Abstract
To study the kinetics of lymphocytes, models have divided the cell population into subpopulations with different turnover rates. These have been called ‘kinetic heterogeneity models’ so as to distinguish them from ‘temporal heterogeneity models’, in which a cell population may have different turnover rates at different times, e.g. when resting versus when activated. We model labelling curves for temporally heterogeneous populations, and predict that they exhibit equal biphasic up- and downslopes. We show when cells divide only once upon activation, these slopes are dominated by the slowest exponent, yielding underestimates of the average turnover rate. When cells undergo more than one division, the labelling curves allow fitting of the two exponential slopes in the temporal heterogeneity model. The same data can also be described with a two-compartment kinetic heterogeneity model. In both instances, the average turnover rate is correctly estimated. Because both models assume a different cell biology but describe the data equally well, the parameters of either model have no simple biological interpretation, as each parameter could reflect a combination of parameters of another biological process. Thus, even if there are sufficient data to reliably estimate all exponentials, one can only accurately estimate an average turnover rate. We illustrate these issues by re-fitting labelling data from healthy and HIV-infected individuals.
Keywords: lymphocyte, turnover, lifespan, modelling
1. Introduction
Despite great advances in immunological research during the last decades, estimating the kinetics of lymphocyte populations has proved quite difficult. There are widely divergent estimates of the production rates, division rates and lifespans of mouse and human lymphocyte populations [1]. As a consequence, it is poorly understood how memory is maintained, how the naive lymphocyte repertoire remains diverse and how homeostasis is brought about. Additionally, it remains poorly understood how viruses such as HIV, and diseases such as rheumatoid arthritis, disturb the normal population dynamics and deteriorate the functioning of the immune system.
Several in vivo labelling techniques have been developed that have enabled the generation of quantitative data on lymphocyte dynamics. Examples of labels are the fluorescent dye carboxy-fluorescein diacetate succinimidyl ester (CFSE), the base analogue 5-bromo-2′-deoxyuridine (BrdU) and the stable isotype deuterium. Major advantages of deuterium labelling are that it is unlikely to change the cellular dynamics, and that, unlike CFSE and BrdU, it can safely be used in humans, and hence provide insights into human lymphocyte turnover in health and disease. Animals and humans have been labelled with deuterated glucose (2H2-glucose) or heavy water (2H2O) for 1 or 2 days [2–6], 5 days [7] to one week [8] or several months [9–11], and have subsequently been followed to study the loss of label during a washout phase. After deuterium labelling, one sorts the cell population of interest, isolates the DNA from the cells, and uses mass spectrometry to determine the enrichment of deuterium in the DNA [12–15]. During the labelling period, some fraction of the hydrogen atoms in the body will be replaced by deuterium, and dividing cells will incorporate both deuterium and hydrogen in newly synthesized DNA molecules. Ultimately, the data correspond to a normalized fraction of labelled DNA, 0 ≤ L(t) < 1, which increases during the labelling phase, and which decreases after deuterium has been withdrawn. We will refer to these two phases of L(t) as the upslope and the downslope, respectively. Adding deuterium should not change the number of T cells, and thus one typically assumes steady state for the total cell population during the labelling experiment.
Although deuterium labelling is increasingly being used, the interpretation of the data in terms of the underlying kinetics of the cells has turned out to be notoriously difficult [1,8,10,16–18]. One major problem is that the estimated turnover rates differ markedly depending on whether 2H2-glucose or 2H2O has been used for the labelling. Another problem is that the upslopes and the downslopes depend on the period of labelling, with steeper slopes for shorter labelling periods [1] (figure 1a). For the downslopes, this is relatively well understood because Asquith et al. [16] demonstrated that whenever the population is kinetically heterogeneous, the labelled fraction will initially be enriched in cells with a higher than average turnover rate. For the upslopes, this is not understood. The initial upslope should reflect the average turnover rate [1,16], and there should be one unique labelling curve that monotonically approaches the asymptote, L(∞) = 1, where all cells are labelled. Thus, in situations where the estimated upslope seems to depend on the labelling period, one either has insufficient data to determine the one common slope, and/or one is fitting the true labelling curve with an incorrect model.
Figure 1.
Examples of memory T cell kinetics inferred from deuterium labelling. (a) Half-lives of memory T cells taken from table 1 in Borghans & De Boer [1]. The half-life is based on the average proliferation rate (i.e., the upslope of labelling) estimated in various deuterium labelling experiments, and is not expected to depend on the length of labelling period [1,16]. The bullets and the squares represent the CD4 and CD8 estimates, and the solid and the dashed lines depict their averages, respectively. The data points at a labelling period of 63 days were obtained with heavy water labelling, all others with d-glucose labelling; see Borghans & De Boer [1] for a discussion the various data sets. The horizontal axis is logarithmic to bring the time points closer together. (b) Raw deuterium enrichment data of the CD4+ (bullets) and CD8+ (squares) memory T cells from subject E in Vrisekoop et al. [10], which is their case with the most dense time sampling. Label was withdrawn at day 63. The vertical axis is logarithmic to better see the biphasic decline. The dashed thin lines depict the extrapolation of the initial downslope between day 63 and 75, and illustrate that late time points remain above those lines. Since the half-life of heavy water is relatively slow, i.e. 5–10 days [10], this simple extrapolation is conservative as some of the dividing cells continue to become labelled between day 63 and 75. Fitting one and two compartment models like equation (1.1) to these data confirms that all memory T cell data from these healthy volunteers [10] show evidence of heterogeneity [19].
One of the major problems for interpreting labelling experiments is that we do not know which mathematical model should be employed to fit the data, and that truly realistic models have so many parameters that one would be overfitting the simple up- and downslopes of deuterium enrichment over time. The appropriate model may also depend on the cells types that have been labelled, as naive T cells, self-renewing memory T cells and clonally expanding effector T cells have very different dynamics. Even if one deals with a single population of sorted cells, the population may still be heterogeneous. Here, we will analyse two types of heterogeneity: heterogeneity in phenotype, i.e. what type of cell; and in history, i.e. what has recently happened to the cell [3], which have been called ‘kinetic heterogeneity’ and ‘temporal heterogeneity’, respectively [20]. Kinetic heterogeneity implies that there exist separate subpopulations of cells that have different turnover rates. Temporal heterogeneity is more subtle and implies that a cell may transiently have a different turnover rate. For example, lymphocytes after being activated may have an increased death rate, a phenomenon called activation-induced cell death.
Currently, there is reasonable consensus on what models to use for describing the labelling of a possibly heterogeneous population of cells that are turning over at different rates. Ganusov et al. [20] demonstrated that the several models that have previously been used for describing kinetically heterogeneous populations [8,16,21] can all be written as a model for n independent subpopulations, each turning over at a rate di,
 |
1.1 |
where αi is the fraction of cells with turnover rate di, and tend defines the end of the labelling phase [20]. The first equation describes the kinetics of label acquisition from which one can derive the upslope. The second equation describes the kinetics of label loss after the end of the labelling period, from which one can derive the downslope. For the case of just one population, n = 1 and no asymptote, α1 = 1, this model is formally identical to the precursor product model that Hellerstein [22] used to describe deuterium data. For the case n = 1 and α1 < 1, the model has been shown [20] to be identical to the model of Asquith et al. [16], and to that used by Mohri et al. [8]. For n → ∞, this model can be used to describe populations with a continuous distribution of turnover rates [20]. The average turnover rate of the model is defined as  . The initial upslope reflects the average turnover rate, whereas the initial downslope is dominated by the subpopulation with the fastest turnover, and hence depends on the period of labelling. The difference between the up- and downslope diminishes when the length of the labelling periods increases [20].
