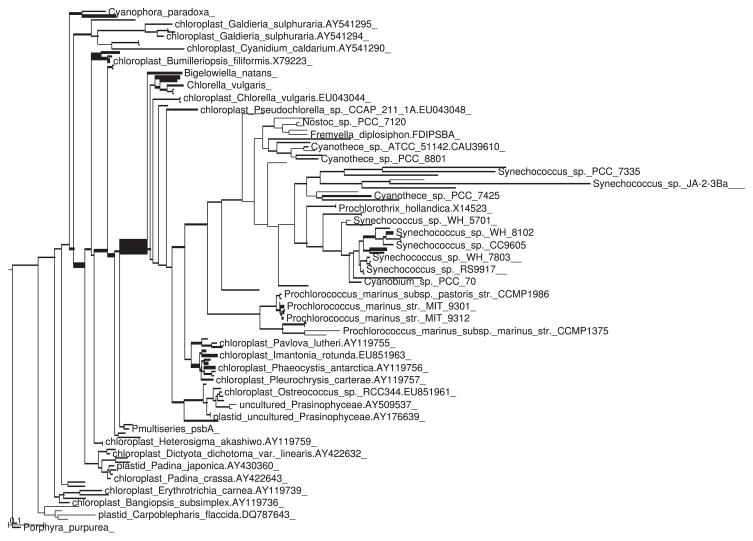
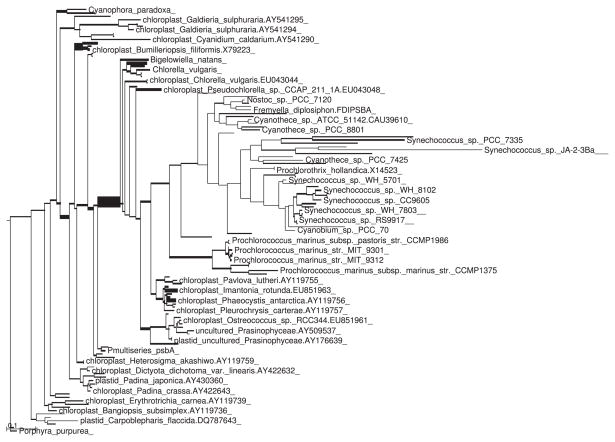
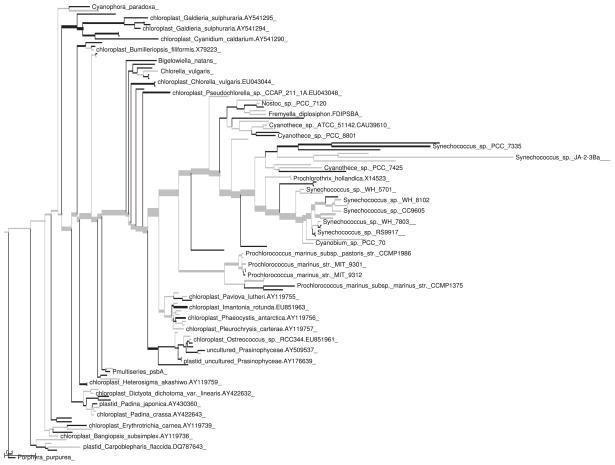
Visual inspection of the trees fattened by number of placements showed the same overall pattern with some minor differences (Figs 1 and 2). However, application of the KR metric revealed a significant difference between the two samples. The value of Z1 for this example (using spread placements and normalizing by total tree length) was 0.006 601. This is far out on the tail of the distribution (Fig. 3), and is in fact larger than any of the 1000 replicates generated via shuffling or the Gaussian-based approximation.
Fig. 1.
Tree with branches thickened as a linear function of the number of placements in the control sample placed on that branch
Fig. 2.
