Abstract
The development of novel biomaterials able to control cell activities and direct their fate is warranted for engineering functional biological tissues, advanced cell culture systems, single-cell diagnosis as well as for cell sorting and differentiation. It is well established that crosstalk at the cell–material interface occurs and this has a profound influence on cell behaviour. However, the complete deciphering of the cell–material communication code is still far away. A variety of material surface properties have been reported to affect the strength and the nature of the cell–material interactions, including biological cues, topography and mechanical properties. Novel experimental evidence bears out the hypothesis that these three different signals participate in the same material–cytoskeleton crosstalk pathway via adhesion plaque formation dynamics. In this review, we present the relevant findings on material-induced cell response along with the description of cell behaviour when exposed to arrays of signals—biochemical, topographical and mechanical. Finally, with the aid of literature data, we attempt to draw unifying elements of the material–cytoskeleton–cell fate chain.
Keywords: cell–material interaction, biomaterials, biomolecular signals, topography, cell adhesion, patterned substrates
1. Introduction
Cell–material interaction has been proved to occur through a combination of biochemical and biophysical signals, including interfacial presentation of molecular, topographic and mechanical cues. Indeed, both biochemical and biophysical material features have been reported to affect and somehow influence cell functions by triggering specific molecular events at the cell–material interface. Cellular activities that are mostly influenced by material properties are adhesion, spreading, migration, proliferation and differentiation. Specifically engineered surfaces displaying selected biofunctional groups or micrometre-scale patterns have been used in order to study signal interactions in a systematic way. These assays proved to be valuable tools to enrich the body of knowledge on cell–material interaction that represents today a solid foundation for further progress. Owing to technological limitations, however, these kinds of substrates allowed only modest control of signal presentation, in terms of signal dose, spatial arrangement and temporal evolution. Advancements in chemistry, material science and nanotechnologies greatly improved the possibility of presenting many different signals, according to predefined spatial patterns or temporal chronoprogrammes. In particular, patterns of biochemical signals, topographies on different length scales and mechanical cues clearly revealed very sophisticated abilities with which cells ‘sense’ (and react to) external stimuli [1–3]. However, the majority of the studies dealt with single signal types and thus individual senses. This contributed to depicting a scenario in which cells ‘taste’ chemical moieties, ‘see’ topographies and ‘touch’ mechanical properties.
To date, a plethora of cell-responsive material properties has been ascertained and catalogued, including hydrophobicity, surface charge, roughness and stiffness [4–8]. These findings have pointed to the attractive prospect of engineering materials able to control and guide specific cellular events. The potential to pattern material properties with nanometric precision paves the way to the next biomaterials generation—referred to as cell instructive materials (CIMs)—with extended functionality and bioactivity. CIMs are envisioned as nanofeatured materials expressly programmed to impart even complex commands or instructions to cells with the aim of directing, guiding and controlling their fate. The realization of these attractive materials relies upon a deep understanding of the mechanisms that regulate cell–material interactions and, in particular, upon the disclosing of the complex molecular machinery of recognition and decoding that occurs at the interface between the cell membrane and materials. However, the mechanism underpinning the process of cell recognition of material property cues—topographical, mechanical, chemical—is poorly understood, and therefore the possibility to correctly display a single signal or an array of signals at the cell–material interface to elicit a given and predefined cell response is still far away. Several research groups are currently focusing their attention on the cell–material interface, with the aim of unravelling the intricate and unidentified principles that regulate cell–material crosstalk, leading to a rapid increase in the basic knowledge of the different stages and phases that occur across the cell membrane as a result of different material properties and distribution density of biological sites. Recent literature has described the phenomenon of crosstalk at the cell–material interface as being mostly influenced by the dynamics of large macromolecular complexes across the cell membrane whose formation and extension depend on material properties. According to this vision, properties such as mechanical, topographical and biochemical influence cell–material interaction by affecting in different ways the dynamics of these macromolecular complexes. Therefore, mechanical, topographical or biochemical signals may not represent separate or different cues, as suggested by some reports, but rather produce a different effect on the macromolecular complex dynamics.
In this paper, we first present a synthesis of basic knowledge of macromolecular complexes forming across the cell membrane at the cell–material interface, then review the most relevant findings on the material-induced cell response along with the most used experimental techniques to quantify the strength of interaction, proceeding with a description of cell behaviour exposed to arrays of signals—biochemical, topographical and mechanical—and finally summarize and discuss the massive literature drawing some unifying elements.
2. Time and space scale in cell–material interaction
Cells and materials interact through different pathways depending upon the distance between the cell membrane and the material surface. Four different phases of cell–material interaction have been described, each occurring at a defined time and distance from the surface. At a distance greater than micrometres, there is no interaction, and cells present a typical spherical shape. As the distance decreases to about 1 μm, a surface recognition activity begins and is mediated by weak non-specific interactions that are established between the pericellular coat and material surfaces [9]. This phase occurs within a time frame of tenths of seconds. Weak and cooperative interactions foster more specific strong contacts leading to the onset of the early attachment stage. In this stage, the distance between the cell membrane and the material surface reaches the hundreds of nanometres and the interactions are mediated by cell membrane proteins—integrins—that recognize specific molecular motifs on the material surface. This phase occurs with a time scale of seconds. Depending on the density of the adhesive motifs, their distribution and their mechanical compliance the cell can start to build larger and more stable molecular complexes to improve the membrane anchorage and reduce the distance from the material surface to tens of nanometres. This stage is called intermediate attachment or membrane adhesion, and involves the cell cytoskeleton assembly and occurs with a time scale of tens of seconds. Finally, the late adhesion or cell spreading phase initiates with the establishment of mature adhesion molecular clusters that mediate a dynamic material–cytoskeleton crosstalk. To discuss the material–cell interactions, here we will focus particularly on this late phase, pointing to the effects that material properties might induce on cytoskeleton assemblies. Therefore, we will not discuss the well-documented mechanisms by which the cytoskeleton affects cell fate, but we will confine our interest to the underlying processes by which cytoskeleton assembly and architecture can be regulated by interaction at the cell–material interface.
3. Cell adhesion to the substrate
The most notable cellular structures that mediate material–cytoskeleton crosstalk are molecular complexes called focal adhesions. These are dynamic structures that involve the recruitment, interaction and turnover of several molecular components. The mechanisms of adhesion formation, maturation and turnover are reported in detail in specialized reviews [10,11]. Here, we highlight the role of relevant molecules in the signal recognition process occurring on functionalized substrates. Among these, integrins connect ligands of the extracellular space to the cytoskeleton, and are responsible for transmitting exogenous stimuli to the cell. They are constituted by α and β subunits. 18α and 8β subunits have been found so far, which can combine to form 24 distinct dimers that interact with specific extracellular matrix (ECM) motifs. Integrin binding to extracellular ligands is closely related to the ruffling activity of the cell membrane, i.e. to the peripheral actin polymerization. During cell spreading, nascent adhesions are formed in the lamellipodium by the binding of integrins to the extracellular ligands. The cytoplasmic domain of these clusters is subsequently stabilized by the incorporation of adhesion proteins such as talin, vinculin, paxillin, α-actinin and focal adhesion kinase (FAK). Nascent adhesion may either disassemble or mature into larger molecular structures called focal complexes. This requires clustering of additional integrin dimers, which increases the dimension of the adhesion assembly and allows the recruitment of more cytoplasmic adhesion proteins that stabilize the construct.
Unlike nascent adhesion nucleation, focal complex maturation is dependent on actin-generated tension through myosin II. Nascent adhesion may engage the retrograde flow of actin through talin. Once coupled, myosin-generated force can induce a conformational change of talin that exposes additional binding sites for vinculin, which in turn stabilizes and recruits additional proteins [12]. Other proteins, such as vinculin, paxillin and p130cas, might exhibit similar behaviour. Provided that integrins have sufficient mobility and space to cluster, the maturation process can continue to generate focal adhesions that are 0.5–2 μm wide and up to 10 μm long.
Several attempts have been made to measure cell adhesion on substrates. The cell spreading area and contour morphology have always been considered good indicators of the interaction with surfaces. Microscopic techniques, such as phase contrast, differential interference contrast and fluorescence, have been used to perform morphometric analyses. The cell spreading area, for example, is usually reported in square micrometres or as a ratio with a reference value. More detailed information might be acquired from the analysis of the cell contour. The number and shape of cell processes and features of lamellipodia are of particular interest when studying the dynamics of cell adhesion and migration, or when analysing cell behaviour on patterned or non-homogeneous substrates. Immunofluorescence and microscopic techniques with improved resolution provide useful tools to visualize and study the formation of surface-bound ligand–cell receptor complexes, making it possible to perform quantitative measurements on their spatial distribution on the plasma membrane. Moreover, cells transfected with plasmids encoding fluorescently labelled receptors have been extensively used to dynamically analyse adhesion and receptor trafficking.
Besides adhesion clustering, cells also exert forces on the extracellular space through the ligand–receptor–cytoskeleton chain. The magnitude of the applied force depends, among other things, on the ligand–receptor affinity, as well as on the number of ligand–receptor complexes. Therefore, assessing the force required to break such complexes provides useful information about cell attachment. In cell-population-based assays, such as spinning discs and flow chambers, a force field with predetermined features (force direction and magnitude) is applied to a cell layer with a known density. Then, one measures the force required to detach a selected number of cells. Other techniques, involving probes with dimensions comparable to those of cells, are able to measure adhesion strength at the single-cell level. Atomic force microscopes equipped with specifically designed tips have been variously used to detect the force to dislodge or to stretch cells. Depending on the substrate properties and cell type, detachment forces of the order of 10–100 nN (cell dislodging) or 0.1–1 nN (cell stretching) have been reported [13,14]. Similar results were obtained by means of micropipettes. In this case, detachment forces are calculated from the pressure required to suck up a cell into the capillary. According to this technique, the adhesion forces are of the order of 1–10 nN [15,16].
Forces that cells exchange with substrates are not homogeneous along their body and may vary considerably over time. For example, lamellipodial regions are those cellular processes that more frequently probe surface features by forming and pulling adhesion structures, while forces exerted by adhesions depend on their spatial position and length [17]. In order to measure the forces exerted by such dynamic subcellular domains, methodologies able to detect receptor–ligand bond strength at the molecular level were developed. Among these, traction force microscopy has been widely used to characterize cell contractility on compliant polymers and hydrogels. Basically, it measures cell forces from the displacements of fiducial markers embedded within the substrates. Despite its relative ease of implementation, the resolution of traction force microscopy is limited by the ability to measure very small displacements, around 1 μm, which, in the case of compliant substrates such as polyacrylamide gels, provides a force sensitivity of the order of 0.1 nN [18,19]. Moreover, extensive computational labour might be required to trace back the force field from the displacements. Higher resolution techniques such as magnetic or laser tweezers can achieve force resolution of the order of 10−5 and 10−4 nN, respectively [20]. Their use requires micrometric beads decorated with the ligand of interest and they are able to detect individual ligand–receptor bond strength. Scanning force microscopes provide high-resolution force measurements. Towards this aim, however, tips need to be specifically designed to engage the receptor or molecule of interest.