. The initial upslope reflects the average turnover rate, whereas the initial downslope is dominated by the subpopulation with the fastest turnover, and hence depends on the period of labelling. The difference between the up- and downslope diminishes when the length of the labelling periods increases [20].
Although equation (1.1) seems like an excellent general model for a kinetically heterogeneous population, it is not clear that it is the best model to describe the possibility of temporal dynamic heterogeneity in lymphocytes. Lymphocytes that have recently divided, and hence are enriched in deuterium, have gone through a process of activation and may transiently experience a faster death rate than a quiescent cell, by a process that is sometimes referred to as activation-induced cell death. This form of temporal heterogeneity is particularly important for lymphocytes that are undergoing a phase of clonal expansion [23], as is typical during an immune response, because clonal expansion is followed by a phase of contraction owing to rapid cell death by apoptosis [24]. Unfortunately, most models for clonal expansion have so many parameters that they would lead to overfitting labelling data [17,18,21,25], and therefore, we still lack a reliable model for cell labelling when temporal heterogeneity is involved.
Another example where temporal heterogeneity could be important is provided by the study of Choo et al. [26], who recently described the process by which CD8+ memory T cells maintain themselves in mice. Clones of memory cells were generated by infecting mice with lymphocytic choriomeningitis virus (LCMV), the cells were labelled with CFSE ex vivo and transferred to naive recipient mice. Drawing blood samples from the recipients at various later time points, it was shown that the clones maintained a stable size, and that cells completed single divisions stochastically with an average interval of 50 days [26]. Although it is not known whether the two daughter cells that result from a rare stochastic division of a cell recruited from an otherwise quiescent population have a transiently increased death rate (which would account for temporal heterogeneity), these data—for the first time—allow us to study what deuterium labelling in a self-renewing population of lymphocytes should look like with a mathematical model that we believe is correct because it captures the underlying biology. Here we present this model, study its properties and generalize it to other situations with temporal heterogeneity.
2. Model
Consider a population of resting cells that are recruited into division at an activation rate a. Cell division yields c daughter cells, where c = 2 corresponds to the single divisions that account for the renewal of an otherwise quiescent population of memory T cells [26], and c > 2 corresponds to a burst of divisions that characterize clonal expansion. Small non-overlapping bursts of clonal expansion may also occur during chronic infection [23], and may play a role in the normal maintenance of part of the CD4+ T cells expressing a memory phenotype [27]. It would obviously be best to model a burst of cell expansion by a cascade indexed by the number of divisions the cells have completed [23,28], but such a model typically has too many parameters to fit to limited experimental data. To keep the analyses tractable, we model clonal expansion as an instantaneous process, but we do not expect that our main results will change if clonal expansion were incorporated as a cascade. Moreover, because we assume steady state in our model, we will limit ourselves to small bursts (c = 32, corresponding to 5 divisions).
The activated daughter cells revert to a resting state at rate r, and resting and activated cells have death rates dR and dA, respectively. Whenever dA > dR, the model accounts for temporal heterogeneity. Without loss of generality, one can scale the total population size to one, so that A + R = 1, and write for the fractions of resting, R, and activated, A, cells
 |
2.1 |
Since we expect that the system is at steady state and that deuterium labelling does not change the dynamics, we consider dR/dt = dA/dt = 0. Using A + R = 1, the steady state dA/dt = 0 gives that the fraction of dividing cells  , and the steady state of dR/dt = 0 can be used to eliminate one parameter, e.g.
, and the steady state of dR/dt = 0 can be used to eliminate one parameter, e.g.
| 2.2 |
Substituting the latter into  gives the fraction of activated cells
gives the fraction of activated cells  , or
, or
 |
2.3 |
The average turnover of the population is defined as  . This model is identical to that of Ribeiro et al. [18], who were modelling two subpopulations with different turnover rates. Equation (2.2) requires that (c − 1)a − dR > 0, which implies that for c = 2 we require dR < a. For biological reasons, we only consider c > 1 and dR < dA. Below we compare populations with different values of c, but with the same average turnover rate
. This model is identical to that of Ribeiro et al. [18], who were modelling two subpopulations with different turnover rates. Equation (2.2) requires that (c − 1)a − dR > 0, which implies that for c = 2 we require dR < a. For biological reasons, we only consider c > 1 and dR < dA. Below we compare populations with different values of c, but with the same average turnover rate  , which can be achieved by keeping (c − 1)a the same when c is changed, thus keeping f constant. Note that the average turnover rate, or the average death rate,
, which can be achieved by keeping (c − 1)a the same when c is changed, thus keeping f constant. Note that the average turnover rate, or the average death rate,  , is not related to the average residence times in the two subpopulations, i.e. 1/(a + dR) and 1/(r + dA), respectively.
, is not related to the average residence times in the two subpopulations, i.e. 1/(a + dR) and 1/(r + dA), respectively.
To model deuterium labelling, it is most convenient to write equations for the unlabelled fractions of resting and activated cells, UR and UA, respectively, during the labelling phase [18], i.e.
 |
2.4 |
where UR + LR = R and UA + LA = A and the total labelled fraction is defined as L = LR + LA = 1 − UR − UA. To obtain the downslope, it is preferable to write equations for the loss of the labelled fractions, and—conveniently—these are identical to those used to obtain the upslope [18] when U and L are interchanged, i.e.
 |
2.5 |
2.1. Upslope
Because the model is linear, Ribeiro et al. [18] were able obtain the analytical solutions. For the labelling phase with the initial condition UA(0) = f and UR(0) = 1 − f, one obtains
| 2.6 |
which, after some algebra, is identical to the upslope of the two-compartment variant of the kinetic heterogeneity model of equation (1.1). Here
 |
2.7 |
and
 |
2.8 |
and r is defined by equation (2.2). The upslope is a combination of e1 and e2, where e1 < e2. The initial upslope of equation (2.6), i.e. dL(t)/dt for t → 0, is the average turnover rate  . Thus, although the structure of this solution is identical to that of equation (1.1) with n = 2, the parameters have completely different interpretations because the exponents e1 and e2 are complicated expressions that do not reflect the turnover rates, dR and dA, of the subpopulations R and A, and the parameter α is not the fraction of activated cells f.
. Thus, although the structure of this solution is identical to that of equation (1.1) with n = 2, the parameters have completely different interpretations because the exponents e1 and e2 are complicated expressions that do not reflect the turnover rates, dR and dA, of the subpopulations R and A, and the parameter α is not the fraction of activated cells f.
2.2. Downslope
The general expression for the downslope is given by Ribeiro et al. [18], and is complicated because it involves the fraction of labelled resting cells, LR(tend), and the fraction of labelled activated cells, LA(tend). We therefore first consider the case where all cells are labelled at the end of labelling, i.e. L(tend) = LR(tend) + LA(tend) = 1, because the solution can then be written as
| 2.9 |
where the parameters are defined as in equation (2.7). The initial downslope reflects  (because we consider the case where all cells are labelled at time tend). For finite labelling periods tend, it can numerically be shown that the general expression for the downslope [18] rapidly approaches the approximate solution
(because we consider the case where all cells are labelled at time tend). For finite labelling periods tend, it can numerically be shown that the general expression for the downslope [18] rapidly approaches the approximate solution
 |
2.10 |
which is indeed equal to equation (2.9) when all cells are labelled at tend, i.e. when  . Thus, the downslope of this temporal heterogeneity model strongly resembles the downslope of equation (1.1) for n = 2, albeit with different parameter interpretation, and for all practical purposes it will be difficult to distinguish between the models.