Tree as in Fig. 1, but for the DMSP-treated sample
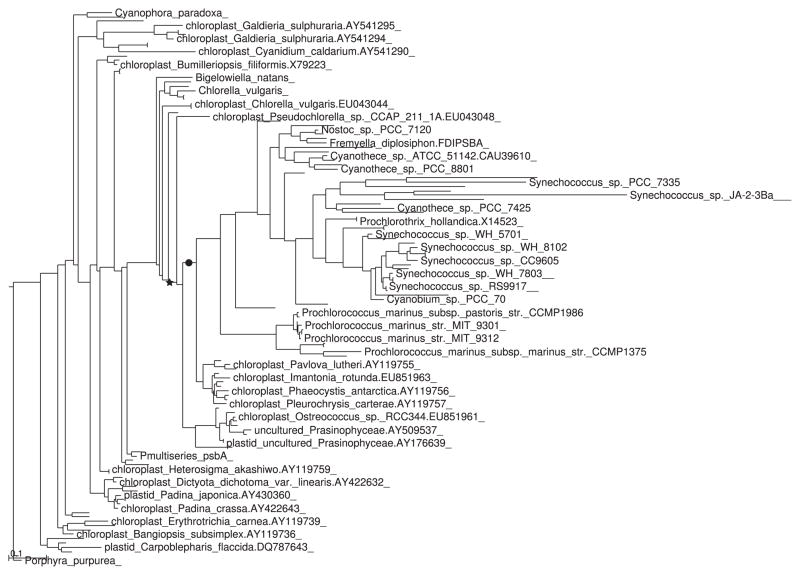
Such a low p-value prompts the question of whether the centre of mass of the two distributions is radically different in the two samples (see Section 5.2). In this case, the answer is no, as the two barycentres are quite close together (Fig. 4; see Section 5.2).
Fig. 4.
Dendrogram with barycenters marked: ●, control sample; ★, sample treated with DMSP
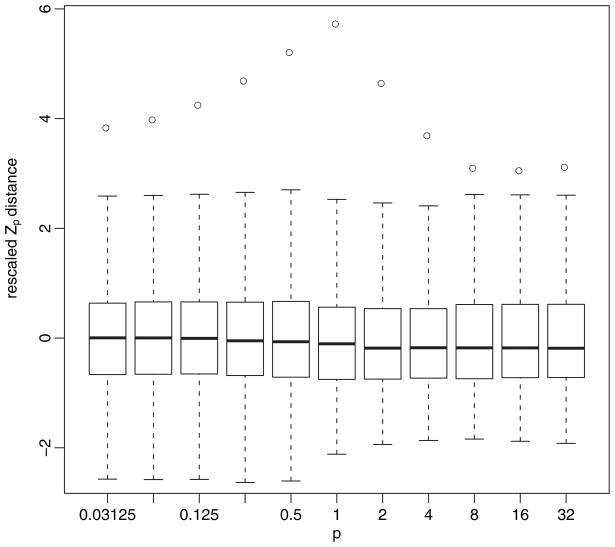
It was not intuitively obvious to us how varying p would affect the distribution of the Zp-distance under the null hypothesis of no clustering. To investigate this question, we plotted the observed distance along with boxplots of the null distribution for a collection of different p (Fig. 5). It is apparent that there is a consistent conclusion over a wide range of values of p.
Fig. 5.
Plot showing sample (○) and randomized ranges (
 ): outliers have been eliminated for clarity; for each p, the distribution was rescaled by subtracting the mean and dividing by the standard deviation
): outliers have been eliminated for clarity; for each p, the distribution was rescaled by subtracting the mean and dividing by the standard deviation
We can also visualize the difference between the two samples by drawing the reference tree with branch thicknesses that represent the minimal amount of ‘mass’ that flows through that branch in the optimal transport of mass implicit in the computation of Z1(P, Q) and with branch shadings that indicate the sign of the movement (Fig. 6).
Fig. 6.
Tree displaying the optimal movement of mass for the KR metric: when moving from the first probability distribution to the second, branches marked in gray have mass moving towards the root, whereas those marked in black have mass moving towards the leaves; thickness shows the quantity of mass moving through that branch
Next we illustrate the effect of simple clustering on randomization tests for the KR metric. The clustering for these tests will be done by rounding placement locations by using two parameters, the mass cut-off C and the number of significant figures S, as follows. Placement locations with low probability mass for a given read are likely to be error prone (Matsen et al., 2010); thus the first step is to throw away those locations that are associated with posterior probability or ‘likelihood weight ratio’ below C. The second step is to round the placement attachment location and pendant branch length by multiplying them by 10S and rounding to the nearest integer. The reads whose placements are identical after this rounding process are then said to cluster together. We shall call the number of reads in a given cluster the ‘multiplicity’ of the cluster.
