Abstract
As neurons age, their survival depends on eliminating a growing burden of damaged, potentially toxic proteins and organelles—a capability that declines owing to aging and disease factors. Here, we review the two proteolytic systems principally responsible for protein quality control in neurons and their important contributions to Alzheimer disease pathogenesis. In the first section, the discovery of paired helical filament ubiquitination is described as a backdrop for discussing the importance of the ubiquitin–proteasome system in Alzheimer disease. In the second section, we review the prominent involvement of the lysosomal system beginning with pathological endosomal–lysosomal activation and signaling at the very earliest stages of Alzheimer disease followed by the progressive failure of autophagy. These abnormalities, which result in part from Alzheimer-related genes acting directly on these lysosomal pathways, contribute to the development of each of the Alzheimer neuropathological hallmarks and represent a promising therapeutic target.
Neurons rely on the proteasome and lysosomal systems to remove damaged proteins and organelles. Their disruption leads to the accumulation of toxic proteins—including amyloid-β and tau.
Cellular aging, the sine qua non for Alzheimer disease development, is associated with cumulative oxidative damage to proteins and membranes, translational errors leading to the synthesis of defective proteins, and various genetic and environmental insults to organelles and proteins (Terman 2001; Roy et al. 2002; Sohal et al. 2002; Troen 2003; Levine et al. 2004). In some aging-related neurodegenerative diseases, mutations of the pathogenic gene(s) may also cause anomalous conformations of the encoded protein or its metabolites, leading to increased proteolytic resistance and accumulation of these potentially toxic proteins (Soto 2003). A neuron’s ability to function in the face of these mounting aging- and disease-related insults is determined in significant part by the efficiency with which it can eliminate this burden of damaged cellular constituents. An array of proteases regulates cell function, but two proteolytic systems are mainly responsible for the turnover of proteins and organelles—the proteasome and lysosomal systems (Grune et al. 2001; Berke et al. 2003; Goldberg 2003; Levine et al. 2004). The proteasome selectively degrades normal proteins (mainly those with short half-lives) and abnormal proteins, which are earmarked for elimination by a process involving their conjugation to ubiquitin (Ub; Goldberg 2003). The lysosomal system, and specifically the autophagic pathway, is the principal mechanism for degrading proteins with long half-lives and is the only system in cells for degrading organelles and large protein aggregates or inclusions. A second route to lysosomes, the endocytic pathway, delivers extracellular material and plasma membrane constituents to lysosomes under the direction of specific targeting signals (Nixon 2004).
Not surprisingly, protein quality control and proteostasis—the appropriate balance of protein synthesis, folding, and turnover in the cell—involve interdependent regulation of these two proteolytic systems. This interdependence is best exemplified by the regulatory influences of the protein ubiquitin. It has recently become appreciated that ubiquitination of proteins by covalent modification tags them for elimination not only through the proteasome (the ubiquitin–proteasome system or UPS) but also through the lysosomal system. Endocytosed receptors are targeted for lysosomal degradation, in part, by ubiquination in late endosomes/multivesicular bodies (MVBs). Recently, protein aggregates and certain organelles have been shown to be tagged with ubiquination for selective removal by autophagy (Narendra et al. 2009; Dikic et al. 2010; Youle et al. 2011), a degradative process previously believed to be only nonselective. In selective autophagy, an adaptor molecule with a ubiquitin-binding domain engages the ubiquitinated structure and couples it to the pre-autophagosomal isolation membrane for subsequent sequestration. The exact types of ubiquitin motifs recognized by the UPS and autophagy may differ and the degree to which ubiquitination drives autophagic protein turnover relative to that by proteasomes is still unclear. Nevertheless, the role of ubiquitin as a tag for targeting substrates for elimination is more universal than earlier imagined, implying that changes in ubiquitin balance may be one way that disease-related perturbations of one proteolytic system are likely to influence the other.