. Thus, the downslope of this temporal heterogeneity model strongly resembles the downslope of equation (1.1) for n = 2, albeit with different parameter interpretation, and for all practical purposes it will be difficult to distinguish between the models.
2.3. No death of quiescent cells
The death rate of resting cells, dR, is the slowest timescale of this model, and for c = 2, we indeed require that dR < a. Thus, it is enlightening to simplify the exponents by assuming dR = 0. This yields
 |
2.11 |
where c′ = c/(c − 1). For c = 2, this simplifies further into
 |
2.12 |
which for low activation rates, a, approaches e1 ≃ a/2 and e2 ≃ a/2 + 2dA. Using the Choo et al. [26] estimate of a = 0.02 d−1, and giving recently divided cells a short lifespan, i.e. choosing dA = 1 d−1, the exponents would be e1 ≃ 0.01 d−1 and e2 ≃ 2.01 d−1.
3. Results and discussion
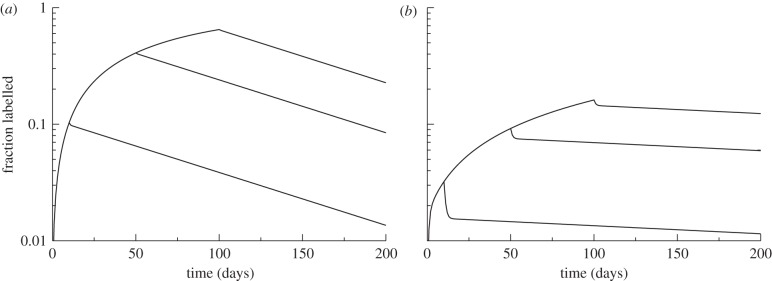
Let us first study what one would expect for a deuterium labelling experiment of LCMV-specific CD8+ memory T cells, as described by Choo et al. [26]. The cells on average divide every 50 days and then do one division, i.e. we set a = 0.02 d−1 and c = 2. Considering long-lived resting cells, e.g. dR = 0.001 d−1, and short-lived activated cells, e.g. dA = 1 d−1, one calculates a reversion rate of r = 1.105 d−1, and a steady-state fraction of activated cells of f = 0.01865, from equations (2.2) and (2.3), respectively. The average turnover rate of the cells is then  d−1 (which is close to fdA because we assume that resting cells are long-lived). We study deuterium labelling experiments in this system by numerically solving equations (2.4) and (2.5) for labelling periods of 10, 50 and 100 days (figure 2a). Although the analytical solutions of this model involve two exponentials, see equations (2.6) and (2.9), the up and downslopes shown in figure 2a are hardly biphasic. For the present case, the parameters of the solution of equation (2.6) are α = 0.995, e1 = 0.01 d−1 and e2 = 2.12 d−1, from which one sees that the fast exponential describes only a minor fraction of the population (i.e. 1 − α ≃ 0.005). This explains why the system is hardly biphasic, and the slow exponential, e1, largely determines the ‘observed’ upslope and downslopes. The downslope barely depends on the labelling period, and even after all cells have been labelled, the ‘observed’ downslope, e1 = 0.01 d−1, is about half of the average turnover rate, as
d−1 (which is close to fdA because we assume that resting cells are long-lived). We study deuterium labelling experiments in this system by numerically solving equations (2.4) and (2.5) for labelling periods of 10, 50 and 100 days (figure 2a). Although the analytical solutions of this model involve two exponentials, see equations (2.6) and (2.9), the up and downslopes shown in figure 2a are hardly biphasic. For the present case, the parameters of the solution of equation (2.6) are α = 0.995, e1 = 0.01 d−1 and e2 = 2.12 d−1, from which one sees that the fast exponential describes only a minor fraction of the population (i.e. 1 − α ≃ 0.005). This explains why the system is hardly biphasic, and the slow exponential, e1, largely determines the ‘observed’ upslope and downslopes. The downslope barely depends on the labelling period, and even after all cells have been labelled, the ‘observed’ downslope, e1 = 0.01 d−1, is about half of the average turnover rate, as  d−1.
d−1.
Figure 2.
Simulating a labelling experiment for the parameters estimated by Choo et al. [26] for 10, 50 and 100 days of labelling. The total labelled fraction is defined as L = LR + LA = 1 − UR − LR. (a) c = 2, a = 0.02 d−1 and hence f = 0.01865, r = 1.105 d−1 and  d−1 (which is close to fdA). (b) c = 32,a = 0.02/31 = 0.0006452 d−1 and hence r = 0.08658 d−1. The fraction of activated cells, f, and the average turnover rate,
d−1 (which is close to fdA). (b) c = 32,a = 0.02/31 = 0.0006452 d−1 and hence r = 0.08658 d−1. The fraction of activated cells, f, and the average turnover rate,  , are the same in both panels. Other parameters: dR = 0.001 d−1 and dA = 1 d−1. Although the up- and downslopes are technically biphasic [18], this is hardly visible when c = 2 (a).
, are the same in both panels. Other parameters: dR = 0.001 d−1 and dA = 1 d−1. Although the up- and downslopes are technically biphasic [18], this is hardly visible when c = 2 (a).
As shown by equations (2.7) and (2.8), the slowest exponential is determined by several parameters of the model, and is complicated to understand in biological terms. Because, in this example, there is a slow death rate of resting cells, one can obtain better intuition about the slowest exponential by letting dR → 0 (see equation (2.12)). For the current parameters, the slowest exponential in equation (2.12) is e1 = 0.00995 d−1, which is very close to the true e1 = 0.01 d−1, and is close to its further approximation e1 ≃ a/2 for low values of a (as a = 0.02 d−1). Thus, for the current parameters, the expected up- or downslopes from this model tend to look like a single exponential, reflecting half of the activation rate, which is here half of the true turnover rate. Summarizing, if one were to perform a deuterium labelling experiment on the LCMV-specific memory CD8+ T cells described by Choo et al. [26], and had somewhat imperfect experimental data that are fitted with equation (2.1), which we believe accurately summarizes the underlying biology, one would have no reason to conclude that the population is heterogeneous, and one would estimate an average turnover rate that is about half of the actual turnover rate.
If one were to fit these data not knowing that the underlying biology is described by the temporal heterogeneity model of equation (2.1), and instead used the general kinetic heterogeneity model assuming that memory T cells are composed of n subpopulations with different turnover rates, i.e. equation (1.1), one would obtain excellent fits to the data. Fitting the three labelling curves in figure 2a simultaneously to the one-compartment version of equation (1.1), i.e. n = 1 and α1 = 1, allowing just tend to differ between the curves, describes the data with excellent quality, and one would estimate that  d−1 (not shown). Note that for n = 1, equation (1.1) is equal to the simple precursor–product model [1,2,20]. Given the monophasic nature of the curves in figure 2a, the high quality of the fit with this simple model seems a natural result. As expected, the exponent that is picked up by this one compartment model is very similar to the slowest exponential, e1, of the solution of the temporal heterogeneity model that underlies the data. As argued earlier, e1 is approximately half the activation rate, and half of the true turnover rate for these parameters. Hence, one would also make a twofold error when fitting these data perfectly with the wrong model.
d−1 (not shown). Note that for n = 1, equation (1.1) is equal to the simple precursor–product model [1,2,20]. Given the monophasic nature of the curves in figure 2a, the high quality of the fit with this simple model seems a natural result. As expected, the exponent that is picked up by this one compartment model is very similar to the slowest exponential, e1, of the solution of the temporal heterogeneity model that underlies the data. As argued earlier, e1 is approximately half the activation rate, and half of the true turnover rate for these parameters. Hence, one would also make a twofold error when fitting these data perfectly with the wrong model.