After clustering, various choices can be made about how to scale the mass distribution according to multiplicity. Again, each cluster has some multiplicity and a distribution of mass across the tree according to likelihood weight. One option (which we call straight multiplicity) is to multiply the mass distribution by the multiplicity. Alternatively, we might forget about multiplicity by distributing a unit of mass for each cluster irrespectively of multiplicity. Or we might do something intermediate by multiplying by a transformed version of multiplicity; in this case we transform by the hyperbolic inverse sine.
We calculated distances and p-values for several clustering parameters and multiplicity uses (Table 1). To randomize a clustered collection of reads, we reshuffled the labels on the clusters, maintaining the groupings of the reads within the clusters; thus, all the placements in a given cluster were assigned to the same pseudosample. The distances do not change very much under different collections of clustering parameters, as there is little redistribution of mass. However, the p-values are different, because under our randomization strategy mass is relabelled cluster by cluster. The different choices that are represented in Table 1 represent different perspectives on what the multiplicities mean. The ‘strict’ multiplicity-based p-value corresponds to interpreting the multiplicity with which reads appear as meaningful, the unit cluster p-value corresponds to interpreting the multiplicities as noise and the inverse hyperbolic sine transformed multiplicity sits somewhere in between. The p-value with no clustering (as above, Z1 = 0.006 601, with a p-value of 0) corresponds to interpreting reads as being sampled one at a time from a distribution.
Table 1.
Distances Z1 and significance levels P for various choices of clustering parameters and multiplicity interpretations described in the text for 10000 randomizations
| S | C | Strict Z1 | Strict p | Inverse hyperbolic sine Z1 | Inverse hyperbolic sine p | Unit Z1 | Unit p |
|---|---|---|---|---|---|---|---|
| 1 | 0.01 | 0.006578 | 0.0087 | 0.007016 | 0.0008 | 0.007054 | 0.0003 |
| 1 | 0.05 | 0.006584 | 0.0218 | 0.006986 | 0.0018 | 0.007036 | 0.0005 |
| 1 | 0.1 | 0.006562 | 0.035 | 0.007214 | 0.001 | 0.007322 | 0.0005 |
| 2 | 0.01 | 0.006601 | 0.0018 | 0.007076 | 0.0003 | 0.007281 | 0.0001 |
| 2 | 0.05 | 0.006587 | 0.0029 | 0.00696 | 0.0005 | 0.007111 | 0.0002 |
| 2 | 0.1 | 0.006592 | 0.0039 | 0.007088 | 0.0003 | 0.007423 | 0 |
| 3 | 0.01 | 0.006601 | 0.0017 | 0.006806 | 0.0005 | 0.006922 | 0.0002 |
| 3 | 0.05 | 0.006602 | 0.0018 | 0.006719 | 0.0003 | 0.006695 | 0.0001 |
| 3 | 0.1 | 0.006612 | 0.0012 | 0.006775 | 0.0003 | 0.006816 | 0.0001 |
The choice of how to use multiplicity information depends on the biological setting. There is no doubt that increased organism abundance increases the likelihood of sampling a read from that organism; however, the relationship is almost certainly non-linear and dependent on species and experimental set-up (Morgan et al., 2010). How multiplicities are interpreted and treated in a specific instance is thus a decision that is best left to the researcher using his or her knowledge of the environment being studied and the details of the experimental procedure.
5. Discussion
5.1. Other approaches
5.1.1. Operational taxonomic units
The methods that are described in this paper are complementary to comparative methods based on ‘operational taxonomic units’ (OTUs). OTUs are groups of reads which are assumed to represent the reads from a single species and are typically heuristically defined by using a fixed percentage sequence similarity cut-off. A comparative analysis then proceeds by comparing the frequency of various OTUs in the different samples. There has been some contention about whether OTU-based methods or phylogenetic-based methods are superior—e.g. Schloss (2008) and Lozupone et al. (2010)—but most studies use a combination of both, and the major software packages implement both. A recent comparative study for distances on OTU abundances can be found in Kuczynski et al. (2010).
5.1.2. Other phylogenetic approaches
There are various ways to compare microbial samples in a phylogenetic context besides the method that is presented here. One popular means of comparing samples is the ‘parsimony test’, by which the most parsimonious assignment of internal nodes of the phylogenetic tree to communities is found; the resulting parsimony score is interpreted as a measure of difference between communities (Slatkin and Maddison, 1989; Schloss and Handelsman, 2006). Another interesting approach is to consider a ‘generalized principal components analysis’ whereby the tree structure is incorporated into the process of finding principal components of the species abundances (Bik et al., 2006; Purdom, 2008). The KR metric complements these methods by providing a means of comparing samples that leverages established statistical methodology, that takes into account uncertainty in read location, and can be visualized directly on the tree.