4. Molecular cue-mediated crosstalk
Since the early years of cell culture, biomaterial interfaces have been endowed with adsorbed ECM proteins, such as fibronectin, vitronectin, laminin and collagen, to support the cells and present an instructive background to guide their behaviour. This approach still remains popular owing to its simplicity, and it has been enriched by using more selective techniques such as self-assembling monolayers (SAMs) to generate a defined surface able to control the nature and the distribution of protein adsorption. However, because cells depend on specific proteins for anchorage and extracellular instruction, the composition of the adsorbed layer is a key factor in cell behaviour. The required proteins, correctly presented, can stimulate a constructive cell response, favouring wound repair and tissue integration, while proteins in an unrecognizable state may indicate foreign materials to be isolated or removed. However, the complexity and the dynamic nature of the involved process strongly limit the control on protein adsorption and impair the long-term stability. Therefore, shorter amino acid sequences present in extracellular proteins have been screened, which proved to elicit analogous stimuli with the advantages of being more stable and much easier to conjugate on material surfaces. Among these sequences, RGD, YIGSR, YKVAV, LGTIPG, PDGSR, LRE, LRGDN and IKLLI from laminin, RGD and DGEA from collagen I and RGD, KQAGDV, REDV and PHSRN from fibronectin have been identified. Probably the most studied peptide is the integrin-binding sequence RGD alone or integrated in a more complex architecture. Today, a considerable variety of peptide sequences containing RGD with different intrinsic abilities to promote various cell responses are known and excellently reviewed elsewhere [21,22]. Different molecular architectures containing the RGD sequence demonstrate different affinities for integrins as determined by the flanking residues, conformation and accessibility towards integrins. Therefore, these observations lead to the general statement that, besides the chemical entity of the biochemical signal, it is the way it is presented at the cell–material interface that plays a central role in regulating the cell response.
There are different approaches in which a bioactive sequence can be presented towards a cell, and the manner in which the peptide is immobilized on a surface can affect the peptide concentration, density, arrangement, conformation and accessibility. The methods of immobilizing peptide moieties on material surfaces can be classified into physical and chemical. Physical methods involve the adsorption or precipitation onto solid material, while, in the chemical approach, the peptide is covalently bound to the materials; the former probably leading to inactive conformations, the latter to be specifically designed [21–23]. Examples of the evaluation of cell adhesion on bioactive substrates are reported in table 1.
Table 1.
Cell adhesion parameters on substrates endowed with matricellular cues (peptides/proteins). The table shows the kinds of matricellular cues, the substrates on which they are presented and the densities at which they are provided. Cell adhesion is measured in terms of detachment force, energy and stress or, alternatively, spreading areas. The table also provides the techniques used to evaluate the adhesion parameter in parenthesis.
| peptide, protein | substrate | density | cell | force, stress or energy | spreading area | references |
|---|---|---|---|---|---|---|
| RGD | dopamine–glass | fibroblasts | 5 × 10−14 J adhesion energy per cell | 2500 μm2 | [24] | |
| GRGDSP | glass | 2.3 × 1012 mol cm–2 | BAECs | 75 dyne cm–2 | [25] | |
| YAVTGRGDS | poly-acrylamide | 1.0 mg ml–1 (1500 mol μm–2) | BAECs | 6000 μm2 | [26] | |
| C(-RGDfK-) | gold nanoparticles | 1473 mol μm–2 (spacing 28 nm) | REF52 | 0.75 nN μm–2 global detachment force density (atomic force microscope-based measurement) | [27] | |
| YAVTGRGDS | poly-acrylamide | 1 mg ml–1 | BAECs | 0.22 dyne (traction force microscopy) | 2.5 × 10−5 cm2 | [28] |
| fibronectin | poly-acrylamide | 200 mol μm–2 | Balb/c 3T3 fibroblasts | 8.7 kdyne cm–2 (traction force microscopy) | 900 μm2 | [29] |
| chitosan | bead | osteoblast (2 × 105 cell ml–1) | 209 nN (atomic force microscope tip) | 650 μm2 | [30] | |
| collagen I | adsorption on a poly-styrene dish | murine fibroblasts L929 | 1500 Pa detachment force per cell area (atomic force microscope tip); 29 pJ detachment surface energy | [31] |
The density of the bioactive molecule immobilized on the material surface is one of the most relevant parameters in controlling cell–material interaction. Several literature reports have addressed this issue by modulating the nominal density of RGD at cell–material interfaces [29,32]. However, the nominal density does not match the actual density of RGD presented and available at the cell membrane, which instead depends on many factors such as the deposited or incorporated amount, spatial arrangement and accessibility of the peptide [33]. The accessibility of the peptide depends upon the chemical–physical and the texture properties of the surface, which, in turn, can be affected by the procedure of peptide conjugation on the surface [34]. Owing to this interplay, the effect on cell behaviour of inserting a peptide on a substrate could be cumulative, cooperative or even competitive. In general, the effect of density of the adhesion peptide on the surface brings about an enhancement of cell adhesion and the spreading area [26], according to a saturation-type profile. Cell adhesion commences to be observed roughly at a density of tenths of fmol cm−2 and reaches full spreading at concentrations of about tens of fmol cm−2. At a surface density of 0.1 fmol cm−2, fibroblast cells were observed to adhere but not to spread, while at a density of 1 fmol cm−2, a single cell spreading was observed on a glass-substrated, conjugated RGD peptide [35]. Clustering of the avb3 receptor was observed only at densities of 10 fmol cm−2 and higher, as was the assembly of a normal actin cytoskeleton. These measures provided a benchmark for the design of practical peptide-incorporating surfaces, as they indicate a minimum RGD oligopepetide density of 10 fmol cm−2. However, as mentioned already, these values or thresholds depend upon the material properties of the substrate. Indeed, for PET-conjugated RGD it has recently been reported that, to observe an enhancement of cell adhesion, a density as high as 1000 fmol cm−2 should be reached [36]. As the RGD concentration increases, focal adhesion extension also increases with a threshold level corresponding to 60 ligands per µm2 [35]. Integrin lateral clustering as a consequence of ligand density and spacing on materials deeply affects focal adhesion assembly and dynamics. This suggests that the local density of integrin receptors plays an essential role in controlling the dynamics of adhesive plaques [37].
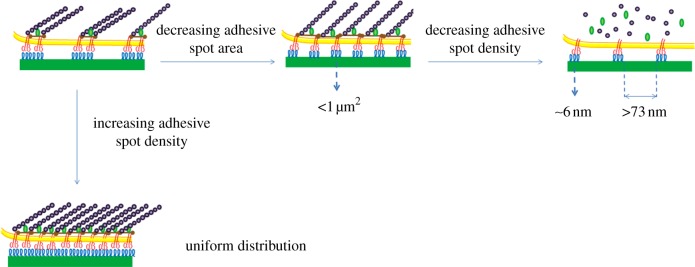
Beyond ligand density, the spatial distribution of the adhesive moieties at the nanoscale is also an important feature that has strong implications for cell–material interaction. In particular, distribution features such as uniformity, clustering and gradient were liberally addressed in the literature. It has been reported that, in the case of a lateral spacing of 73 nm between integrin binding sites, focal contact formation is inhibited and cells do not spread. For spacing lower than 58 nm, in contrast, focal contacts and actin stress fibres form and cells adopt a well-spread, pancake-like shape [38]. Therefore, the presentation of a clustered integrin–ligand format may result in more efficient building of the adhesive complexes that constitute the focal adhesion, resulting in enhancement of cell spreading and an anticipation of the onset of cytoskeleton assembly [39] (figure 1).
Figure 1.
Adhesion formation and cytoskeletal assemblies on patterns of biological signals. Decreasing the size of RGD-functionalized gold squares under 1 μm2 results in poor recruitment of α5β1-integrin, altered actin assembly and a decreased rate of cell migration [40]. Decreasing adhesive spot density until a lateral spacing of 73 nm between integrin binding sites (RGD-functionalized gold nanoparticles 6 nm in diameter) inhibits focal contact formation [41]. Uniform distribution of RGD on functionalized substrates allows for cell adhesion when overcoming a threshold level depending on materials, bioconjugation and cell type. A minimum cluster of six RGD–gold nanoparticles (6 nm in diameter), each separated by 100 nm, was found to be the minimal number to activate cell adhesion, paxillin accumulation and subsequent focal adhesion formation [38]. (Online version in colour.)
The molecular tether occurring between substrate and the peptide sequences is also an important parameter that affects cell–material interaction [42]. Because the ligand must be reached by cell-surface integrin, the presence of a molecular tether and its flexibility has been reported to have an interesting effect on the time of adhesion. As the length of the tether increases, the time required for cell spreading increases, but the final spreading area is not affected by the length of the tether [43]. Presumably, the kinetics of focal adhesion formation are retarded by a long tether owing to an overlapping of the dynamics of sequestration and release of RGD with the focal adhesion formation dynamics.
It is worth pointing out once more that generalization of the threshold values discussed above is not possible owing to their strong dependence on the substrate properties. Textural and mechanical features of the surface could determine different thresholds levels. Some consensus seems to exist for solid and rigid materials, such as glass, assuming that the amount of peptide is evenly distributed over the surface of the substrates. Indeed, it is generally considered that only a modification of the surface is achieved and that the reagent penetration depth is the same as the integrin accessible depth of 10 nm, treating the results in a two-dimensional manner. In the case of polymers, instead, the depth of the surface modification after treatment will depend upon the polymer crystallinity, the size of the pore and the swelling capability of the polymer at the surface [44] (figure 2a–c).
Figure 2.
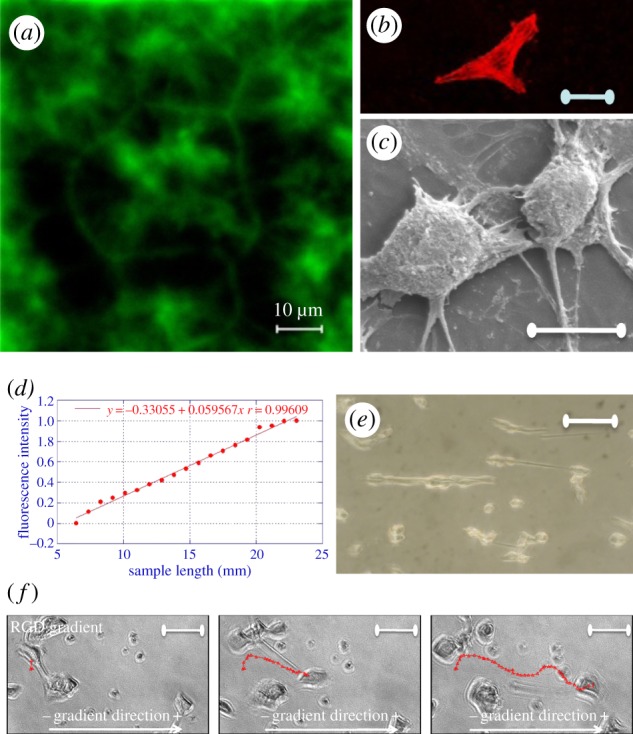
RGD motif on polymeric platforms. Spatial distribution of GRGDY on polycaprolactone (PCL) substrates [PCL-glutaraldehyde-(GA)-GRGDY] obtained by aminolysis in a 10% (w/w) 1,6-hexane-diamine/isopropanol solution and bioconjugation with 2% glutaraldehyde in 10 mM phosphate buffer (a). Confocal image for phalloidin staining of microfilaments (b) and scanning electron microscopy image (c) of NIH cells on PCL-GA-GRGDY substrates; a corrected surface density of about 1 mM cm−1 was estimated as the number of molecules per area available sufficient for the integrin engagement with a characteristic rose-like clustering of the immobilized peptide at the interface. Profile of the variation of the RGD–polyethylene glycol (PEG)–acrylate concentration along the RGD gradient length (d); NIH3T3 cells adhered (e) and frames (f) of NIH3T3 migration on the RGD–PEG–acrylate scaffold (slope = 1 mM cm−1; average RGD concentration = 1.5 mM). These results highlight the profound effect of spatial distribution of matricellular signals on the dynamics of focal formation and their directionality and orientation and ultimately on the shape and organization of the cytoskeleton. Therefore, the engineered gradient of adhesive molecules is another important ingredient of cell–material crosstalk because it can be instrumental to tuning cytoskeleton assembly and mechanics and therefore controlling cell functions and fate. (Online version in colour.)