Interdependence of the proteasome and lysosomal system is also suggested by observations that, when proteasome activity is inhibited, proteins accumulate that become substrates for autophagy (Fortun et al. 2003). Inhibiting autophagy by genetically deleting components of the sequestration machinery causes ubiquitinated protein aggregates to appear in neurons, reflecting additional negative effects on the UPS (Korolchuk et al. 2009a,b). Besides ubiquitin, proteins identified to play regulatory roles in the UPS and autophagy are being increasingly identified (Zhao et al. 2007). For example, p62, an adaptor protein for autophagy, also influences proteasomal degradation, whereas VCP/p97 acting through p62 and ubiquitin regulates both the proteasome-dependent endoplasmic reticulum–associated degradation (ERAD) pathway and aspects of autophagosome maturation (Tresse et al. 2010). The E3 ligase Parkin, a protein implicated in Parkinson's disease, creates an autophagy signal on mitochondria and also tags proteins elsewhere for proteasomal degradation (Yoshii et al. 2011). Interestingly, proteasomal subunits may be degraded by lysosomes (Cuervo et al. 1995), hinting at an additional level of cross talk between these proteolytic systems.
Proteolysis has been an active area of Alzheimer disease research over the past two decades mainly in relation to the processing of specific proteins, like the β-amyloid precursor protein. A broader appreciation is emerging, however, of the roles of proteolytic systems in other crucial aspects of Alzheimer disease (AD) pathogenesis related to protein clearance, neural plasticity, and neurodegenerative mechanisms. In this article, we focus on the involvement of the UPS and the lysosomal system (endosomal–lysosomal pathway and autophagy) in AD, their contributions to disease development and progression, and the possibilities for targeting these systems in the design of innovative therapies for AD.
PART I: THE UBIQUITIN–PROTEASOME SYSTEM IN AD
Discovery of Ubiquitin in Paired Helical Filaments: A Personal Retrospective by Yasuo Ihara
Polyclonal antibodies to the classical paired helical filaments (PHFs) found in the neurofibrillary tangles and dystrophic neurites of AD were first raised in about 1982, allowing exploration of the component(s) of PHFs using immunochemical approaches (Ihara et al. 1983). This strategy was useful because PHFs were resistant to conventional protease digestion, and it was difficult to obtain reproducible and distinct limited-digest protein fragments of PHFs for sequencing. Sodium dodecyl sulfate (SDS)-insoluble material was extracted from typical AD brains, and PHFs were partially purified from the SDS-insoluble material by differential centrifugation and sucrose density gradient centrifugation (Ihara et al. 1983). The PHFs obtained by these procedures were not yet sufficiently pure, as shown later by sequencing of fragments of PHF proteins (Kondo et al. 1988). The polyclonal PHF antisera that were initially raised also reacted with lipofuscin-like granules and other SDS-insoluble materials in the PHF-rich immunogen. A major frustration with the antisera was that, although they intensely stained neurofibrillary tangles (NFTs), dystrophic neurites, and neuropil threads in AD brain sections, they did not label a distinct band(s) on immunoblots of AD brain homogenates, instead giving peculiar smears running from very high to low molecular mass (Ihara et al. 1983). These immunoreactive smears were highly characteristic of AD brain homogenates and never seen in control brain homogenates. Thus, the indirect immunochemical approach to identify the PHF component proteins seemed unsuccessful at first. However, it succeeded a few years later when fetal or neonatal brain homogenates were probed with anti-PHF antisera: A somewhat diffuse band around 50 kDa was intensely labeled in immunoblots of fetal brain homogenates. This protein reactive with the initial anti-PHF sera was soon identified as tau, a microtubule-associated protein (MAP), based on its molecular weight, isoform change during development, microtubule-binding activity, and heat stability (Kosik et al. 1986; Nukina and Ihara 1986; see also Brion et al. 1985; Grundke-Iqbal et al. 1986; Wood et al. 1986; discovery of tau in PHF is reviewed in Mandelkow and Mandelkow 2011).