There are other metrics that could be used to compare probability distributions on a phylogenetic tree. The metric on probability distributions that is most familiar to statisticians other than the total variation distance is probably the Prohorov metric and so they may feel more comfortable using it rather than the KR metric. However, the Prohorov metric is defined in terms of an optimization that does not appear to have a closed form solution on a tree and, in any case, for a compact metric space there are results that bound the Prohorov metric above and below by functions of the KR metric (see problem 3.11.2 of Ethier and Kurtz (1986)) so the two metrics incorporate very similar information about the differences between a pair of distributions.
5.2. Barycentre of a probability distribution on a phylogenetic tree
It can be useful to compare probability distributions on a metric space by calculating a suitably defined centre of mass that provides a single point summary for each distribution. Recall the standard fact that, if P is a probability distribution on a Euclidean space such that ∫ |y − x|2 P(dy) is finite for some (and hence all) x, then the function x ↦ ∫ |y − x|2 P(dy) has a unique minimum at x0 = ∫ y P(dy). A probability distribution P on an arbitrary metric space (S, r) has a centre of mass or barycentre at x0 if ∫r(x, y)2P(dy) is finite for some (and hence all x) and the function x ↦ ∫ r(x, y)2 P(dy) has a unique minimum at x0. In terms of the concepts that were introduced above, the barycentre is the point x that minimizes the Z2-distance between the point mass δx and P.
Barycentres need not exist for general metric spaces. However, it is well known that barycentres do exist for probability distributions on Hadamard spaces. A Hadamard space is a simply connected complete metric space in which there is a suitable notion of the length of a path in the space, the distance between two points is the infimum of the lengths of the paths joining the points and the space has non-positive curvature in an appropriate sense—see Burago et al. (2001). Equivalently, a Hadamard space is a complete CAT(0) space in the sense of Bridson and Haefliger (1999).
It is a straightforward exercise to check that a tree is a Hadamard space—see example II.1.15(4) of Bridson and Haefliger (1999) and note the remark after definition II.1.1 of Bridson and Haefliger (1999) that a Hadamard space is the same thing as a complete CAT(0) space. Note that CAT(0) spaces have already made an appearance in phylogenetics in the description of spaces of phylogenetic trees (Billera et al., 2001).
The existence of barycentres on the tree (T, d) may also be established directly as follows. As a continuous function on a compact metric space, the function f: T → ℝ+ defined by f(x):= ∫T d(x, y)2 P(dy) achieves its infimum. Suppose that the infimum is achieved at two points x′ and x″. Define a function γ: [0, d(x′, x″)] → [x′, x″], where [x′, x″] ⊆ T is the arc between x′ and x″, by the requirement that γ(t) is the unique point in [x′, x″] that is distance t from x′. It is straightforward to check that the composition f ◦ γ is strongly convex, i.e.
for 0 < α < 1 and r, s ∈ [0, d(x′, x″)]. In particular, f[γ(d{x′, x″)/2}] = (f ◦ γ){d(x′, x″)/2} < {f(x′) + f(x″)}/2, contradicting the definitions of x′ and x″. Thus, a probability distribution on a tree has a barycentre in the above sense.
We next consider how to compute the barycentre of a probability distribution P on the tree (T, d). For each point u ∈ T there is the associated set of directions in which it is possible to proceed when leaving u. There is one direction for every connected component of T \ {u}. Thus, just one direction is associated with a leaf, two directions associated with a point in the interior of a branch and k associated with a vertex of degree k. Given a point u and a direction δ, write T(u, δ) for the subset of T consisting of points ν ≠ u such that the unique path connecting u and v departs u in the direction δ, set
and note that
where the limit is taken over ν → u, ν ∈ T(u, δ). If u is in the interior of a branch [a, b] and b is in the direction δ from u, u is in the direction α from a, and u is in the direction β from b, then
If, for some vertex u of the reference tree, D(u, δ) is greater than 0 for all directions δ associated with u, then u is the barycentre (this case includes the trivial case in which u is a leaf and all the mass of P is concentrated on u). If there is no such vertex, then there must be a unique pair of neighbouring vertices a and b such that D(a, α) < 0 and D(b, β) < 0, where α is the direction from a pointing towards b and β is the direction from b pointing towards a. In that case, the barycentre must lie on the branch between a and b, and it follows from the calculations above that the barycentre is the point u ∈ (a, b) such that d(a, u) = −D(a, α).