The same data are obviously also well described by the two compartment variant of the kinetic heterogeneity model of equation (1.1) because adding two parameters to a model that already describes the data well can only make it better. Once sufficient data points are provided, the two compartment variant will fit the data in figure 2a significantly better than the n = 1 variant. Fitting the model to the 600 daily values of L(t) in figure 2a, we estimate that α1 = 0.995, d1 = 0.01 d−1, d2 = 2.13 d−1 and an average turnover  d−1, which is very similar to the parameters of the true solution (see earlier text). This excellent fit was to be expected because the temporal heterogeneity model has the same solution for the upslope, equation (2.6), and a very similar solution for the downslope, equation (2.10), as those given in equation (2.1). The average turnover rate would therefore be estimated correctly, but the values of α1, d1 and d2 would not describe the kinetics of any subpopulation in the data, which have turnover rates of dR = 0.001 d−1 and dA = 1 d−1, respectively. One can sample the data in figure 2a every 3 days, and add 10 per cent noise by multiplying L(t) with a factor chosen from a normal distribution with mean μ = 1 and standard deviation σ = 0.1 to mimic a real labelling experiment. As expected, the information on the minor contributions (less than 0.5%) to the initial rapid up- and downslopes is lost from that sampled data, and fitting that data with equation (1.1) with n = 2 compartments gives excellent fits to the data, but no longer allows for estimating all three parameters correctly (not shown). The slowest exponent is the only parameter that is reliably estimated from the sampled data, i.e. with a 95% CI of 0.0102 ≤ d1 ≤ 0.0108 d−1, where the ranges denote 95% CI based on 500 simulations bootstrapping the residuals [29]. For α, the 95% CI, 0.98 ≤ α1 ≤ 1, includes one, which would correspond to the n = 1 model. The fast exponent d2, and the average turnover rate
d−1, which is very similar to the parameters of the true solution (see earlier text). This excellent fit was to be expected because the temporal heterogeneity model has the same solution for the upslope, equation (2.6), and a very similar solution for the downslope, equation (2.10), as those given in equation (2.1). The average turnover rate would therefore be estimated correctly, but the values of α1, d1 and d2 would not describe the kinetics of any subpopulation in the data, which have turnover rates of dR = 0.001 d−1 and dA = 1 d−1, respectively. One can sample the data in figure 2a every 3 days, and add 10 per cent noise by multiplying L(t) with a factor chosen from a normal distribution with mean μ = 1 and standard deviation σ = 0.1 to mimic a real labelling experiment. As expected, the information on the minor contributions (less than 0.5%) to the initial rapid up- and downslopes is lost from that sampled data, and fitting that data with equation (1.1) with n = 2 compartments gives excellent fits to the data, but no longer allows for estimating all three parameters correctly (not shown). The slowest exponent is the only parameter that is reliably estimated from the sampled data, i.e. with a 95% CI of 0.0102 ≤ d1 ≤ 0.0108 d−1, where the ranges denote 95% CI based on 500 simulations bootstrapping the residuals [29]. For α, the 95% CI, 0.98 ≤ α1 ≤ 1, includes one, which would correspond to the n = 1 model. The fast exponent d2, and the average turnover rate  , cannot be estimated as they have large confidence limits, i.e. 0.074 ≤ d2 ≤ 85.7 d−1, and
, cannot be estimated as they have large confidence limits, i.e. 0.074 ≤ d2 ≤ 85.7 d−1, and  d−1.
d−1.
3.1. Clonal expansion
Using CFSE labelling, Choo et al. [26] demonstrated that both CD8+ LCMV-specific memory T cells and CD8+ memory T cells in non-immunized mice typically perform only one division per renewal event, and that these events are on average 50 days apart. Conversely, using BrdU labelling, Ki67 staining and TCR sequencing, Younes et al. [27] show that both in conventional non-immunizied mice and in germ-free mice, CD4+ T cells expressing a memory phenotype have an almost fivefold faster turnover, and that division sometimes involves clonal bursts. We now expect that labelling cells performing single divisions with deuterium would give labelling curves that can be described well with a single exponential. Deuterium labelling curves from the whole CD45RO+ effector/memory population of CD4+ or CD8+ memory T cells in humans [10] and mice [30] tend to be biphasic, and hence reflect some form of heterogeneity (figure 1b). This heterogeneity could be temporal, kinetic or both, and it would seem very reasonable that effector/memory cells on average perform more than one division when they are activated, because some of them will be specific for persistent infections and environmental antigens.
We investigate the impact of clonal expansion by setting c > 2 without changing the average turnover rate and the fraction of activated cells. Choosing c = 32 we scale a, i.e. choose a = 0.02/31 = 0.0006452 d−1, and compute from equation (2.2) that r = 0.08659 d−1 (f, dR, dA and  remain the same). Simulating the same three labelling experiments in a system having exactly the same average turnover rate now gives strongly biphasic curves with a markedly lower accrual of label (figure 2b). The slower enrichment can intuitively be understood from the fact that most labelled cells die before they return to the resting state. Substituting the parameter values into equation (2.7), the true solution is now described by α = 0.983, e1 = 0.0016 d−1 and e2 = 1.09 d−1, implying that the contribution of the fast exponential, (1 − α), has increased from 0.5 to about 2 per cent, and that both the slow and the fast exponents decreased. Note that the approximation of equation (2.12) no longer holds because now a < dR, and dR is no longer the slowest time scale. The labelling period has hardly any effect on the downslopes, which run reasonably parallel, but has a large effect on the initial contribution of the fast exponent to the downslope (see the magnitude of the initial fall in labelling in figure 2b). After a short labelling period, the label is not yet equally divided over the resting and activated compartments, and the activated compartment will contribute more to the total loss of label.
remain the same). Simulating the same three labelling experiments in a system having exactly the same average turnover rate now gives strongly biphasic curves with a markedly lower accrual of label (figure 2b). The slower enrichment can intuitively be understood from the fact that most labelled cells die before they return to the resting state. Substituting the parameter values into equation (2.7), the true solution is now described by α = 0.983, e1 = 0.0016 d−1 and e2 = 1.09 d−1, implying that the contribution of the fast exponential, (1 − α), has increased from 0.5 to about 2 per cent, and that both the slow and the fast exponents decreased. Note that the approximation of equation (2.12) no longer holds because now a < dR, and dR is no longer the slowest time scale. The labelling period has hardly any effect on the downslopes, which run reasonably parallel, but has a large effect on the initial contribution of the fast exponent to the downslope (see the magnitude of the initial fall in labelling in figure 2b). After a short labelling period, the label is not yet equally divided over the resting and activated compartments, and the activated compartment will contribute more to the total loss of label.
The initial upslopes in figure 2a,b are identical, i.e. for t → 0 the slope  , but at later times the label accrual occurs slower when there is clonal expansion (figure 2b). Estimating turnover rates by fitting the whole labelling curve with a one-compartment model, or from the peak value at the end of the labelling phase, would thus be incorrect. Fitting these data with models other than the true equations (2.6) and (2.9) clearly requires a model with more than one exponential, and because equation (1.1) with n = 2 has an almost identical solution it almost perfectly describes the data with the same three parameters (i.e. α1 = 0.983, d1 = 0.0016 d−1 and d2 = 1.08 d−1). Hence, having biphasic data, a kinetic heterogeneity model would get the average turnover rate correct, but the individual parameter estimates would not be related to the size and turnover of its subpopulations, as one might have expected.