In contrast to our initial PHF antisera, we assumed that a monoclonal antibody that recognizes a highly defined epitope on PHF would not give rise to the smearing pattern on immunoblots that greatly puzzled us. In 1985, we became aware of a hybrid system (Pike et al. 1982) for raising monoclonal antibodies to glycolipids. Lewis rats responded well to immunizations with partially purified PHF preparations, yielding high titers of antisera to PHFs. These rats provided splenocytes for fusion with mouse myeloma cells. We (Hiroshi Mori and Yasuo Ihara) employed a two-step screening procedure to identify monoclonals in the resultant hybridoma supernatants: first by enzyme-linked immunosorbent assay (ELISA) using plates coated with partially purified PHFs, and second by immunostaining of NFTs that were prepared from AD brains under nondenaturing conditions. Only two confirmed and stable cell lines emerged, the antibodies from which had very similar specificities on the blots: Besides the characteristic “PHF smear,” they also labeled a very small protein of ∼8 kDa. Hiroshi Mori named these monoclonal antibodies DF (Dementia Filament) 1 and 2, of which the latter (DF2) was used for subsequent characterization (Mori et al. 1987).
In contrast to our PHF antisera, DF2 produced immunoreactive smears even in control brain homogenates (presumably representing Ub-conjugated proteins). In addition, DF2 gave a distinct band at 8 kDa on blots of aged human brain homogenates, whereas the PHF antisera had not given distinct bands. We were puzzled as to why the two antibodies labeled different bands, but hypothesized that they recognized different components of PHFs. Optical diffraction analyses carried out by Wischik et al. (1985) had suggested that the core of the PHF may be composed of a subunit around ∼100 kDa. We reasoned that one protein might make up the core framework of PHFs, whereas another might occur on a peripheral portion.
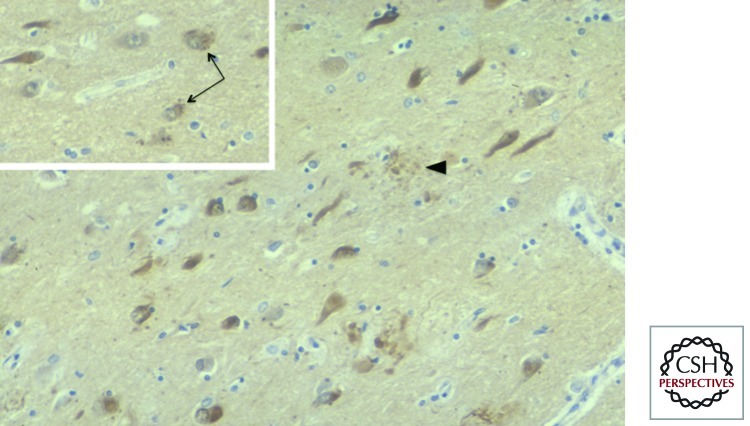
In AD cortical sections, we observed that NFTs and dystrophic neurites (Fig. 1) and, unexpectedly, granulovacuolar changes (Fig. 1, inset) were intensely immunolabeled by the DF2 monoclonal. When mild fixation conditions were used, innumerable neuropil threads were also detected. (One peculiar characteristic was an apparent “background” staining of AD brain sections, whereas widely used Ub polyclonal antibodies [Haas and Bright 1985] gave almost no background staining. This discrepancy was resolved in 1996: DF2 is specific for the conjugated form of Ub, whereas the Ub polyclonal antibodies are specific for free Ub [Morimoto et al. 1996].)
Figure 1.
DF2 immunostaining of a tissue section from Alzheimer disease (AD) hippocampus. Neurofibrillary tangles and dystrophic neurites (arrowhead) are intensely labeled, whereas the neuropil (background) is uniformly stained, simulating nonspecific background staining. However, this was found to be true staining of ubiquitin-conjugated proteins in the neuropil (Morimoto et al. 1996). (Inset) Some neurons in CA1 undergo granulovacuolar changes (arrows) and are also intensely stained with DF2.
What was the small protein at 8 kDa that was strongly labeled by DF2? Purifying the protein was not difficult with high-performance liquid chromatography (HPLC), which was beginning to be used for protein fractionation. The sequence of the DF2-reactive protein up to approximately 30 residues exactly corresponded to that of ubiquitin (Mori et al. 1987). This was probably the first description of Ub protein modification in the field of neuropathology. Two seminal papers describing the significance of protein ubiquitination had recently been published by the Varshavsky laboratory (Ciechanover et al. 1984; Finley et al. 1984). Despite reading these papers, we could not understand why Ub is present on PHFs. Our understanding was that Ub is a tag for protein degradation: A Ub-tagged protein should be recognized and degraded by the proteasome, an unusually large cytoplasmic protease. However, Ub was found to have other functional roles; for example, a proportion of histone (H)2B is ubiquitinated in a way that has a role in the transcription.