5.3. and analysis of variance
In this section we demonstrate how can be interpreted as a difference between the pooled average of pairwise distances and the average for each sample individually.
As above, let π1, …, πm and πm+1, …, πm+n respectively be the placements in the first and second sample, so that each πk is a probability distribution on the tree T, and . Set
Recall the T -valued random variables X′, X″, Y′ and Y″ that appeared in equation (6). If I′ and I″ are {0, 1}-valued random variables with ℙ{I′ = 1} = ℙ{I″ = 1} = m/(m + n) and X′, X″, Y′, Y″, I′ and I″ are independent, then defining Z′ and Z″ by Z′ = X′ on the event {I′ = 1} (and Z″ = X″ on the event {I″ = 1}) and Z′ = Y′ on the event {I′ = 0} (and Z″ = Y″ on the event {I″= 0}) gives two T -valued random variables with common distribution R.
It follows readily from equation (6) that
Thus, gives an indication of the ‘variability’ in the pooled collection πk, 1 ≤ k ≤ m + n, that is over the variability in the two collections πi, 1 ≤ i ≤ m, and pj, m + 1 ≤ j ≤ m + n.
6. Conclusion
As sequencing becomes faster and less expensive, it will become increasingly common to have a collection of large data sets for a given gene. Phylogenetic placement can furnish an evolutionary context for query sequences, resulting in each data set being represented as a probability distribution on a phylogenetic tree. The KR metric is a natural means to compare those probability distributions. In this paper we showed that the weighted UniFrac metric is the phylogenetic KR metric for point placements. We explored Zolotarev-type generalizations of the KR metric, showed how to approximate the limiting distribution and made connections with analysis of variance.
The phylogenetic KR metric and its generalizations can be used any time that we want to compare two probability distributions on a tree. However, our software implementation is designed with metagenomic and metatranscriptomic investigations in mind; for this reason it is tightly integrated with the phylogenetic placement software pplacer (Matsen et al., 2010). With more than two samples, principal components analysis and hierarchical clustering can be applied to the pairwise distances to cluster environments on the basis of the KR distances as has been done with UniFrac (Lozupone and Knight, 2005; Lozupone et al., 2008; Hamady et al., 2009). We have recently developed versions of these techniques which leverage the special structure of these data (Matsen and Evans, 2011).
Another potential future extension which was not explored here is to partition the tree into subsets in a principal components fashion for a single data set. Recall that equation (8) gives a formula for the eigenfunctions of the covariance kernel Γ given the eigenvectors of M. For any k, we could partition the tree into subsets on the basis of the sign of the product of the first k eigenfunctions, which would be analogous to partitioning Euclidean space by the hyperplanes that are associated with the first k eigenvectors in traditional principal components analysis.
Future methods will also need to take details of the DNA extraction procedure into account. Recent work shows that current laboratory methodology cannot recover absolute mixture proportions owing to differential ease of DNA extraction between organisms (Morgan et al., 2010). However, relative abundance between samples for a given organism with a fixed laboratory protocol potentially can be measured, assuming that consistent DNA extraction protocols are used. An important next step is to incorporate such organism-specific biases into the sort of analysis that was described here.
Acknowledgments
The first author was supported in part by National Science Foundation grant DMS-0907630. The second author was supported by the Miller Institute for Basic Research in Science, University of California at Berkeley, by Fred Hutchinson Cancer Research Center start-up funds, and National Institutes Health grant HG005966-01.
The authors are grateful to Robin Kodner for her psbA alignment, the Armbrust laboratory for advice and for the use of their computing cluster, Mary Ann Moran and her laboratory for allowing us to use her metagenomic sample from the DMSP experiment, David Donoho for an interesting suggestion, Steve Kembel for helpful conversations and Aaron Gallagher for programming support.
The manuscript was greatly improved by suggestions from The Joint editor, Associate Editor and two reviewers.