, but at later times the label accrual occurs slower when there is clonal expansion (figure 2b). Estimating turnover rates by fitting the whole labelling curve with a one-compartment model, or from the peak value at the end of the labelling phase, would thus be incorrect. Fitting these data with models other than the true equations (2.6) and (2.9) clearly requires a model with more than one exponential, and because equation (1.1) with n = 2 has an almost identical solution it almost perfectly describes the data with the same three parameters (i.e. α1 = 0.983, d1 = 0.0016 d−1 and d2 = 1.08 d−1). Hence, having biphasic data, a kinetic heterogeneity model would get the average turnover rate correct, but the individual parameter estimates would not be related to the size and turnover of its subpopulations, as one might have expected.
3.2. Activation
Because the model was parameterized based on the Choo et al. [26] data on memory cells undergoing infrequent divisions, the fraction of activated cells in the model is small, i.e. f ≃ 0.018. To study labelling in systems with more activation, we increase the activation rate by 25-fold to a = 0.5 d−1 and again compare single divisions (i.e. c = 2) with clonal expansion (i.e. c = 32). Now the fraction of activated cells is f = 0.3329 and the average turnover is  (figure 3). For c = 2, the parameters of the solutions are α = 0.94, e1 = 0.22 d−1 and e2 = 2.28 d−1; for c = 32, they are α = 0.69, e1 = 0.017 d−1 and e2 = 1.03 d−1. Owing to the fast activation, labelling accrual occurs rapidly. Even when a large fraction of the cells has recently divided, and transiently have a markedly increased death rate, the labelling curves show little evidence for heterogeneity when cells undertake one division at a time (figure 3a). The three labelling curves for c = 2 are well described with the n = 1 variant of equation (1.1) (not shown), but with an average turnover rate of d1 = 0.23 d−1, which agrees with the e1 ≃ a/2 of equation (2.12) as a = 0.5, but which is only about 70 per cent of the true average turnover rate,
(figure 3). For c = 2, the parameters of the solutions are α = 0.94, e1 = 0.22 d−1 and e2 = 2.28 d−1; for c = 32, they are α = 0.69, e1 = 0.017 d−1 and e2 = 1.03 d−1. Owing to the fast activation, labelling accrual occurs rapidly. Even when a large fraction of the cells has recently divided, and transiently have a markedly increased death rate, the labelling curves show little evidence for heterogeneity when cells undertake one division at a time (figure 3a). The three labelling curves for c = 2 are well described with the n = 1 variant of equation (1.1) (not shown), but with an average turnover rate of d1 = 0.23 d−1, which agrees with the e1 ≃ a/2 of equation (2.12) as a = 0.5, but which is only about 70 per cent of the true average turnover rate,  .
.
Figure 3.
Deuterium labelling curves expected for the temporal heterogeneity model with (a) c = 2 and (b) c = 32 after 10, 50 or 100 days of labelling. (a) c = 2, a = 0.5 d−1, and hence f = 0.3329, r = 1.004 d−1 ≃ dA d−1, and  d−1 (which is close to fdA). (b) c = 32 with a = 0.5/31 d−1, and hence r = 0.03422 d−1 ≪dA. The fraction of activated cells, f, and the average turnover rate,
d−1 (which is close to fdA). (b) c = 32 with a = 0.5/31 d−1, and hence r = 0.03422 d−1 ≪dA. The fraction of activated cells, f, and the average turnover rate,  , are the same in both panels. Other parameters: dR = 0.001 d−1 and dA = 1 d−1.
, are the same in both panels. Other parameters: dR = 0.001 d−1 and dA = 1 d−1.
The curves become biphasic when we allow for clonal expansion by setting c = 32 (figure 3b). The period of labelling again hardly affects the downslopes (which run approximately parallel for the three labelling periods), and largely affects the contribution of the fast exponent. Given the similar solutions, fitting the data with the two compartment model of equation (1.1) gives an excellent fit with very similar parameters i.e. α = 0.69, d1 = 0.017 d−1 and d2 = 1.03 d−1 (not shown). This excellent fit would again provide no information on the turnover of subpopulations, and would only get the average turnover rate right.
3.3. Memory T cell turnover in man
Healthy volunteers and HIV-infected patients have been labelled with deuterated glucose [2,8] and with deuterated water [9,10,19]. The resulting T cell labelling curves tend to be somewhat biphasic. Ribeiro et al. [17] interpreted the labelling data from a 7 day labelling experiment with deuterated glucose [8] using a variation of equation (2.1) as a model for kinetic heterogeneity with a slow and fast subpopulation of the total T cell pool. For instance, the resting and activated subpopulations could correspond to naive and memory T cells, respectively. The main difference is that instead of using a clonal expansion parameter, c, they explicitly allowed activated cells to proliferate by adding a pA term to equation (2.1). Since neither c nor p appears in equations (2.4) and (2.5) for the labelled fractions, one should be able to fit these data in exactly the same manner with the temporal heterogeneity model presented here (and, as a matter of fact, with the kinetic heterogeneity model of equation (1.1)).
Ribeiro et al. [17] fitted that data by assuming that on the timescale of the experiment, resting cells hardly die; i.e. they set dR = 0. This seems a reasonable assumption because the peak enrichment in healthy volunteers after about nine weeks of labelling with heavy water is approximately 20 per cent, and fitting the up- and downslope with the kinetic heterogeneity model of Asquith et al. [16], the average lifespan of naive and memory T cells in volunteers was estimated to be several years and several months, respectively [10]. Adopting the dR = 0 assumption, we computed the corresponding parameters of the temporal heterogeneity model from the parameter estimates obtained by fitting the two compartment model of Ribeiro et al. [17] to the deuterated glucose data of Mohri et al. [8] (table 1 and figure 4).
Table 1.
The values of dA, f and a in columns 2–4 are copied from table 1 in Ribeiro et al. [17]. The value of r in column 5 is computed from r = a(1 − f)/f given in [17]. The sixth column is the average turnover rate  (because dR = 0). The values of α, e1 and e2 are computed from equations (2.7) and (2.8). The three free parameters of the temporal heterogeneity model, i.e. dA, a and c, are solved numerically from the definitions of α, e1 and e2. Finally, r and f are solved from equations (2.2) and (2.3), respectively. When a = 0 in table 1 in Ribeiro et al. [17], we have set it to a = 0.0004. The asterisks in the ratio rows denote that the two means (i.e., of the volunteers C1 to C4 and the HIV-1 infected patients P1 to P7) differ significantly by a Wilcoxon test with p < 0.05.
(because dR = 0). The values of α, e1 and e2 are computed from equations (2.7) and (2.8). The three free parameters of the temporal heterogeneity model, i.e. dA, a and c, are solved numerically from the definitions of α, e1 and e2. Finally, r and f are solved from equations (2.2) and (2.3), respectively. When a = 0 in table 1 in Ribeiro et al. [17], we have set it to a = 0.0004. The asterisks in the ratio rows denote that the two means (i.e., of the volunteers C1 to C4 and the HIV-1 infected patients P1 to P7) differ significantly by a Wilcoxon test with p < 0.05.