Ubiquitin appeared to represent a novel constituent of PHFs; DF2 strongly stained SDS-stripped NFTs, suggesting that Ub was an integral component. Nevertheless, we felt that the immunochemical identification of the component was not sufficient to fully confirm the presence of the molecule on PHFs. At the time, there was some confusion about the PHF protein constituents. Using well-characterized antibodies to various MAPs as well as PHF polyclonal antibodies, tau had recently been established as a major component of PHFs (see above and Mandelkow and Mandelkow 2011). However, monoclonal antibodies to neurofilament (NF) proteins also strongly immunolabeled PHF (Anderton et al. 1982), and these were widely applied to AD brain sections. This confusion about PHF components was clarified when Nobuyuki Nukina and coworkers found significant cross-reactivities of certain NF monoclonal antibodies with phosphorylated epitopes of tau (Nukina et al. 1987). In light of such observations, it became essential to show unambiguously that Ub was a component of PHF. In 1987, we finally obtained definitive evidence using protein chemical techniques that Ub was integral to the PHF (Mori et al. 1987).
Ubiquitinated Tau in PHFs: K48-Linked Mono- and Polyubiquitin Modification
In the mid 1980s, the distinction between mono- and polyubiquitination was not yet clear, and the role of the multiubiquitin chain as a strong degradative signal was just emerging. A fraction of H2B was shown to be monoubiquitinated, and lymphocyte homing receptor was similarly found to be ubiqutinated, but the significance of monoubiquitination was entirely unclear. In our research on Ub in PHFs, we were unable to identify the Ub-conjugated protein by direct sequencing. It was most likely to be tau, but we had no definitive evidence. In the period 1989–1992, the Ihara laboratory was mostly involved in determining the phosphorylation sites on tau proteins in PHFs. We became aware that ion spray mass spectroscopy (MS) was an excellent method for determining phosphorylation sites on PHF-tau (Hasegawa et al. 1992). This provided an opportunity to reanalyze purified PHFs from AD cortex and identify the Ub-targeted protein.
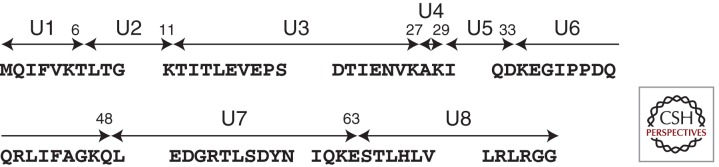
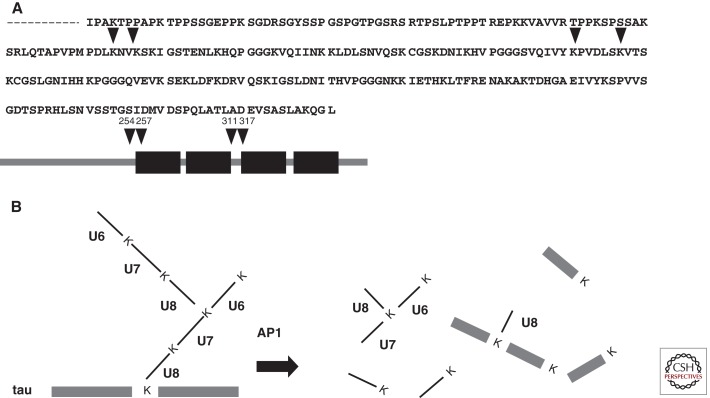
We had previously raised antibodies to the carboxy-terminal region (Gly-76) of Ub, which was predicted to be conjugated with ε-amino groups of lysines of other proteins targeted for proteasomal degradation. However, despite repeated trials using these antibodies, we were unable to identify a particular HPLC peak containing the Ub carboxyl terminus. In our hands, Ub reactivity was detected exclusively in the insoluble AD brain fractions displaying so-called “PHF smears” on immunoblots. However, Ub itself was not responsible for this smear pattern, because we also observed Ub-negative/tau-positive smears, in addition to Ub-positive/tau-positive smears (Morishima-Kawashima et al. 1993). The guanidine HCl-solubilized, Ub- and tau-reactive material purified from AD cortex by HPLC was subjected to peptide mapping using Achromobacter lyticus protease I (AP1), which is highly specific for Lys-X bonds. This yielded peaks corresponding to API-generated fragments of Ub (Fig. 2), in addition to carboxy-terminal fragments of tau, whereas virtually no fragments derived from the amino-terminal portion of tau were recovered. As expected, fragments U1–U7 (see Fig. 2) were found in our digests, but U8 (the carboxy-terminal