Patterning molecular adhesive islands on the substrate has proved to be very effective in guiding the formation of adhesive plaques and therefore affecting cell adhesion, shape and migration [45–47]. Technologies, such as microcontact printing, atomic force microscope lithography and micromoulding in capillaries were successfully used to transfer patterns of adhesive proteins with predefined geometric features onto synthetic substrates. These techniques fostered studies on the effect of adhesion confinement on cell behaviour. Since the pioneering work by Chen et al. [48], which first demonstrated how different patterns of adhesive regions could induce endothelial cell apoptosis or growth, several other scientific reports have addressed the effects of adhesive islands with well-defined geometric features on cell fate. These studies indicate strong cell phenotype dependence in response to adhesive island patterning. The same patterns do not trigger the same material–cytoskeleton crosstalk and may elicit very different behaviour on different cell types. Stem cells, for instance, are very sensitive to the shape and dimensions of adhesive islands and commit to their fate according to specific patterns [49,50]. Stem cells can be directed towards osteogenesis or adipogenesis lineage by modulating their shape and controlling the patterning of adhesion regions on the substrate [51]. Hence, it can be hypothesized that patterns of adhesive regions dictate cell fate through the modulation of material–cytoskeleton interaction. The finding that stem cells' commitment is mediated through the activity of the RhoA, in particular through its effects on Rho-associated protein kinase (ROCK)-mediated cytoskeletal tension, supports this hypothesis [49].
Recently, continuous gradients of immobilized adhesive molecules on material surfaces have also been investigated [52,53]. Graded distribution of covalently bound bioactive peptides have been realized in two and three dimensions by using different techniques [54–56]. Cells react to the gradient by exhibiting a highly elongated and stretched shape polarized along the gradient direction with the degree of stretching depending upon gradient intensity and local average density of adhesive cues [57]. Adhesion complexes at the cell–material interface grow faster and become more stable with higher adhesion molecule density [58]; therefore, focal adhesion plaques form preferentially towards higher density, leading to cell polarization along the gradient.
The most striking effect of gradient is probably on cell migration. Cells preferentially move towards the direction of increasing RGD concentration with the speed depending on the gradient intensity and local density [58]. At a low density of adhesion molecules, cell speed increases almost linearly with gradient slope, while at a high density it levels off and becomes unaffected by the gradient [52]. Moreover, cells move along the direction of the gradient with a speed higher than that on a uniform distribution of RGD [57] (figure 2d–f).
5. Topo-cue-mediated crosstalk
The topographical textures of material surfaces have been reported to affect cell function and activity [59,60], and this has led to an increasing belief that cells can actually feel and react to micro- and nanomaterial corrugation, possibly through membrane deformation and stretching. Hence, according to this assumption, topographical features start to play a role in cell–material interaction through the regulation of material–cytoskeleton crosstalk via membrane alteration. Topographical signals, however, are not to be considered in vitro artefacts because the effects of topographical patterns on cell activity are also present in an in vivo context. Examples of native topographic micro- and nanopatterns are found in fibrils and fibre bundles (collagen and fibrin), rough surfaces (crystal deposit in bone) and porous membranes (basement membranes).
In recent times, the development of micro- and nanofabrication technologies made it possible to pattern surfaces with very detailed features significantly favouring the study of the role of topography in cell–material interaction. Lithographic techniques such as soft lithography [61,62], electron beam lithography [63] and nano-imprint lithography [64] can imprint topographic patterns with spatial resolution of a few nanometres. Other methodologies, such as polymer phase separation and polymer electrospinning, are in principle faster, allowing large areas to be patterned with limited costs. However, they may lack homogeneity, and the materials thus produced usually do not exhibit long-range order.
The most extensively studied topographies are grooves and grids, protrusions and pit arrays. Three characteristic dimensions, namely ridge (or groove) width, inter-feature length (or pitch) and feature depth (or height), define these surfaces. Neglecting, in the first instance, the chemical/physical properties of the material surface, which actually plays an important role, there are countless combinations of topographic signals that can be presented to cells. However, topographies exhibiting characteristic dimensions that surpass those of cells (tens of micrometres) at most provide a geometric confinement to the cell and are of little interest in this context. The most relevant effects are observed as soon as feature dimensions approximate those of the cell's sensorial organelles, i.e. focal complexes and adhesions. Several works have reported that 70 nm seems to be a limiting inter-feature dimension under which topographic signals begin to be less effective [65,66]. Within such a dimensional range, i.e. more than 70 nm and less than 2–5 μm, many studies have investigated the effects of nanometric features on cell adhesion, morphology, migration, biosynthesis and differentiation.
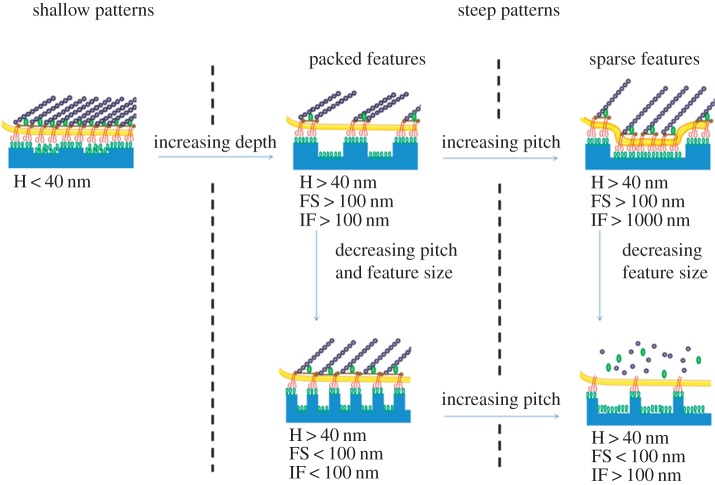
Most of the studies reported in the literature are mainly focused on the investigation of the effects of the topography itself, without additional adhesive signals that are tethered to the nanotextured surface. Cell attachment is usually provided by a layer of adhesive proteins adsorbed on the surface, including the serum proteins fibronectin, vitronectin or collagen. Because the presence of nanometric features alters the surface properties of materials, such as the surface energy and wettability, the preferred protein adsorption might result. Furthermore, topographic patterns create surfaces that are readily accessible for cell lamellipodia and filopodia, such as the top of ridges or pillars, as well as impervious recesses, such as the bottom of grooves and pits. The accessibility of a given region to the cell membrane and its protrusion structures depend on the geometric characteristic of the pattern, i.e. the feature depth and pitch. When these parameters are such that cell membranes cannot accommodate surface recesses, the cell is suspended on the top of the pattern, and adhesion is limited to specific parts. Generally, this is achieved when features are densely packed and their height is more than 40 nm. In shallower patterns, serum proteins can clog up grooves, pits and recesses, thus creating an adhesive ‘carpet’ for the integrins. Provided that features are sufficiently deep, integrin clustering and adhesion formation is observed on the top of pillars with diameter larger than 70 nm, even though focal adhesions are usually smaller for those observed on flat surfaces [66,67]. On the contrary, weak adhesions are observed on smaller features. These observations are consistent with the data reported by Choi et al. [68], who observed that nascent adhesions reach a plateau dimension before their disassembly, which is in the range of a hundred nanometres. Therefore, it is reasonable to expect that exposing cells to ridges or protrusions smaller than 100 nm strongly interferes with adhesion maturation and subsequent stabilization. Presumably, further reduction of inter-feature distance may restore integrin clustering and stabilization by allowing intracellular adhesion proteins (e.g. talin) to connect to adjacent integrin domains. Broadening inter-feature spacing would cause the cell basal membrane to come in close contact with grooves or pit surfaces, thus enabling the formation of additional focal complexes (figure 3).
Figure 3.
Illustration of cell–nanotopography interactions. Different combinations of feature sizes and depths might reduce the surface available for the cell membrane to establish adhesions. Looking at the pattern from the cell side, five integrins are necessary to connect the cytoskeleton to the extracellular environment [69], which requires integrin clusters of approximately 40 nm. Therefore, topographic features exceeding this dimension can be recognized by cells as ‘adhesive’. From the material side, ligand spacings below 60 nm proved to be sufficient to ensure cell-substrate binding [37]. In this case, inter-feature size exceeding this dimension might impair the formation of adhesive complexes. Moreover, surfaces are generally coated with serum proteins, which account for a macromolecular layer of 20–30 nm. Therefore, in order to make a surface not available for the establishment of cell adhesions, this has to be at least 40 nm away from the cell membrane, which comprises the distance of the protein layer and the extracellular domain of the integrins (H, height; FS, feature size; IF, inter-feature dimension). (Online version in colour.)
Increasing feature height, while keeping constant inter-feature length, causes the space between adjacent grooves to become impervious to the cell membrane and integrins and therefore a decrease in adhesion stability occurs [70,71]. Analogous results have been observed on substrates presenting arrays of nanometric pits. In particular, when pit depth approaches 100 nm, focal adhesions seem to be confined to the inter-pit area, indicating that pit density and inter-feature size became crucial in controlling integrin clustering and focal adhesion formation. Generally, reduced adhesion and spreading, accompanied by a less organized cytoskeleton, are observed on arrays of hundred nanometre wide pits, with an inter-feature distance of approximately 300 nm [72–75].
Cells cultivated on nanogrooved substrates exhibit similar behaviour. However, ridges provide a longitudinal direction for the integrins to cluster and adhesions to form. Topographic characteristic dimensions regulate whether the cell membrane can conform to grooves or bridge between the ridges. According to Teixeira et al. [70], membrane processes of epithelial cells can adhere to the surface of grooves that are 150 nm deep and 2100 nm wide, while they anchor to the ridge top for 330–950 nm wide grooves. Moreover, they observed that focal adhesions and actin fibres co-align with the ridge direction, eventually leading to cell alignment (a phenomenon usually called contact guidance). Along this line, Loseberg et al. [65] and Lamers et al. [66] examined a broad range of groove/ridge dimensions and their effects on contact guidance. Although they used different cells, fibroblasts and osteoblasts, respectively, they reported comparable results in terms of minimum size that is able to induce cell alignment, i.e. 80–100 nm for ridge/groove width and, at least, 35 nm in height. Yet, this general trend in some cases is dependent on cell type, material properties and culturing conditions. For example, Teixeira et al. [76] demonstrated that epithelial cells cultured on narrow ridges (70 nm and 400 nm pitch) can assume a spindle-like morphology parallel or orthogonal to the pattern direction, according to the culturing conditions. The molecular mechanisms governing cell alignment are not fully characterized, although it is hypothesized that filopodial activity perpendicular to the pattern direction is decreased owing to unfavoured stress tension [77]. Table 2 provides a summary of the more relevant data presented so far on cell topography interactions.