| dA | f | a | r |  |
α | e1 | e2 | dA | f | a | r | c | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4 | |||||||||||||
| C1 | 0.043 | 0.127 | 0.003 | 0.021 | 0.005 | 0.944 | 0.00205 | 0.063 | 0.041 | 0.132 | 0.003 | 0.021 | 3.01 |
| C2 | 0.132 | 0.043 | 0.003 | 0.067 | 0.006 | 0.981 | 0.00188 | 0.211 | 0.143 | 0.040 | 0.003 | 0.067 | 3.13 |
| C3 | 0.013 | 0.219 | 0 | 0.001 | 0.003 | 0.806 | 0.00040 | 0.013 | 0.012 | 0.246 | 0 | 0.001 | 9.40 |
| C4 | 0.039 | 0.105 | 0.001 | 0.009 | 0.004 | 0.931 | 0.00081 | 0.048 | 0.039 | 0.104 | 0.001 | 0.008 | 5.62 |
| mean | 0.057 | 0.124 | 0.002 | 0.025 | 0.005 | 0.916 | 0.00129 | 0.084 | 0.059 | 0.130 | 0.002 | 0.024 | 5.29 |
| P1 | 0.109 | 0.312 | 0.005 | 0.011 | 0.034 | 0.746 | 0.00452 | 0.120 | 0.109 | 0.312 | 0.005 | 0.011 | 10.9 |
| P2 | 0.142 | 0.249 | 0.004 | 0.012 | 0.035 | 0.790 | 0.00368 | 0.154 | 0.142 | 0.249 | 0.004 | 0.012 | 12.8 |
| P3 | 0.056 | 0.178 | 0.003 | 0.014 | 0.010 | 0.891 | 0.00231 | 0.073 | 0.058 | 0.171 | 0.003 | 0.014 | 5.17 |
| P4 | 0.081 | 0.488 | 0.009 | 0.009 | 0.040 | 0.620 | 0.00801 | 0.091 | 0.081 | 0.491 | 0.009 | 0.009 | 9.58 |
| P5 | 0.303 | 0.066 | 0.006 | 0.085 | 0.020 | 0.960 | 0.00461 | 0.394 | 0.308 | 0.065 | 0.006 | 0.085 | 4.62 |
| P6 | 0.190 | 0.100 | 0.004 | 0.036 | 0.019 | 0.929 | 0.00340 | 0.224 | 0.187 | 0.102 | 0.004 | 0.036 | 6.20 |
| P7 | 0.073 | 0.307 | 0.006 | 0.014 | 0.022 | 0.790 | 0.00498 | 0.088 | 0.073 | 0.305 | 0.006 | 0.014 | 6.40 |
| mean | 0.136 | 0.242 | 0.005 | 0.026 | 0.026 | 0.818 | 0.00450 | 0.163 | 0.137 | 0.242 | 0.005 | 0.026 | 7.95 |
| ratio | 2.4 | 2.0 | 3.0* | 1.1 | 5.7* | 0.9 | 3.5* | 2.0 | 2.3 | 1.9 | 2.9* | 1.1 | 1.5 |
| CD8 | |||||||||||||
| C1 | 0.078 | 0.053 | 0.001 | 0.018 | 0.004 | 0.964 | 0.00084 | 0.093 | 0.075 | 0.055 | 0.001 | 0.018 | 5.20 |
| C2 | 0.019 | 0.120 | 0 | 0.003 | 0.002 | 0.899 | 0.00040 | 0.019 | 0.016 | 0.142 | 0 | 0.003 | 6.62 |
| C3 | 0.010 | 0.212 | 0 | 0.001 | 0.002 | 0.821 | 0.00040 | 0.010 | 0.009 | 0.249 | 0 | 0.001 | 7.01 |
| C4 | 0.042 | 0.054 | 0.003 | 0.053 | 0.002 | 0.990 | 0.00139 | 0.091 | 0.036 | 0.062 | 0.003 | 0.052 | 1.70 |
| mean | 0.037 | 0.110 | 0.001 | 0.019 | 0.003 | 0.918 | 0.00076 | 0.053 | 0.034 | 0.127 | 0.001 | 0.019 | 5.13 |
| P1 | 0.075 | 0.490 | 0.010 | 0.010 | 0.037 | 0.638 | 0.00869 | 0.086 | 0.075 | 0.493 | 0.010 | 0.010 | 8.20 |
| P2 | 0.096 | 0.189 | 0.002 | 0.009 | 0.018 | 0.842 | 0.00183 | 0.105 | 0.096 | 0.188 | 0.002 | 0.009 | 12.2 |
| P3 | 0.046 | 0.229 | 0.004 | 0.013 | 0.011 | 0.869 | 0.00307 | 0.060 | 0.046 | 0.231 | 0.004 | 0.013 | 4.39 |
| P4 | 0.033 | 0.721 | 0 | 0 | 0.024 | 0.282 | 0.00040 | 0.033 | 0.033 | 0.724 | 0 | 0 | 216. |
| P5 | 0.053 | 0.528 | 0 | 0 | 0.028 | 0.476 | 0.00040 | 0.053 | 0.053 | 0.532 | 0 | 0 | 149. |
| P6 | 0.045 | 0.538 | 0.005 | 0.004 | 0.024 | 0.571 | 0.00445 | 0.051 | 0.046 | 0.530 | 0.005 | 0.004 | 11.5 |
| P7 | 0.027 | 0.612 | 0 | 0 | 0.017 | 0.394 | 0.00040 | 0.027 | 0.027 | 0.618 | 0 | 0 | 108. |
| mean | 0.054 | 0.472 | 0.003 | 0.005 | 0.023 | 0.582 | 0.00275 | 0.059 | 0.053 | 0.474 | 0.003 | 0.005 | 72.8 |
| ratio | 1.4 | 4.3* | 3.0 | 0.3 | 9.1* | 0.6* | 3.6 | 1.1 | 1.6 | 3.7* | 2.3 | 0.3 | 14.2* |
Figure 4.
Deuterium labelling curves of (a) CD4+ and (b) CD8+ T cells in the first healthy volunteer (C1) and the first HIV-1-infected patient (P1) in the study of Mohri et al. [8]. The bullets are the data from the volunteer and the boxes are those from the patient. The lines were obtained by recomputing the parameters given in table 1 in Ribeiro et al. [17] into those of the temporal heterogeneity model (table 1). The fit is therefore identical to that in Ribeiro et al. [17].
The labelling curves in HIV-1-infected patients in Mohri et al. [8], Ribeiro et al. [17], and the two examples in figure 4, are much more biphasic than those in healthy volunteers. Comparing the means over all individuals, the average turnover rate  of CD4+ and CD8+ T cells increases 5.7-fold and 9.1-fold, respectively, in infected individuals compared with uninfected controls (table 1). Ribeiro et al. [17] found that in CD4+ T cells HIV infection increases the activation rate a (threefold), and the proliferation/death rate, dA (2.4-fold). In CD8+ T cells, these parameters were not changed significantly by HIV infection (despite the threefold increase in the activation rate a), and the main difference was the size of activated compartment, f (see the asterisks in table 1). The increased death rate of CD4+ T cells could be attributed to the killing of CD4+ T cells by HIV-1, which would help us to explain that the fraction of activated CD4+ T cells, f, was not significantly increased (the twofold increase in f was borderline significant, and much less than the 4.3-fold increase of f in CD8+ T cells) [17].
of CD4+ and CD8+ T cells increases 5.7-fold and 9.1-fold, respectively, in infected individuals compared with uninfected controls (table 1). Ribeiro et al. [17] found that in CD4+ T cells HIV infection increases the activation rate a (threefold), and the proliferation/death rate, dA (2.4-fold). In CD8+ T cells, these parameters were not changed significantly by HIV infection (despite the threefold increase in the activation rate a), and the main difference was the size of activated compartment, f (see the asterisks in table 1). The increased death rate of CD4+ T cells could be attributed to the killing of CD4+ T cells by HIV-1, which would help us to explain that the fraction of activated CD4+ T cells, f, was not significantly increased (the twofold increase in f was borderline significant, and much less than the 4.3-fold increase of f in CD8+ T cells) [17].