Contributor Information
Steven N. Evans, University of California at Berkeley, USA
Frederick A. Matsen, Fred Hutchinson Cancer Research Center, Seattle, USA
References
- Ambrosio L, Gigli N, Savaré G. Gradient Flows in Metric Spaces and in the Space of Probability Measures. 2. Basel: Birkhäuser; 2008. [Google Scholar]
- Baker B, Banfield J. Microbial communities in acid mine drainage. FEMS Microbiol Ecol. 2003;44:139–152. doi: 10.1016/S0168-6496(03)00028-X. [DOI] [PubMed] [Google Scholar]
- Berger S, Stamatakis A. Evolutionary placement of short sequence reads. Submitted to Syst Biol. 2010 doi: 10.1093/sysbio/syr010. (Available from http://arxiv.org/abs/0911.2852.) [DOI] [PMC free article] [PubMed]
- Bik E, Eckburg P, Gill S, Nelson K, Purdom E, Francois F, Perez-Perez G, Blaser M, Relman D. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natn Acad Sci USA. 2006;103:732. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billera L, Holmes S, Vogtmann K. Geometry of the space of phylogenetic trees. Adv Appl Math. 2001;27:733–767. [Google Scholar]
- Bogachev VI. Gaussian Measures. Providence: American Mathematical Society; 1998. [Google Scholar]
- Bridson MR, Haefliger A. Metric Spaces of Non-positive Curvature. Berlin: Springer; 1999. [Google Scholar]
- Burago D, Burago Y, Ivanov S. A Course in Metric Geometry. Providence: American Mathematical Society; 2001. [Google Scholar]
- Caporaso J, Kuczynski J, Stombaugh J, Bittinger K, Bushman F, Costello E, Fierer N, Peña A, Goodrich J, Gordon J. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnues C, Rodriguez-Brito B, Rayhawk S, Kelley S, Tran T, Haynes M, Liu H, Furlan M, Wegley L, Chau B. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- Eddy S. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Edgington ES, Onghena P. Randomization Tests. 4. Boca Raton: Chapman and Hall—CRC; 2007. [Google Scholar]
- Ethier SN, Kurtz TG. Markov Processes: Characterization and Convergence. New York: Wiley; 1986. [Google Scholar]
- Felsenstein J. Inferring Phylogenis. Sunderland: Sinauer; 2004. [Google Scholar]
- Fierer NK, Hamady M, Lauber C, Knight R. The influence of sex handedness and washing on the diversity of hand surface bacteria. Proc Natn Acad Sci USA. 2008;105:17994. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The Design of Experiment. New York: Hafner; 1935. [Google Scholar]
- Frank D, St Amand A, Feldman R, Boedeker E, Harpaz N, Pace N. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natn Acad Sci USA. 2007;104:13780. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gideon RA, Gurland J. Series expansions for quadratic forms in normal variables. J Am Statist Ass. 1976;71:227–232. [Google Scholar]
- Gill S, Pop M, DeBoy R, Eckburg P, Turnbaugh P, Samuel B, Gordon J, Relman D, Fraser-Liggett C, Nelson K. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good P. Permutation, Parametric and Boostrap Tests of Hypotheses. 3. New York: Springer; 2005. [Google Scholar]
- Gurland J. Distribution of definite and of indefinite quadratic forms. Ann Math Statist. 1955;26:122–127. [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2009;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman A, Riddle S, McPhillips T, Ludaescher B, Eisen J. WATERS: a workflow for the alignment, taxonomy, and ecology of ribosomal sequences. BMC Bioinform. 2010;11:317. doi: 10.1186/1471-2105-11-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang CR. Gaussian measure of large balls in a Hilbert space. Proc Am Math Soc. 1980;78:107–110. [Google Scholar]
- Jain NC, Marcus MB. Probability on Banach Spaces. New York: Dekker; 1978. Continuity of sub-Gaussian processes; pp. 81–196. [Google Scholar]
- Kallenberg O. Foundations of Modern Probability. 2. New York: Springer; 2001. [Google Scholar]
- Kosakovsky Pond S, Scheffler K, Gravenor M, Poon A, Frost S. Evolutionary fingerprinting of genes. Molec Biol Evol. 2010;27:520. doi: 10.1093/molbev/msp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotz S, Johnson NL, Boyd DW. Series representations of distributions of quadratic forms in normal variables: I, Central case. Ann Math Statist. 1967;38:823–837. [Google Scholar]