Table 2.
Effects of topographic cues on cell behaviour. A general reduction in cell spreading and focal adhesion length is observed when the feature size is in the range of 100 nm. In the case of nanogrooved substrates, a cell pattern co-alignment is observed when the feature depth is above 50–100 nm. Poly-L-lactide (PLLA); polycarbonate (PC); polystyrene (PS); polymethylmethacrylate (PMMA); polycaprolactone (PCL).
| topographic feature | material | cell type | feature size (nm) | pitch (nm) | depth (nm) | effects | reference |
|---|---|---|---|---|---|---|---|
| pits | PCL | rat fibroblasts | 50 | 75–300 (centre to centre) | 100 | reduced spreading; reduced adhesion length | [72] |
| grooves | Si | human corneal epithelium | 70–1900 | 400–4000 | 150–600 | enhanced cell/groove co-alignment on steep patterns; reduced spreading | [70] |
| grooves | Si | human corneal epithelium | 70–1900 | 400–4000 | 600 | perpendicularly oriented cells on FS = 70 nm with oblique focal adhesions; co-aligned cells for FS > 850 nm with oblique focal adhesions. | [70] |
| pits | PCL | human fibroblasts | 35–75–120 | 100–200–300 | 50–100–100 | reduced spreading; disorganized cytoskeleton; mature focal contacts at cell periphery; increased filopodia | [73] |
| pillars | PLLA | human fibroblasts | 700–800; 500–550 | 600–700; 250–300 | 620; 590 | increased adhesion; adhesion observed on top and between pillars | [78] |
| posts and gratings | Si | human fibroblasts | 10 (apex radius) | 230 | 50–600 | reduced spreading; constant proliferation on shallow patterns; reduced proliferation on deep patterns | [79] |
| pits | PC | human osteo-progenitor cells | 120 | 300 | — | reduced spreading; increased filopodia density; small and sparse adhesions; less developed cytoskeleton | [75] |
| grooves | PS | rat fibroblasts | 20–500 | 40–1000 | 4.4–158 | cell/groove co-alignment for FS > 150 nm and D > 35 nm | [65] |
| pits | PMMA | human fibroblasts | 120 | 300 | 100 | increased filopodia; increased endocytosis; disorganized cytoskeleton | [74] |
| grooves | Si | osteoblast-like | 90–500 | 90–500 | 300 | reduced spreading; increased length; actin/groove co-alignment; vinculin/groove co-alignment; oblong nuclear shape | [80] |
| pillars | Ti | human osteo-progenitor cells | 28–41–55 | 40–74–115 | 15–55–100 | normal spreading and cytoskeleton on 15 nm deep pillars; reduced spreading and disorganized cytoskeleton on steep pillars; low matrix deposition on steep pillars | [67] |
| grooves | PS | rat osteoblasts | 10–750 | 40–1000 | 11–153 | cell/grove co-alignment (FS > 75 nm and D > 33 nm); focal adhesion/groove co-alignment (for FS > 150 nm and D > 120 nm); aligned CaP deposits; upregulation of osteospecific genes | [81] |
| pillars | PS | rat osteoblasts | 60–600 | 60–600 | 8–160 | constant spreading; reduced FA length (for FS > 200 nm and D > 30 nm) | [66] |
| grooves | PS | rat osteoblasts | 20–750 | 40–1000 | 4.4–158 | increased cell elongation (FS > 200 nm and D > 77.4 nm); increased cell/groove co-alignment (FS > 160 nm and D > 52 nm); constant spreading; reduced focal adhesion length | [66] |
Nanostructured features have also been shown to be a powerful tool to trigger and regulate stem cell differentiation. In a landmark study by Dalby et al. [82], nanostructured surfaces proved to be sufficient per se to induce stem cell differentiation. In particular, adult skeletal stem cells not only proved to perceive nanometric features, but also to be very sensitive towards their spatial configuration, i.e. order/disorder. In a different study, Yim et al. [83] demonstrated that nanograted substrata were able to induce transdifferentiation of human mesenchymal stem cells (MSCs) in neurons.
As expected, cell migration is also deeply influenced by nanometric topographies, in terms of both persistence and speed. Most of the studies concerning the effects of nanostructures on migration have been performed on nanograted substrates. A large variety of cells such as epithelial [84], endothelial [85], glial [86], fibroblasts [87] and osteoblasts [66] exhibit a biased motion along the nanometric groove direction. Such a biased migration might arise from predominant orientation along the pattern as well as from specific intracellular assemblies of cytoskeletal components. As observed by Yim et al. [62], topographies alter the intracellular distributions of the marker for directional motion, i.e. microtubule organization centres, with respect to conventional planar surfaces. No clear conclusion on the effect of topography on cell speed has been drawn so far, which probably depends on the cell type, culturing conditions and material properties.
Taken all together, these results clearly indicate the potency of topographic features in regulating the cell–material crosstalk and in directing and guiding cellular activity and fate. Similarly to that depicted for patterning of a biochemical adhesive, the topographical pattern regulates the material–cytoskeleton crosstalk by influencing the dynamics of formation of adhesion complexes. Indeed, grooves, grids, pits or pillars can all be seen as spatial confiners for growth and remodelling of adhesion plaques. Because the formation of macromolecular transmembrane complexes, underpinning the material–cytoskeleton crosstalk, may only occur in a defined region, the patterning of topography at the nanoscale offers the possibility to control their macromolecular assembly and associated dynamics.
It is still to be ascertained whether topography is an independent cell signal that is directly recognized by the cell or whether it prompts a pattern in the distribution of adhesive proteins, which, in turn, effect material–cytoskeleton crosstalk. The first hypothesis requires that the cells possess membrane sensors for topography recognition. This may be envisaged to occur through deformation and local stretching of the membrane occurring at corners or squared or sharp curvature changes that it encounters along the patterned topographical profile of the surface. However, especially in the case of nanotopographic patterns, it is unlikely that the membrane can bend in nanocavities to closely follow the material profile. On the other hand, it is likely that topographical patterns influence the distribution of serum adhesive protein, which, in turn, triggers pattern recognition via the well-known integrin recognition pathways.
6. Mechano-cue-mediated crosstalk
Cell adhesion implies formation of adhesion plaques, on which actin stress fibres anchor to build the mechanical integrity of cytoskeleton [88]. The pulling force acting on the adhesion plaques triggers plaque growth, which, in turn, allows thickening of the stress fibres [89]. Therefore, material–cytoskeleton crosstalk ineluctably involves force exchange between cells and substrates. The ability of cells to react to the mechanical properties of substrata is generally referred to as mechanosensing and implies both the action of the material on the cell and the action of the cell on the mechanical properties of the material [90]. The first experimental evidence of mechanosensing dates back more than 30 years when Harris et al. [91] reported and measured cell contractile forces on flexible rubber membranes. Numerous other studies have followed this first report especially focused on developing systems to analyse the dynamics of cell-generated forces [92–94]. These forces fall within the 1 pN–1 nN range that is sufficient to deform ECM components, because native tissues possess a range of stiffness spanning from 10−3 to 103 MPa. These studies greatly contributed to elucidating the mechanics of cell-generated forces, making it possible to identify the most relevant molecular players that are involved in this process. Notably, cells are likely to exert higher forces on compliant substrates than on stiff substrates [95]. These observations corroborated the existence of a regulatory mechanism for the interplay between cytoskeleton-generated forces and the mechanical properties of materials that represent the foundation of mechanosensing. Therefore, the mechanical properties of the material represent an additional facet of the material–cytoskeleton crosstalk that intervenes in controlling the dynamic reciprocity between adhesion plaque growth and substrate deformation. According to this view, patterning of mechanical properties may be used to direct and guide cell activity. Indeed, cells cultivated on collagen-coated polyacrylamide gels display very different adhesions in response to the mechanical stiffness of the substrate [3]. In particular, mature and stable focal adhesions are observed on rigid gels, whereas shorter and more dynamic complexes are observed on soft gels. The cytoskeleton is weakly assembled in the cytoplasm, being predominantly cortical when cells are seeded on soft gels, while on stiff gels larger and well-defined bundles are observed. However, it must be pointed out that not all cell types respond consistently to substrate stiffness [96]. Mechanical forces also have a large impact on cell proliferation and migration. Higher proliferation rates have been observed on stiffer gels [97]. Moreover, cells migrate with higher speed on soft gels while they are more stationary on stiff substrates [3]. However, these phenomena are strictly related to the culturing conditions and cell types [98,99]. Interestingly, cells cultured on gels or substrates presenting gradients of stiffness migrate preferentially from the soft to the stiff region, a phenomenon referred to as durotaxis [100]. Micrometric patterns of mechanical properties have been realized using various materials and techniques [101–103]. Such patterns proved to be adequate in confining cell motion without the use of geometrical barriers such as channels and pillars.
The effects of mechanical cues are not limited to cell adhesion and migration. Mechanical cues, as observed in the case of biochemical and topographic patterns, can be responsible for stem cell commitment to diverse lineages. In a landmark study, Engler et al. [104] reported that substrate stiffness is sufficient to commit MSC differentiation towards different lineages. In particular, MSCs cultivated on polyacrylamide gels have stiffness similar to that of brain, muscle or bone differentiated into neurons, myoblasts and osteoblasts, respectively. In a more recent study, Winer et al. [105] showed that MSCs remained quiescent on substrates whose stiffness matches that of fat or bone marrow.
In summary, the experimental evidence of the cell response to the mechanical properties of the substrate shares surprising similarities with that obtained by culturing undifferentiated cells on biochemical or topographic patterns: the modulation of cell adhesion, and consequently cell shape, induces changes in cell fate. This happens irrespective of the method with which cell adhesion patterns are established, suggesting that, probably, the way cells feel and react to the substrate properties, i.e. the material–cytoskeleton crosstalk, may share common pathways.