Recomputing these parameter estimates into those of the temporal heterogeneity model, we find very similar results. The major new finding is the large increase in the clonal expansion of CD8+ T cells with HIV infection (table 1), i.e. c increases 14.2-fold, which seems a very natural result, given the specific immune responses to the virus. The absence of an increase in c in CD4+ T cells could be due to the fact that CD4+ T cells expand less during clonal expansion [31], and that clonally expanding CD4+ T cells are preferentially infected during HIV-1 infection [32,33]. Note the slow reversion rates of recently divided cells, r, with corresponding long expected residence times in the activated state of 1/r ≃ 50 days. This could be due to the fact that the clonal expansion phase in the temporal heterogeneity model is instantaneous, and that r would become faster if we were modelling clonal expansion with a cascade of equations [23,28]. Fitting a cascade model to labelling data is not straightforward because (i) one typically needs additional parameters, and (ii) one needs to estimate the optimal integer number of divisions that cells complete during clonal expansion.
Summarizing, reinterpreting the two compartment model in terms of temporal heterogeneity suggests that differences in activation and clonal expansion can equally well explain the observed differences in the labelling of volunteers and HIV-1-infected patients. The interpretation of the CD4+ T cell data remains similar. Since we now find evidence for a significant increase in clonal expansion of CD8+ T cells, it remains difficult to have a mechanistic understanding of the several-fold increase in the average turnover rate of both CD4+ and CD8+ T cells, and how an increase in the average turnover rate determines the depletion of CD4+ T cells, and not CD8+ T cells, in HIV-1-infected patients.
4. Conclusion
We have shown that a realistic model for temporal heterogeneity has solutions that are practically indistinguishable from those of a general model for kinetic heterogeneity. Because we typically do not know whether heterogeneity is temporal or kinetic, one should be careful with interpreting the compartment sizes, and the turnover rates estimated with kinetic heterogeneity models. The most reliable estimate that one can obtain from deuterium labelling is the average turnover rate. Interestingly, we have reached the same conclusion about the modelling of BrdU data [21,25]. In reality, there will be both kinetic and temporal heterogeneity, and a model can easily be written splitting each population of the kinetic heterogeneity model into the resting, R, and activated A fractions of equation (2.1). One would have to expand equation (2.1) with an index i, and allow each subpopulation, Ri + Ai, to comprise a certain fraction αi of the total population, i.e. ∑αi(Ri + Ai) = 1. A model with two kinetically different subpopulations would have four exponentials, which are too many to identify from the typical labelling curves that are currently available.
A second problem that we identified is that the average turnover rate will be underestimated if there is heterogeneity, especially if the data contain too little evidence of the fast exponentials because of their small contribution. In such a case, one would typically fit equation (1.1) to the data, and use the F-test to decide how many compartments, n, are required for an optimal fit to the data. Our results suggest that one should also test how the estimated average turnover rate,  , depends on the number of compartments. If increasing n allows
, depends on the number of compartments. If increasing n allows  to converge to a particular value, this could be the best estimate for the average turnover rate, even though the F-test would argue that one is overfitting the data. One should therefore estimate
to converge to a particular value, this could be the best estimate for the average turnover rate, even though the F-test would argue that one is overfitting the data. One should therefore estimate  and its confidence limits for several values of n, to see how the average turnover rate depends on the number of compartments.
and its confidence limits for several values of n, to see how the average turnover rate depends on the number of compartments.
The issues discussed here should also apply to other labelling techniques, such as CFSE and BrdU, although each of these have specificities that make a full discussion beyond the scope of the current study. In particular, the underestimation of the average cell cycle time by modelling of CFSE data has been discussed before: different death rates during the different phases of the cell cycle can give quite different estimates of the cell cycle time when compared with models assuming a constant death rate [34,35]. Likewise, data obtained with BrdU labelling have been interpreted with different models, and because the values of their parameters strongly depended on the choice of the model, it was proposed to just estimate an average turnover rate from BrdU data [21,25]. Thus, none of the current labelling methods provides sufficient information to accurately describe the kinetics of the possible subpopulations of the population of cells studied in a labelling experiment. To understand the kinetics of human T cells, where deuterium labelling remains the main source of data, we have shown that the estimation of the kinetics of memory T cell populations remains difficult because of its expected kinetic and temporal heterogeneity: memory T cell data obtained with deuterium labelling tend to provide too little information for estimating anything more than just the average turnover rate of this quite heterogeneous population.
Acknowledgments
Portions of this work were done under the auspices of the U. S. Department of Energy under contract DE-AC52-06NA25396 and supported by NIH grants AI028433, P20-RR018754 and National Center for Research Resources and the Office of Research Infrastructure Programs (ORIP) through grant 8R01-OD011095-21 (ASP). R.J.D.B. thanks the Santa Fe Institute for their support and hospitality during a visit when this work was initiated.
References
- 1.Borghans J. A. M., De Boer R. J. 2007. Quantification of T-cell dynamics: from telomeres to DNA labelling. Immunol. Rev. 216, 35–47 [DOI] [PubMed] [Google Scholar]
- 2.Hellerstein M., et al. 1999. Directly measured kinetics of circulating T lymphocytes in normal and HIV-1-infected humans. Nat. Med. 5, 83–89 10.1038/4772 (doi:10.1038/4772) [DOI] [PubMed] [Google Scholar]
- 3.Macallan D. C., et al. 2003. Measurement and modeling of human T cell kinetics. Eur. J. Immunol. 33, 2316–2326 10.1002/eji.200323763 (doi:10.1002/eji.200323763) [DOI] [PubMed] [Google Scholar]
- 4.Macallan, et al. 2003. Rapid turnover of T cells in acute infectious mononucleosis. Eur. J. Immunol. 33, 2655–2665 10.1002/eji.200324295 (doi:10.1002/eji.200324295) [DOI] [PubMed] [Google Scholar]
- 5.Macallan D. C., Wallace D., Zhang Y., De Lara C., Worth A. T., Ghattas H., Griffin G. E., Beverley P. C., Tough D. F. 2004. Rapid turnover of effector-memory CD4+ T cells in healthy humans. J. Exp. Med. 200, 255–260 10.1084/jem.20040341 (doi:10.1084/jem.20040341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace D. L., et al. 2004. Direct measurement of T cell subset kinetics in vivo in elderly men and women. J. Immunol. 173, 1787–1794 [DOI] [PubMed] [Google Scholar]