- Kuczynski J, Liu Z, Lozupone C, McDonald D. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Meth. 2010;7:10. doi: 10.1038/nmeth.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Cantarel B, Coutinho P, Henrissat B, Gordon J, Knight R. The convergence of carbohydrate active gene repertoires in human gut microbes. Proc Natn Acad Sci USA. 2008;105:15076. doi: 10.1073/pnas.0807339105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Kelley S, Knight R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2010;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen F, Evans S. Edge principal components and squash clustering: using the special structure of phylogenetic placement data for sample comparison. 2011. To be published. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen F, Kodner R, Armbrust E. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinform. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Claverie J, Ogata H. Taxonomic distribution of large DNA viruses in the sea. Genome Biol. 2008;9:R106. doi: 10.1186/gb-2008-9-7-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan J, Darling A, Eisen J. Metagenomic sequencing of an in vitro-simulated microbial community. PLOS ONE. 2010;5:article e10209. doi: 10.1371/journal.pone.0010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachares J. Note on the distribution of a definite quadratic form. Ann Math Statist. 1955;26:128–131. [Google Scholar]
- Pitman EJG. Significance tests which may be applied to samples from any populations. J R Statist Soc Suppl. 1937a;4:119–130. [Google Scholar]
- Pitman EJG. Significance tests which may be applied to samples from any population: II, The correlation coefficient test. J R Statist Soc Suppl. 1937b;4:225–232. [Google Scholar]
- Pitman E. Significance tests which may be applied to samples from any population: III, The analysis of variance test. Biometrika. 1938;29:322–335. [Google Scholar]
- Purdom E. Analyzing data with graphs: metagenomic data and the phylogenetic tree. 2008. [Google Scholar]
- Rachev ST. Probability Metrics and the Stability of Stochastic Models. Chichester: Wiley; 1991. [Google Scholar]
- Rachev ST, Rüschendorf L. Probability and Its Applications. I. New York: Springer; 1998. Mass Transportation Problems. [Google Scholar]
- Rawls J, Mahowald M, Ley R, Gordon J. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127:423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintala H, Pitkäranta M, Toivola M, Paulin L, Nevalainen A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 2008;8:56. doi: 10.1186/1471-2180-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins H, Pitman EJG. Application of the method of mixtures to quadratic forms in normal variates. Ann Math Statistics. 1949;20:552–560. [Google Scholar]
- Ruben H. Probability content of regions under spherical normal distributions: IV, The distribution of homogeneous and non-homogeneous quadratic functions of normal variables. Ann Math Statist. 1962;33:542–570. [Google Scholar]
- Schloss P. Evaluating different approaches that test whether microbial communities have the same structure. ISME J. 2008;2:265–275. doi: 10.1038/ismej.2008.5. [DOI] [PubMed] [Google Scholar]
- Schloss P, Handelsman J. Introducing TreeClimber a test to compare microbial community structures. Appl Environ Microbiol. 2006;72:2379. doi: 10.1128/AEM.72.4.2379-2384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P, Westcott S, Ryabin T, Hall J, Hartmann M, Hollister E, Lesniewski R, Oakley B, Parks D, Robinson C, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Maddison WP. A Cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Vila-Costa M, Rinta-Kanto J, Sun S, Sharma S, Poretsky R, Moran M. Transcriptomic analysis of a marine bacterial community enriched with dimethylsulfoniopropionate. ISME J. 2010;4:1410–1420. doi: 10.1038/ismej.2010.62. [DOI] [PubMed] [Google Scholar]
- Villani C. Topics in Optimal Transportation. Providence: American Mathematical Society; 2003. [Google Scholar]
- Villani C. Optimal Transport. Berlin: Springer; 2009. [Google Scholar]
- Von Mering C, Hugenholtz P, Raes J, Tringe S, Doerks T, Jensen L, Ward N, Bork P. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science. 2007;315:1126. doi: 10.1126/science.1133420. [DOI] [PubMed] [Google Scholar]
- White J, Navlakha S, Nagarajan N, Ghodsi MR, Kingsford C, Pop M. Alignment and clustering of phylogenetic markers—implications for microbial diversity studies. BMC Bioinformatics. 2010;11(1) doi: 10.1186/1471-2105-11-152. [DOI] [PMC free article] [PubMed] [Google Scholar]