7. Cell instructive materials
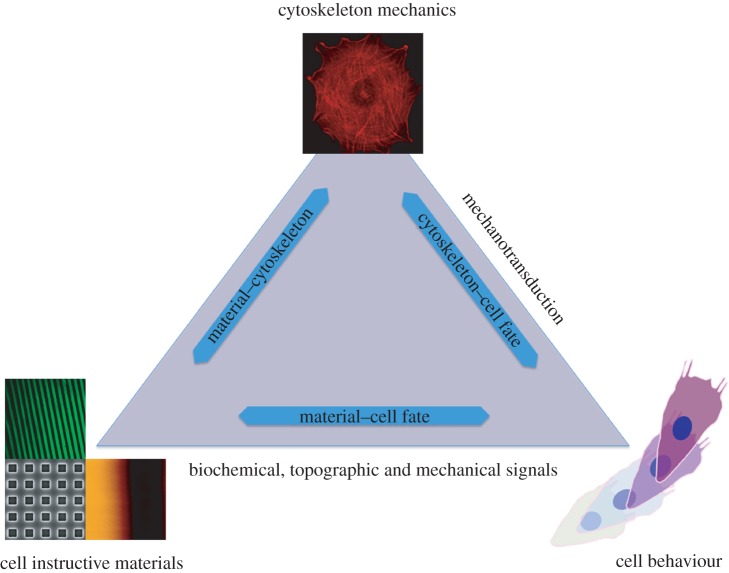
The recent literature has significantly elucidated the mechanisms by which cells sense biochemical, topographic and mechanical signals, translating them into commands that regulate activity and fate by triggering specific intracellular pathways. Most of the studies, however, deal specifically with the effect of a single signal type. This has also led to the tacit acknowledgement that different signal types are recognized by cells through independent pathways and therefore different routes can be pursued to govern material–cytoskeleton crosstalk. Yet, experimental evidence suggests that a common leitmotif connecting the effects of different types of signals on cell response might exist. Cells cultured on large bioadhesive areas, or on shallow nanotopographies or on stiff substrates, exhibit a mature cytoskeleton, with large actin bundles and large focal adhesions. In contrast, cells on small adhesive islands, or on deep nanotopographies or on compliant substrates, display a diffuse cytoskeleton and small adhesions. These observations demonstrate how cell characteristics that might appear so disparate, such as cell shape, cytoskeletal forces and differentiation, are in turn intimately connected, suggesting a ubiquitous role for cell adhesions and cytoskeletal forces in regulating signalling pathways and ultimately cell fate (figure 4). Despite a limited number of studies that have dealt with a synergistic presentation of diverse signals, the experimental evidence suggests that cells are able to integrate and react to different signal types that are presented simultaneously. In particular, Tzvetkova-Chevolleau et al. [106] reported different cellular dynamics, in terms of spreading and motility, of normal and cancer fibroblasts when exposed to combinatorial topographic and mechanical stimuli. Moreover, normal fibroblasts exhibited persistent migration, whereas cancerous cells had a more random, but fast, motility irrespective of the topography and such a response was enhanced on rigid surfaces. The interplay between biochemical and topographic signals on endothelial cells was investigated by Le Saux et al. [107]. They reported a biphasic response of cell adhesion in which cells on flat or nanoscale rough substrates required a higher density of RGD ligands for optimal adhesion, with respect to microscale topography. On the contrary, cell spreading was affected only by the RGD density and not by the topography. Altogether, these findings suggest that topography governs early adhesion events, whereas surface chemistry becomes dominant in cell spreading.
Figure 4.
Material–cytoskeleton crosstalk affects cell behaviour and fate with adhesion localization and composition, and cell-generated forces. Cell instructive materials aim at exploiting these characteristics in order to impart cell-specific orders by modulating adhesion and cytoskeletal assemblies through the delivery of biological, topographical and mechanical signals. (Online version in colour.)
An extensive literature has demonstrated the effects of cytoskeleton-generated forces on cell fate, which are a part of a wider process conventionally referred to as mechanotransduction. This process, i.e. the mechanisms by which mechanical forces are converted into biochemical functions, may occur at different levels. Of particular interest within the context of material–cytoskeleton cross talk are those events occurring at the adhesion and nucleus level.
The formation of focal adhesions and complexes is responsible for proper cell attachment, and specific molecules present in the adhesion plaque regulate the activity of downstream signalling pathways that are involved in cell proliferation and differentiation. In particular, phosphorylation of FAK can lead to stimulation of a cascade that ultimately activates the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) pathway [108], which is known to be central in regulating stem cell differentiation [109]. It is therefore expected that controlling the formation of adhesion structures (either focal complexes or adhesions) might be a potent strategy to control a cell. Nanometric patterns, both biochemical and topographic, proved to be very effective for this goal. In fact, they have a strong ability to interfere with integrin clustering and consequently adhesion formation. In particular, the high spatial resolution of topographic patterns makes these suitable to modulate adhesion length, density and spatial positioning. For example, Biggs et al. [110] observed reduced adhesion maturation and spreading of osteoprogenitor cells cultivated on nano-pit arrays correlated with impaired osteospecific differentiation through a depression of ERK signalling and downregulation of osteocalcin synthesis. Genetic and proteomic studies of osteoprogenitor cells on disordered nano-pit arrays indicated the involvement of integrin and cytoskeletal signalling along with the ERK1/2 pathway in osteogenesis [111,112].
Similar findings were observed by constraining cell shape using patterns of adhesive islands, which proved to be a very effective means of modulating cell contractility. McBeath et al. [49] underlined the role of the RhoA pathway and cell contractility to govern stem cell differentiation. In particular, RhoA and ROCK activity was found to be dependent on cell shape with higher values found on spread cells which underwent osteogenesis. Similarly, Kilian et al. [51] reported enhanced osteogenesis, as a consequence of increased myosin contractility, through MAPK pathways and Wnt signalling, whereas disruption of contractility promotes adipogenesis. Non-muscle myosin II has also been implicated in MSC lineage specification induced by substrate stiffness [104], highlighting, again, the role of actin-generated forces in regulating cell fate.
Adhesion-mediated signalling pathways are not the only factors responsible for regulating cell functions, in particular stem cell differentiation. Differently from such a signalling, which indirectly regulates signalling pathways through kinases, mechanotransduction acts in a direct manner by exerting cytoskeleton-generated traction forces directly to the nucleus. Indeed, the nucleus is connected with the cytoskeleton through lamins and nesprins [113]. Therefore, it is reasonable to expect that cytoskeletal forces are transmitted to the chromosomes, thus altering transcriptional events owing to modified accessibility of genes. Again, topography may regulate the magnitude and direction of such cell-generated forces by modulating adhesion length and spatial position. It is not clear whether one mechanism, either direct (nuclear level) or indirect (adhesion mediated), dominates the other. However, owing to the complexity and simultaneity of signals that are constantly broadcasted to cells in vivo, it is likely that the two mechanisms act in concert.
8. Conclusions
Material–cytoskeleton crosstalk occurs in three different phases, namely integrin–ligand engagement, plaque dynamics and growth and cytoskeleton assemblies. These phases occur at different material surface length scales. Integrin engagement occurs at a length of tens of nanometres, plaque growth occurs at hundreds of nanometres, while cytoskeletal assemblies occur at the micrometre scale. Ligand engagement is the prerequisite for the recruitment of the intracellular building blocks for plaque construction and a group of three to five integrins constitutes the elementary unit for adhesion plaques. This unit, 60 nm long, starts to build macromolecular complexes that secure the cell membrane on the underlying substrate. Finally, mechanically active cytoskeleton structures develop when this plaque reaches submicrometric dimensions. Material processing techniques, which enable local modifications of material chemical and physical features on different length scales, are now available and spatial patterns of these features may interfere with each or all of the phases that dictate the assembly of the cytoskeleton. Because it is well accepted that cytoskeletal organization and structure govern cell fate through mechanotransduction mechanisms, this might result in material surface textures being designed to impart specific commands or instructions to cells. CIMs directly stem from these principles, and the first attempts along this line have already been reported [114,115]. As Nature provides cells with multiple signal typologies that vary in time and space according to highly accurate schemes, the next generation of biomaterials has to replicate this complexity by integrating the combinatorial presentation on multiple length scales with a dynamic presentation of signals. This would pave the way towards the development of physiologically relevant platforms to analyse or alter cell behaviour. In principle, one might envisage material programmes to stretch cells in predefined shapes, to transport them along predefined paths, sort them according to cytoskeleton mechanics, guide their division and proliferation, stimulate and direct biosynthetic events and modulate lineage specifications.