- 7.Kovacs J. A., et al. 2005. Induction of prolonged survival of CD4+ T lymphocytes by intermittent IL-2 therapy in HIV-infected patients. J. Clin. Invest. 115, 2139–2148 10.1172/JCI23196 (doi:10.1172/JCI23196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohri H., et al. 2001. Increased turnover of T lymphocytes in HIV-1 infection and its reduction by antiretroviral therapy. J. Exp. Med. 194, 1277–1288 10.1084/jem.194.9.1277 (doi:10.1084/jem.194.9.1277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellerstein M. K., Hoh R. A., Hanley M. B., Cesar D., Lee D., Neese R. A., McCune J. M. 2003. Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. J. Clin. Invest. 112, 956–966 10.1172/JCI17533 (doi:10.1172/JCI17533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vrisekoop N., et al. 2008. Sparse production but preferential incorporation of recently produced naive T cells in the human peripheral pool. Proc. Natl Acad. Sci. USA 105, 6115–6120 10.1073/pnas.0709713105 (doi:10.1073/pnas.0709713105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ladell K., Hellerstein M. K., Cesar D., Busch R., Boban D., McCune J. M. 2008. Central memory CD8+ T cells appear to have a shorter lifespan and reduced abundance as a function of HIV disease progression. J. Immunol. 180, 7907–7918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macallan D. C., Fullerton C. A., Neese R. A., Haddock K., Park S. S., Hellerstein M. K. 1998. Measurement of cell proliferation by labeling of DNA with stable isotope-labelled glucose: studies in vitro, in animals, and in humans. Proc. Natl Acad. Sci. USA 95, 708–713 10.1073/pnas.95.2.708 (doi:10.1073/pnas.95.2.708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Busch R., Neese R. A., Awada M., Hayes G. M., Hellerstein M. K. 2007. Measurement of cell proliferation by heavy water labeling. Nat. Protoc. 2, 3045–3057 10.1038/nprot.2007.420 (doi:10.1038/nprot.2007.420) [DOI] [PubMed] [Google Scholar]
- 14.Voogt J. N., Awada M., Murphy E. J., Hayes G. M., Busch R., Hellerstein M. K. 2007. Measurement of very low rates of cell proliferation by heavy water labeling of DNA and gas chromatography/pyrolysis/isotope ratio-mass spectrometric analysis. Nat. Protoc. 2, 3058–3062 10.1038/nprot.2007.421 (doi:10.1038/nprot.2007.421) [DOI] [PubMed] [Google Scholar]
- 15.Busch R., Siah I. M., Gee T. A., Hellerstein M. K. 2008. Heavy water labeling of DNA for measurement of cell proliferation and recruitment during primary murine lymph node responses against model antigens. J. Immunol. Methods 337, 24–34 10.1016/j.jim.2008.05.014 (doi:10.1016/j.jim.2008.05.014) [DOI] [PubMed] [Google Scholar]
- 16.Asquith B., Debacq C., Macallan D. C., Willems L., Bangham C. R. 2002. Lymphocyte kinetics: the interpretation of labelling data. Trends Immunol. 23, 596–601 10.1016/S1471-4906(02)02337-2 (doi:10.1016/S1471-4906(02)02337-2) [DOI] [PubMed] [Google Scholar]
- 17.Ribeiro R. M., Mohri H., Ho D. D., Perelson A. S. 2002. In vivo dynamics of T cell activation, proliferation, and death in HIV-1 infection: why are CD4+ but not CD8+ T cells depleted? Proc. Natl Acad. Sci. USA 99, 15 572–15 577. 10.1073/pnas.242358099 (doi:10.1073/pnas.242358099) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribeiro R. M., Mohri H., Ho D. D., Perelson A. S. 2002. Modeling deuterated glucose labeling of T-lymphocytes. Bull. Math. Biol. 64, 385–405 10.1006/bulm.2001.0282 (doi:10.1006/bulm.2001.0282) [DOI] [PubMed] [Google Scholar]
- 19.Vrisekoop N., et al. Submitted Qualitative changes in T cell turnover during HIV-1 infection. [Google Scholar]
- 20.Ganusov V. V., Borghans J. A., De Boer R. J. 2010. Explicit kinetic heterogeneity: mathematical models for interpretation of deuterium labeling of heterogeneous cell populations. PLoS. Comput. Biol. 6, e1000666. 10.1371/journal.pcbi.1000666 (doi:10.1371/journal.pcbi.1000666) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Boer R. J., Mohri H., Ho D. D., Perelson A. S. 2003. Turnover rates of B cells, T cells, and NK cells in simian immunodeficiency virus-infected and uninfected rhesus macaques. J. Immunol. 170, 2479–2487 [DOI] [PubMed] [Google Scholar]
- 22.Hellerstein M. 2002. HIV tropism and CD4+ T-cell depletion. Nat. Med. 8, 537–538 10.1038/nm0602-537b (doi:10.1038/nm0602-537b) [DOI] [PubMed] [Google Scholar]
- 23.Grossman Z., Herberman R. B., Dimitrov D. S. 1999. T cell turnover in SIV infection. Science 284, 555a–555b 10.1126/science.284.5414.555a (doi:10.1126/science.284.5414.555a) [DOI] [Google Scholar]
- 24.Antia R., Ganusov V. V., Ahmed R. 2005. The role of models in understanding CD8+ T-cell memory. Nat. Rev. Immunol. 5, 101–111 10.1038/nri1550 (doi:10.1038/nri1550) [DOI] [PubMed] [Google Scholar]
- 25.De Boer R. J., Mohri H., Ho D. D., Perelson A. S. 2003. Estimating average cellular turnover from BrdU measurements. Proc. R. Soc. Lond. B 270, 849–858 10.1098/rspb.2002.2316 (doi:10.1098/rspb.2002.2316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choo D. K., Murali-Krishna K., Anita R., Ahmed R. 2010. Homeostatic turnover of virus-specific memory CD8 T cells occurs stochastically and is independent of CD4 T cell help. J. Immunol. 185, 3436–3444 10.4049/jimmunol.1001421 (doi:10.4049/jimmunol.1001421) [DOI] [PubMed] [Google Scholar]
- 27.Younes S. A., Punkosdy G., Caucheteux S., Chen T., Grossman Z., Paul W. E. 2011. Memory phenotype CD4 T cells undergoing rapid, nonburst-like, cytokine-driven proliferation can be distinguished from antigen-experienced memory cells. PLoS. Biol. 9, e1001171. 10.1371/journal.pbio.1001171 (doi:10.1371/journal.pbio.1001171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones L. E., Perelson A. S. 2005. Opportunistic infection as a cause of transient viremia in chronically infected HIV patients under treatment with HAART. Bull. Math. Biol. 67, 1227–1251 10.1016/j.bulm.2005.01.006 (doi:10.1016/j.bulm.2005.01.006). [DOI] [PubMed] [Google Scholar]
- 29.Efron B., Tibshirani R. 1986. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat. Sci. 1, 54–77 [Google Scholar]
- 30.Den Braber I., et al. 2012. Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity 36, 288–297 10.1214/ss/1177013815 (doi:10.1214/ss/1177013815) [DOI] [PubMed] [Google Scholar]
- 31.De Boer R. J., Homann D., Perelson A. S. 2003. Different dynamics of CD4+ and CD8+ T cell responses during and after acute lymphocytic choriomeningitis virus infection. J. Immunol. 171, 3928–3935 10.1016/j.immuni.2012.02.006 (doi:10.1016/j.immuni.2012.02.006) [DOI] [PubMed] [Google Scholar]
- 32.Brenchley J. M., Ruff L. E., Casazza J. P., Koup R. A., Price D. A., Douek D. C. 2006. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. J. Virol. 80, 6801–6809 10.1128/JVI.00070-06 (doi:10.1128/JVI.00070-06). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geldmacher C., et al. 2010. Preferential infection and depletion of Mycobacterium tuberculosis-specific CD4 T cells after HIV-1 infection. J. Exp. Med. 207, 2869–2881 10.1084/jem.20100090 (doi:10.1084/jem.20100090). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilyugin S. S., Ganusov V. V., Murali-Krishna K., Ahmed R., Antia R. 2003. The rescaling method for quantifying the turnover of cell populations. J. Theor. Biol. 225, 275–283 10.1016/S0022-5193(03)00245-5 (doi:10.1016/S0022-5193(03)00245-5) [DOI] [PubMed] [Google Scholar]
- 35.Ganusov V. V., Pilyugin S. S., De Boer R. J., Murali-Krishna K., Ahmed R., Antia R. 2005. Quantifying cell turnover using CFSE data. J. Immunol. Methods. 298, 183–200 10.1016/j.jim.2005.01.011 (doi:10.1016/j.jim.2005.01.011) [DOI] [PubMed] [Google Scholar]