References
- 1.Lehnert D., Wehrle-Haller B., David C., Weiland U., Ballestrem C., Imhof B. A., Bastmeyer M. 2004. Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. J. Cell Sci. 117, 41–52 10.1242/jcs.00836 (doi:10.1242/jcs.00836) [DOI] [PubMed] [Google Scholar]
- 2.Flemming R. G., Murphy C. J., Abrams G. A., Goodman S. L., Nealey P. F. 1999. Effects of synthetic micro- and nano-structured surfaces on cell behavior. Biomaterials 20, 573–588 10.1016/S0142-9612(98)00209-9 (doi:10.1016/S0142-9612(98)00209-9) [DOI] [PubMed] [Google Scholar]
- 3.Pelham R. J., Jr, Wang Y. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility. Proc. Natl Acad. Sci. USA 94, 13 661–13 665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Kooten T. G., Spijker H. T., Busscher H. J. 2004. Plasma-treated polystyrene surfaces: model surfaces for studying cell–biomaterial interactions. Biomaterials 25, 1735–1747 10.1016/j.biomaterials.2003.08.071 (doi:10.1016/j.biomaterials.2003.08.071) [DOI] [PubMed] [Google Scholar]
- 5.Dowling D. P., Miller I. S., Ardhaoui M., Gallagher W. M. 2011. Effect of surface wettability and topography on the adhesion of osteosarcoma cells on plasma-modified polystyrene. J. Biomater. Appl. 26, 327–347 10.1177/0885328210372148 (doi:10.1177/0885328210372148) [DOI] [PubMed] [Google Scholar]
- 6.Hallab N. J., Bundy K. J., O'Connor K., Clark R., Moses R. L. 1995. Cell adhesion to biomaterials: correlations between surface charge, surface roughness, adsorbed protein, and cell morphology. J. Long Term Eff. Med. Implants 5, 209–231 [PubMed] [Google Scholar]
- 7.Ponsonnet L., Reybier K., Jaffrezic N., Comte V., Lagneau C., Lissac M., Martelet C. 2003. Relationship between surface properties (roughness, wettability) of titanium and titanium alloys and cell behaviour. Mater. Sci. Eng. C 23, 551–560 10.1016/S0928-4931(03)00033-X (doi:10.1016/S0928-4931(03)00033-X) [DOI] [Google Scholar]
- 8.Yeung T., et al. 2005. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton 60, 24–34 10.1002/cm.20041 (doi:10.1002/cm.20041) [DOI] [PubMed] [Google Scholar]
- 9.Sackmann E., Bruinsma R. F. 2002. Cell adhesion as wetting transition? Chemphyschem 3, 262–269 (doi:10.1002/1439-7641(20020315)3:3<262::AID-CPHC262>3.0.CO;2-U) [DOI] [PubMed] [Google Scholar]
- 10.Geiger B., Yamada K. M. 2011. Molecular architecture and function of matrix adhesions. Cold Spring Harb. Perspect. Biol. 3, 1–21 10.1101/cshperspect.a005033 (doi:10.1101/cshperspect.a005033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsons J. T., Horwitz A. R., Schwartz M. A. 2010. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11, 633–643 10.1038/nrm2957 (doi:10.1038/nrm2957) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. 2009. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 10.1126/science.1162912 (doi:10.1126/science.1162912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sagvolden G., Giaever I., Pettersen E. O., Feder J. 1999. Cell adhesion force microscopy. Proc. Natl Acad. Sci. USA 96, 471–476 10.1073/pnas.96.2.471 (doi:10.1073/pnas.96.2.471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weder G., Vörös J., Giazzon M., Matthey N., Heinzelmann H., Liley M. 2009. Measuring cell adhesion forces during the cell cycle by force spectroscopy. Biointerphases 4, 27–34 10.1116/1.3139962 (doi:10.1116/1.3139962) [DOI] [PubMed] [Google Scholar]
- 15.Qin T. W., Yang Z. M., Wu Z. Z., Xie H. Q., Qin J., Cai S. X. 2005. Adhesion strength of human tenocytes to extracellular matrix component-modified poly(DL-lactide-co-glycolide) substrates. Biomaterials 26, 6635–6642 10.1016/j.biomaterials.2005.04.023 (doi:10.1016/j.biomaterials.2005.04.023) [DOI] [PubMed] [Google Scholar]
- 16.Gao Z., Wang S., Zhu H., Su C., Xu G., Lian X. 2008. Using selected uniform cells in round shape with a micropipette to measure cell adhesion strength on silk fibroin-based materials. Mater. Sci. Eng. C 28, 1227–1235 10.1016/j.msec.2007.11.003 (doi:10.1016/j.msec.2007.11.003) [DOI] [Google Scholar]
- 17.Stricker J., Aratyn-Schaus Y., Oakes P. W., Gardel M. L. 2011. Spatiotemporal constraints on the force-dependent growth of focal adhesions. Biophys. J. 100, 2883–2893 10.1016/j.bpj.2011.05.023 (doi:10.1016/j.bpj.2011.05.023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sabass B., Gardel M. L., Waterman C. M., Schwarz U. S. 2008. High resolution traction force microscopy based on experimental and computational advances. Biophys. J. 94, 207–220 10.1529/biophysj.107.113670 (doi:10.1529/biophysj.107.113670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stricker J., Sabass B., Schwarz U. S., Gardel M. L. 2010. Optimization of traction force microscopy for micron-sized focal adhesions. J. Phys. Condens. Matter. 22, 194 104–194 114 10.1088/0953-8984/22/19/19410 (doi:10.1088/0953-8984/22/19/19410) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustamante C., Macosko J. C., Wuite G. J. 2000. Grabbing the cat by the tail: manipulating molecules one by one. Nat. Rev. Mol. Cell Biol. 1, 130–136 10.1038/35040072 (doi:10.1038/35040072) [DOI] [PubMed] [Google Scholar]
- 21.Hersel U., Dahmen C., Kessler H. 2003. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials 24, 4385–4415 10.1016/S0142-9612(03)00343-0 (doi:10.1016/S0142-9612(03)00343-0) [DOI] [PubMed] [Google Scholar]
- 22.Perlin L., MacNeil S., Rimmer S. 2008. Production and performance of biomaterials containing RGD peptides. Soft Matter 4, 2331–2349 10.1039/B801646A (doi:10.1039/B801646A) [DOI] [Google Scholar]
- 23.Bhadriraju K., Hansen L. K. 2000. Hepatocyte adhesion, growth and differentiated function on RGD-containing proteins. Biomaterials 21, 267–272 10.1016/S0142-9612(99)00175-1 (doi:10.1016/S0142-9612(99)00175-1) [DOI] [PubMed] [Google Scholar]
- 24.He T., Shi Z. L., Fang N., Neoh K. G., Kang E. T., Chan V. 2009. The effect of adhesive ligands on bacterial and fibroblast adhesions to surfaces. Biomaterials 30, 317–326 10.1016/j.biomaterials.2008.09.049 (doi:10.1016/j.biomaterials.2008.09.049) [DOI] [PubMed] [Google Scholar]
- 25.Xiao Y., Truskey G. A. 1996. Effect of receptor–ligand affinity on the strength of endothelial cell adhesion. Biophys. J. 71, 2869–84 10.1016/S0006-3495(96)79484-5 (doi:10.1016/S0006-3495(96)79484-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinhart-King C. A., Dembo M., Hammer D. A. 2005. The dynamics and mechanics of endothelial cell spreading. Biophys. J. 89, 676–689 10.1529/biophysj.104.054320 (doi:10.1529/biophysj.104.054320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selhuber-Unkel C., Erdmann T., Lopez-Garcia M., Kessler H., Schwarz U. S., Spatz J. P. 2010. Cell adhesion strength is controlled by intermolecular spacing of adhesion receptors. Biophys. J. 98, 543–551 10.1016/j.bpj.2009.11.001 (doi:10.1016/j.bpj.2009.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinhart-King C. A., Dembo M., Hammer D. A. 2003. Endothelial cell traction forces on RGD-derivatized polyacrylamide substrata. Langmuir 19, 1573–1579 10.1021/la026142j (doi:10.1021/la026142j) [DOI] [Google Scholar]
- 29.Rajagopalan P., Marganski W. A., Brown X. Q., Wong J. Y. 2004. Direct comparison of the spread area, contractility, and migration of balb/c 3T3 fibroblasts adhered to fibronectin- and RGD-modified substrata. Biophys. J. 87, 2818–2827 10.1529/biophysj.103.037218 (doi:10.1529/biophysj.103.037218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsiao S. W., Thien D. V. H., Ho M. H., Hsieh H. J., Li C. H., Hung C. H., Li H. H. 2010. Interactions between chitosan and cells measured by AFM. Biomed. Mater. 5, 1–8 10.1088/1748-6041/5/5/054117 (doi:10.1088/1748-6041/5/5/054117) [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto A., Mishima S., Maruyama N., Sumita M. 2000. Quantitative evaluation of cell attachment to glass, polystyrene and fibronectin- or collagen-coated polystyrene by measurement of cell adhesive shear force and cell detachment energy. J. Biomed. Mater. Res. 50, 114–124 (doi:10.1002/(SICI)1097-4636(200005)50:2<114::AID-JBM4>3.0.CO;2-6) [DOI] [PubMed] [Google Scholar]
- 32.Gobin A. S., West J. L. 2002. Cell migration through defined, synthetic ECM analogs. FASEB J. 16, 751–753 10.1096/fj.01-0759fje (doi:10.1096/fj.01-0759fje) [DOI] [PubMed] [Google Scholar]
- 33.Craig W. S., Cheng S., Mullen D. G., Blevitt J., Pierschbacher M. D. 1995. Concept and progress in the development of RGD-containing peptide pharmaceuticals. Biopolymers 37, 157–175 10.1002/bip.360370209 (doi:10.1002/bip.360370209) [DOI] [PubMed] [Google Scholar]
- 34.Wheeldon I., Farhadi A., Bick A. G., Jabbari E., Khademhosseini A. 2011. Nanoscale tissue engineering: spatial control over cell–materials interactions. Nanotechnology 22, 212001. 10.1088/0957-4484/22/21/212001 (doi:10.1088/0957-4484/22/21/212001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Massia S. P., Hubbell J. A. 1991. An RGD spacing of 440 nm is sufficient for integrin alpha V beta 3-mediated fibroblast spreading and 140 nm for focal contact and stress fibre formation . J. Cell Biol. 114, 1089–1100 10.1083/jcb.114.5.1089 (doi:10.1083/jcb.114.5.1089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chollet C., Chanseau C., Remy M., Guignandon A., Bareille R., Labrugère C., Bordenave L., Durrieu M. C. 2009. The effect of RGD density on osteoblast and endothelial cell behaviour on RGD-grafted polyethylene terephthalate surfaces. Biomaterials 30, 711–720 10.1016/j.biomaterials.2008.10.033 (doi:10.1016/j.biomaterials.2008.10.033) [DOI] [PubMed] [Google Scholar]
- 37.Cavalcanti-Adam E. A., Volberg T., Micoulet A., Kessler H., Geiger B., Spatz J. P. 2007. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 92, 2964–2974 10.1529/biophysj.106.089730 (doi:10.1529/biophysj.106.089730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arnold M., Schwieder M., Blümmel J., Cavalcanti-Adam E. A., López-Garcia M., Kessler H., Geiger B., Spatz J. P. 2009. Cell interactions with hierarchically structured nano-patterned adhesive surfaces. Soft Matter. 5, 72–77 10.1039/B815634D (doi:10.1039/B815634D) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maheshwari G., Brown G., Lauffenburger D. A., Wells A., Griffith L. G. 2000. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 113, 1677–1686 [DOI] [PubMed] [Google Scholar]
- 40.Lutz R., Pataky K., Gadhari N., Marelli M., Brugger J., Chiquet M. 2011. Nano-stenciled RGD-gold patterns that inhibit focal contact maturation induce lamellipodia formation in fibroblasts. PLoS ONE 6, e25459. 10.1371/journal.pone.0025459 (doi:10.1371/journal.pone.0025459) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cavalcanti-Adam E. A., Micoulet A., Blümmel J., Auernheimer J., Kessler H., Spatz J. P. 2006. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur. J. Cell. Biol. 85, 219–224 10.1016/j.ejcb.2005.09.011 (doi:10.1016/j.ejcb.2005.09.011) [DOI] [PubMed] [Google Scholar]
- 42.Beer J. H., Springer K. T., Coller B. S. 1992. Immobilized Arg-Gly-Asp (RGD) peptides of varying lengths as structural probes of the platelet glycoprotein IIb/IIIa receptor. Blood. 79, 117–128 [PubMed] [Google Scholar]
- 43.Kuhlman W., Taniguchi I., Griffith L. G., Mayes A. M. 2007. Interplay between PEO tether length and ligand spacing governs cell spreading on RGD-modified PMMA-g-PEO comb copolymers. Biomacromolecules 8, 3206–3213 10.1021/bm070237o (doi:10.1021/bm070237o) [DOI] [PubMed] [Google Scholar]
- 44.Causa F., Battista E., Della Moglie R., Guarnieri D., Iannone M., Netti P. A. 2010. Surface investigation on biomimetic materials to control cell adhesion: the case of RGD conjugation on PCL. Langmuir 26, 9875–9884 10.1021/la100207q (doi:10.1021/la100207q) [DOI] [PubMed] [Google Scholar]
- 45.Chen C. S., Alonso J. L., Ostuni E., Whitesides G. M., Ingber D. E. 2003. Cell shape provides global control of focal adhesion assembly. Biochem. Biophys. Res. Commun. 307, 355–361 10.1016/S0006-291X(03)01165-3 (doi:10.1016/S0006-291X(03)01165-3) [DOI] [PubMed] [Google Scholar]
- 46.Xia N., Thodeti C. K., Hunt T. P., Xu Q., Ho M., Whitesides G. M., Westervelt R., Ingber D. E. 2008. Directional control of cell motility through focal adhesion positioning and spatial control of Rac activation. FASEB J. 22, 1649–1659 10.1096/fj.07-090571 (doi:10.1096/fj.07-090571) [DOI] [PubMed] [Google Scholar]
- 47.Ventre M., Valle F., Bianchi M., Biscarini F., Netti P. A. 2012. Cell fluidics: producing cellular streams on micropatterned synthetic surfaces. Langmuir 28, 714–721 10.1021/la204144k (doi:10.1021/la204144k) [DOI] [PubMed] [Google Scholar]
- 48.Chen C. S., Mrksich M., Huang S., Whitesides G. M., Ingber D. E. 1997. Geometric control of cell life and death. Science 276, 1425–1428 10.1126/science.276.5317.1425 (doi:10.1126/science.276.5317.1425) [DOI] [PubMed] [Google Scholar]
- 49.McBeath R., Pirone D. M., Nelson C. M., Bhadriraju K., Chen C. S. 2004. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev. Cell 6, 483–495 10.1016/S1534-5807(04)00075-9 (doi:10.1016/S1534-5807(04)00075-9) [DOI] [PubMed] [Google Scholar]
- 50.Peng R., Yao X., Ding J. 2011. Effect of cell anisotropy on differentiation of stem cells on micropatterned surfaces through the controlled single cell adhesion. Biomaterials 32, 8048–8057 10.1016/j.biomaterials.2011.07.035 (doi:10.1016/j.biomaterials.2011.07.035) [DOI] [PubMed] [Google Scholar]
- 51.Kilian K. A., Bugarija B., Lahn B. T., Mrksich M. 2010. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl Acad. Sci. USA 107, 4872–4877 10.1073/pnas.0903269107 (doi:10.1073/pnas.0903269107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith J. T., Elkin J. T., Reichert W. M. 2006. Directed cell migration on fibronectin gradients: effect of gradient slope. Exp. Cell Res. 312, 2424–2432 10.1016/j.yexcr.2006.04.005 (doi:10.1016/j.yexcr.2006.04.005) [DOI] [PubMed] [Google Scholar]
- 53.Acharya A. P., Dolgova N. V., Moore N. M., Xia C. Q., Clare-Salzler M. J., Becker M. L., Gallant N. D., Keselowsky B. G. 2010. The modulation of dendritic cell integrin binding and activation by RGD-peptide density gradient substrates. Biomaterials 31, 7444–7454 10.1016/j.biomaterials.2010.06.025 (doi:10.1016/j.biomaterials.2010.06.025) [DOI] [PubMed] [Google Scholar]
- 54.Chaw K. C., Manimaran M., Tay F. E., Swaminathan S. 2006. A quantitative observation and imaging of single tumor cell migration and deformation using a multi-gap microfluidic device representing the blood vessel. Microvasc. Res. 72, 153–160 10.1016/j.mvr.2006.06.003 (doi:10.1016/j.mvr.2006.06.003) [DOI] [PubMed] [Google Scholar]
- 55.Jeong G. S., Kwon G. H., Kang A. R., Jung B. Y., Park Y., Chung S., Lee S. H. 2011. Microfluidic assay of endothelial cell migration in 3D interpenetrating polymer semi-network HA-collagen hydrogel. Biomed. Microdevices 13, 717–723 10.1007/s10544-011-9541-7 (doi:10.1007/s10544-011-9541-7) [DOI] [PubMed] [Google Scholar]
- 56.Lamb B. M., Park S., Yousaf M. N. 2010. Microfluidic permeation printing of self-assembled monolayer gradients on surfaces for chemoselective ligand immobilization applied to cell adhesion and polarization. Langmuir 26, 12 817–12 823 10.1021/la1022642 (doi:10.1021/la1022642) [DOI] [PubMed] [Google Scholar]
- 57.Guarnieri D., De Capua A., Ventre M., Borzacchiello A., Pedone C., Marasco D., Ruvo M., Netti P. A. 2010. Covalently immobilized RGD gradient on PEG hydrogel scaffold influences cell migration parameters. Acta Biomater. 6, 2532–2539 10.1016/j.actbio.2009.12.050 (doi:10.1016/j.actbio.2009.12.050) [DOI] [PubMed] [Google Scholar]
- 58.DeLong S. A., Gobin A. S., West J. L. 2005. Covalent immobilization of RGDS on hydrogel surfaces to direct cell alignment and migration. J. Control. Release 109, 139–148 10.1016/j.jconrel.2005.09.020 (doi:10.1016/j.jconrel.2005.09.020) [DOI] [PubMed] [Google Scholar]
- 59.Curtis A., Wilkinson C. 1997. Topographical control of cells. Biomaterials 18, 1573–1583 10.1016/S0142-9612(97)00144-0 (doi:10.1016/S0142-9612(97)00144-0) [DOI] [PubMed] [Google Scholar]
- 60.Lim J. Y., Donahue H. J. 2007. Cell sensing and response to micro- and nanostructured surfaces produced by chemical and topographic patterning. Tissue Eng. 13, 1879–1891 10.1089/ten.2006.0154 (doi:10.1089/ten.2006.0154) [DOI] [PubMed] [Google Scholar]
- 61.den Braber E. T., de Ruijter J. E., Ginsel L. A., von Recum A. F., Jansen J. A. 1998. Orientation of ECM protein deposition, fibroblast cytoskeleton, and attachment complex components on silicone microgrooved surfaces. J. Biomed. Mater. Res. 40, 291–300 (doi:10.1002/(SICI)1097-4636(199805)40:2<291::AID-JBM14>3.0.CO;2-P) [DOI] [PubMed] [Google Scholar]
- 62.Yim E. K., Reano R. M., Pang S. W., Yee A. F., Chen C. S., Leong K. W. 2005. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials 26, 5405–5413 10.1016/j.biomaterials.2005.01.058 (10.1016/j.biomaterials.2005.01.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dalby M. J., Biggs M. J., Gadegaard N., Kalna G., Wilkinson C. D., Curtis A. S. 2007. Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. J. Cell. Bichem. 100, 326–338 10.1002/jcb.21058 (doi:10.1002/jcb.21058) [DOI] [PubMed] [Google Scholar]
- 64.Gaubert H. E., Frey W. 2007. Highly parallel fabrication of nanopatterned surfaces with nanoscale orthogonal biofunctionalization imprint lithography. Nanotechnology 18, 135 101–135 107 10.1088/0957-4484/18/13/135101 (doi:10.1088/0957-4484/18/13/135101) [DOI] [PubMed] [Google Scholar]
- 65.Loesberg W. A., te Riet J., van Delft F. C., Schön P., Figdor C. G., Speller S., van Loon J. J., Walboomers X. F., Jansen J. A. 2007. The threshold at which substrate nanogroove dimensions may influence fibroblast alignment and adhesion. Biomaterials 28, 3944–3951 10.1016/j.biomaterials.2007.05.030 (doi:10.1016/j.biomaterials.2007.05.030) [DOI] [PubMed] [Google Scholar]
- 66.Lamers E., van Horssen R., te Riet J., van Delft F. C., Luttge R., Walboomers X. F., Jansen J. A. 2010. The influence of nanoscale topographical cues on initial osteoblast morphology and migration. Eur. Cell Mater. 20, 329–343 [DOI] [PubMed] [Google Scholar]
- 67.Sjöström T., Dalby M. J., Hart A., Tare R., Oreffo R. O., Su B. 2009. Fabrication of pillar-like titania nanostructures on titanium and their interactions with human skeletal stem cells. Acta Biomater. 5, 1433–1441 10.1016/j.actbio.2009.01.007 (doi:10.1016/j.actbio.2009.01.007) [DOI] [PubMed] [Google Scholar]
- 68.Choi C. K., Vicente-Manzanares M., Zareno J., Whitmore L. A., Mogilner A., Horwitz A. R. 2008. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 10.1038/ncb1763 (doi:10.1038/ncb1763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Coussen F., Choquet D., Sheetz M. P., Erickson H. P. 2002. Trimers of the fibronectin cell adhesion domain localize to actin filament bundles and undergo rearward translocation. J. Cell Sci. 115, 2581–2590 [DOI] [PubMed] [Google Scholar]
- 70.Teixeira A. I., Abrams G. A., Bertics P. J., Murphy C. J., Nealey P. F. 2003. Epithelial contact guidance on well-defined micro- and nanostructured substrates. J. Cell Sci. 116, 1881–1892 10.1242/jcs.00383 (doi:10.1242/jcs.00383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fraser S. A., Ting Y. H., Mallon K. S., Wendt A. E., Murphy C. J., Nealey P. F. 2008. Sub-micron and nanoscale feature depth modulates alignment of stromal fibroblasts and corneal epithelial cells in serum-rich and serum-free media. J. Biomed. Mater. Res. A 86, 725–735 10.1002/jbm.a.31519 (doi:10.1002/jbm.a.31519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gallagher G. O., McGhee K. F., Wilkinson C. D. W., Riehle M. O. 2002. Interaction of animal cells with ordered nanotopography. IEEE Trans. Nanobioscience 1, 24–28 10.1109/TNB.2002.806918 (doi:10.1109/TNB.2002.806918) [DOI] [PubMed] [Google Scholar]
- 73.Dalby M. J., Gadegaard N., Riehle M. O., Wilkinson C. D., Curtis A. S. 2004. Investigating filopodia sensing using arrays of defined nano-pits down to 35 nm diameter in size. Int. J. Biochem. Cell Biol. 36, 2005–2015 10.1016/j.biocel.2004.03.001 (doi:10.1016/j.biocel.2004.03.001) [DOI] [PubMed] [Google Scholar]
- 74.Dalby M. J., Gadegaard N., Wilkinson C. D. 2008. The response of fibroblasts to hexagonal nanotopography fabricated by electron beam lithography. J. Biomed. Mater. Res. A 84, 973–999 10.1002/jbm.a.31409 (doi:10.1002/jbm.a.31409) [DOI] [PubMed] [Google Scholar]
- 75.Hart A., Gadegaard N., Wilkinson C. D., Oreffo R. O., Dalby M. J. 2007. Osteoprogenitor response to low-adhesion nanotopographies originally fabricated by electron beam lithography. J. Mater. Sci. Mater. Med. 18, 1211–1218 10.1007/s10856-007-0157-7 (doi:10.1007/s10856-007-0157-7) [DOI] [PubMed] [Google Scholar]
- 76.Teixeira A. I., McKie G. A., Foley J. D., Bertics P. J., Nealey P. F., Murphy C. J. 2006. The effect of environmental factors on the response of human corneal epithelial cells to nanoscale substrate topography. Biomaterials 27, 3945–3954 10.1016/j.biomaterials.2006.01.044 (doi:10.1016/j.biomaterials.2006.01.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fujita S., Ohshima M., Iwata H. 2009. Time-lapse observation of cell alignment on nanogrooved patterns. J. R. Soc. Interface 6(Suppl. 3), S269–S277 10.1098/rsif.2008.0428.focus (doi:10.1098/rsif.2008.0428.focus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Milner K. R., Siedlecki C. A. 2007. Submicron poly(L-lactic acid) pillars affect fibroblast adhesion and proliferation. J. Biomed. Mater. Res. A 82, 80–91 10.1002/jbm.a.31049 (doi:10.1002/jbm.a.31049) [DOI] [PubMed] [Google Scholar]
- 79.Choi C. H., Hagvall S. H., Wu B. M., Dunn J. C., Beygui R. E., Kim C. J. 2007. Cell interaction with three-dimensional sharp-tip nanotopography. Biomaterials 28, 1672–1679 10.1016/j.biomaterials.2006.11.031 (doi:10.1016/j.biomaterials.2006.11.031) [DOI] [PubMed] [Google Scholar]
- 80.Yang J. Y., Ting Y. C., Lai J. Y., Liu H. L., Fang H. W., Tsai W. B. 2009. Quantitative analysis of osteoblast-like cells (MG63) morphology on nanogrooved substrata with various groove and ridge dimensions. J. Biomed. Mater. Res. A 90, 629–640 10.1002/jbm.a.32130 (doi:10.1002/jbm.a.32130) [DOI] [PubMed] [Google Scholar]
- 81.Lamers E., Walboomers X. F., Domanski M., te Riet J., van Delft F. C., Luttge R., Winnubst L. A., Gardeniers H. J., Jansen J. A. 2010. The influence of nanoscale grooved substrates on osteoblast behavior and extracellular matrix deposition. Biomaterials 31, 3307–3316 10.1016/j.biomaterials.2010.01.034 (doi:10.1016/j.biomaterials.2010.01.034) [DOI] [PubMed] [Google Scholar]
- 82.Dalby M. J., Gadegaard N., Tare R., Andar A., Riehle M. O., Herzyk P., Wilkinson C. D., Oreffo R. O. 2007. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 6, 997–1003 10.1038/nmat2013 (doi:10.1038/nmat2013) [DOI] [PubMed] [Google Scholar]
- 83.Yim E. K., Pang S. W., Leong K. W. 2007. Synthetic nanostructures inducing differentiation of human mesenchymal stem cells into neuronal lineage. Exp. Cell Res. 313, 1820–1829 10.1016/j.yexcr.2007.02.031 (doi:10.1016/j.yexcr.2007.02.031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Diehl K. A., Foley J. D., Nealey P. F., Murphy C. J. 2005. Nanoscale topography modulates corneal epithelial cell migration. J. Biomed. Mater. Res. A 75, 603–611 10.1002/jbm.a.30467 (doi:10.1002/jbm.a.30467) [DOI] [PubMed] [Google Scholar]
- 85.Bettinger C. J., Zhang Z., Gerecht S., Borenstein J. T., Langer R. 2008. Enhancement of in vitro capillary tube formation by substrate nanotopography. Adv. Mater. 20, 99–103 10.1002/adma.200702487 (doi:10.1002/adma.200702487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu B., Zhang Q., Lu Q., Xu Y., Yin J., Hu J., Wang Z. 2004. Nanotopographical guidance of C6 glioma cell alignment and oriented growth. Biomaterials 25, 4215–4223 10.1016/j.biomaterials.2003.11.020 (10.1016/j.biomaterials.2003.11.020) [DOI] [PubMed] [Google Scholar]
- 87.Hamilton D. W., Oates C. J., Hasanzadeh A., Mittler S. 2010. Migration of periodontal ligament fibroblasts on nanometric topographical patterns: influence of filopodia and focal adhesions on contact guidance. PLoS ONE 5, e15129. 10.1371/journal.pone.0015129 (doi:10.1371/journal.pone.0015129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ingber D. E. 1997. Tensegrity: the architectural basis of cellular mechanotransduction. Annu. Rev. Physiol. 59, 575–599 10.1146/annurev.physiol.59.1.575 (doi:10.1146/annurev.physiol.59.1.575) [DOI] [PubMed] [Google Scholar]
- 89.Goffin J. M., Pittet P., Csucs G., Lussi J. W., Meister J. J., Hinz B. 2006. Focal adhesion size controls tension-dependent recruitment of alpha-smooth muscle actin to stress fibres. J. Cell Biol. 172, 259–268 10.1083/jcb.200506179 (doi:10.1083/jcb.200506179) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vogel V., Sheetz M. 2006. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7, 265–275 10.1038/nrm1890 (doi:10.1038/nrm1890) [DOI] [PubMed] [Google Scholar]
- 91.Harris A. K., Wild P., Stopak D. 1980. Silicone rubber substrata: a new wrinkle in the study of cell locomotion. Science 208, 177–179 10.1126/science.6987736 (doi:10.1126/science.6987736) [DOI] [PubMed] [Google Scholar]
- 92.Pelham R. J., Jr, Wang Y. 1999. High resolution detection of mechanical forces exerted by locomoting fibroblasts on the substrate. Mol. Biol. Cell 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Balaban N. Q., et al. 2001. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3, 466–472 10.1038/35074532 (doi:10.1038/35074532) [DOI] [PubMed] [Google Scholar]
- 94.du Roure O., Saez A., Buguin A., Austin R. H., Chavrier P., Silberzan P., Ladoux B. 2005. Force mapping in epithelial cell migration. Proc. Natl Acad. Sci. USA 102, 2390–2395 10.1073/pnas.0408482102 (doi:10.1073/pnas.0408482102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Saez A., Buguin A., Silberzan P., Ladoux B. 2005. Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys. J. 89, L52–54 10.1529/biophysj.105.071217 (doi:10.1529/biophysj.105.071217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Georges P. C., Janmey P. A. 2004. Cell type-specific response to growth on soft materials. J. Appl. Physiol. 98, 1547–1553 10.1152/japplphysiol.01121.2004 (doi:10.1152/japplphysiol.01121.2004) [DOI] [PubMed] [Google Scholar]
- 97.Wang H. B., Dembo M., Wang Y. L. 2000. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. Am. J. Physiol. Cell Physiol. 279, C1345–C1350 [DOI] [PubMed] [Google Scholar]
- 98.Lee J. N., Jiang X., Ryan D., Whitesides G. M. 2004. Compatibility of mammalian cells on surfaces of poly(dimethylsiloxane). Langmuir 20, 11684–11691 10.1021/la048562+ (doi:10.1021/la048562+) [DOI] [PubMed] [Google Scholar]
- 99.Peyton S. R., Putnam A. J. 2005. Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion. J. Cell. Physiol. 204, 198–209 10.1002/jcp.20274 (doi:10.1002/jcp.20274) [DOI] [PubMed] [Google Scholar]
- 100.Lo C. M., Wang H. B., Dembo M., Wang Y. L. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79, 144–152 10.1016/S0006-3495(00)76279-5 (doi:10.1016/S0006-3495(00)76279-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gray S. D., Tien J., Chen C. S. 2003. Repositioning of cells by mechanotaxis on surfaces with micropatterned Young's modulus. J. Biomed. Mater. Res. A 66A, 605–614 10.1002/jbm.a.10585 (doi:10.1002/jbm.a.10585) [DOI] [PubMed] [Google Scholar]
- 102.Cortese B., Gigli G., Riehle M. 2009. Mechanical gradient cues for guided cell motility and control of cell behavior on uniform substrates. Adv. Funct. Mater. 19, 2961–2968 10.1002/adfm.200900918 (doi:10.1002/adfm.200900918) [DOI] [Google Scholar]
- 103.Diez M., et al. 2011. Molding micropatterns of elasticity on PEG-based hydrogels to control cell adhesion and migration. Adv. Eng. Mater. 13, B395–B404 10.1002/adem.201080122 (doi:10.1002/adem.201080122) [DOI] [Google Scholar]
- 104.Engler A. J., Sen S., Sweeney H. L., Discher D. E. 2006. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 10.1016/j.cell.2006.06.044 (doi:10.1016/j.cell.2006.06.044) [DOI] [PubMed] [Google Scholar]
- 105.Winer J. P., Janmey P. A., McCormick M. E., Funaki M. 2009. Bone marrow-derived human mesenchymal stem cells become quiescent on soft substrates but remain responsive to chemical or mechanical stimuli. Tissue Eng. Part A. 15, 147–154 10.1089/ten.tea.2007.0388 (doi:10.1089/ten.tea.2007.0388) [DOI] [PubMed] [Google Scholar]
- 106.Tzvetkova-Chevolleau T., Stéphanou A., Fuard D., Ohayon J., Schiavone P., Tracqui P. 2008. The motility of normal and cancer cells in response to the combined influence of the substrate rigidity and anisotropic microstructure. Biomaterials 29, 1541–1551 10.1016/j.biomaterials.2007.12.016 (doi:10.1016/j.biomaterials.2007.12.016) [DOI] [PubMed] [Google Scholar]
- 107.Le Saux G., Magenau A., Böcking T., Gaus K., Gooding J. J. 2011. The relative importance of topography and RGD ligand density for endothelial cell adhesion. PLoS ONE 6, e21869. 10.1371/journal.pone.0021869 (doi:10.1371/journal.pone.0021869) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mitra S. K., Hanson D. A., Schlaepfer D. D. 2005. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 10.1038/nrm1549 (doi:10.1038/nrm1549) [DOI] [PubMed] [Google Scholar]
- 109.Ge C., Xiao G., Jiang D., Franceschi R. T. 2007. Critical role of the extracellular signal-regulated kinase-MAPK pathway in osteoblast differentiation and skeletal development. J. Cell Biol. 176, 709–718 10.1083/jcb.200610046 (doi:10.1083/jcb.200610046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Biggs M. J., Richards R. G., Gadegaard N., Wilkinson C. D., Oreffo R. O., Dalby M. J. 2009. The use of nanoscale topography to modulate the dynamics of adhesion formation in primary osteoblasts and ERK/MAPK signalling in STRO-1+ enriched skeletal stem cells. Biomaterials 30, 5094–5103 10.1016/j.biomaterials.2009.05.049 (doi:10.1016/j.biomaterials.2009.05.049) [DOI] [PubMed] [Google Scholar]
- 111.Biggs M. J., Richards R. G., Gadegaard N., McMurray R. J., Affrossman S., Wilkinson C. D., Oreffo R. O., Dalby M. J. 2008. Interactions with nanoscale topography: adhesion quantification and signal transduction in cells of osteogenic and multipotent lineage. J. Biomed. Mater. Res. A. 91, 195–208 10.1002/jbm.a.32196 (doi:10.1002/jbm.a.32196) [DOI] [PubMed] [Google Scholar]
- 112.Kantawong F., Burgess K. E., Jayawardena K., Hart A., Burchmore R. J., Gadegaard N., Oreffo R. O., Dalby M. J. 2009. Whole proteome analysis of osteoprogenitor differentiation induced by disordered nanotopography and mediated by ERK signalling. Biomaterials 30, 4723–4731 10.1016/j.biomaterials.2009.05.040 (doi:10.1016/j.biomaterials.2009.05.040) [DOI] [PubMed] [Google Scholar]
- 113.Xu R., Boudreau A., Bissell M. J. 2009. Tissue architecture and function: dynamic reciprocity via extra- and intra-cellular matrices. Cancer Metastasis Rev. 28, 167–176 10.1007/s10555-008-9178-z (doi:10.1007/s10555-008-9178-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Place E. S., Evans N. D., Stevens M. M. 2009. Complexity in biomaterials for tissue engineering. Nat. Mater. 8, 457–470 10.1038/nmat2441 (doi:10.1038/nmat2441) [DOI] [PubMed] [Google Scholar]
- 115.Lutolf M. P., Blau H. M. 2009. Artificial stem cell niches. Adv. Mater. 21, 3255–3268 10.1002/adma.200802582 (doi:10.1002/adma.200802582) [DOI] [PMC free article] [PubMed] [Google Scholar]