portion of Ub) was undetectable at the expected position of the nonconjugated U8. Instead, we observed several unusual HPLC peaks that neither corresponded to U1–U8 nor occurred in the digests of Ub-negative/tau-positive smears. By protein sequencing and MS, we identified four Ub-conjugated sites on tau (Fig. 3) and further identified K48-linked Ub chains (Morishima-Kawashima et al. 1993). Crude estimates suggested that most of the products were derived from monoubiquitination and only a minority from polyubiquitination.
Figure 2.
Schematic illustration of 76-residue ubiquitin. Ubiquitin has seven lysine residues and Achromobacter lyticus protease 1 (AP1) cleaves ubiquitin into eight fragments, U1–U8. G76 is conjugated with ε-amino groups of lysine in the target protein. K48-linked ubiquitin chain is a strong degradation signal. Besides this, K6-, 11-, 27-, 29-, and 63-linked multiubiquitin chains are found in the cell (Xu and Peng 2006). It is likely that each type of chain has a distinct role in the cellular metabolism.
Figure 3.
Ubiquitination sites on amino terminally processed tau. As shown in A, four ubiquitin conjugation sites were identified. These are located close to and in the microtubule-binding domain, K254, 257, 311, and 317 according to the numbering of the 441 amino acid human tau isoform. A size exclusion–purified paired helical filament PHF smear was further purified by reverse-phase HPLC, giving three broad (overlapped) peaks that existed only in smear fractions from AD brain. The last-eluting fraction contained a ubiquitin (Ub)-positive, tau-positive smear, which was subjected to AP1 digestion and reverse HPLC fractionation, followed by amino acid sequencing and mass spectrometric analysis. Several late-eluting peaks were found to contain branched fragments, mostly consisting of tau fragments and U8; a minority of U8 conjugated with Lys-48 of U6–U7, derived from the polyubiquitin chain (Morishima-Kawashima et al. 1993). Dysfunction of autophagic and endocytic pathways to lysosomes driven by relevant genes and other risk factors in Alzheimer disease (expressed on the left side of the diagram) causes or promotes pathophysiology critical to the development and progression of the disease (outlined on the right side of the diagram). See the text for further details.
These Ub- and tau-reactive smears were recovered exclusively from the buffer-insoluble fraction of AD cortex, suggesting that ubiquitination might occur after tau deposits into aggregates. At that time, a simple model showed that cells conjugated Ub to abnormal proteins that were to be selectively degraded by the proteasome. If this was correct, minute amounts of abnormal PHF proteins tagged by Ub and targeted for the proteasome might be observed in the buffer-soluble fraction of cytoplasm, but we were unable to confirm this. However, recent work by Cripps et al. (2006) successfully used MC1 (a monoclonal specific for an abnormal PHF-like conformation of tau) to affinity purify soluble full-length tau from PHF-rich extracts of AD cortex and subject it to liquid chromatography–tandem MS (LC–MS/MS) analysis. The presence of K6, K11, and K48- linked polyubiquitinations—in addition to monoubiquitination—was observed. Among these polyubiquitination sites, the K48-linked site was predominant. It is thus possible that a portion of MC1-reactive PHF-tau is in the soluble fraction and exists as free molecules in the cytoplasm, but this has not yet been confirmed.
Interestingly, immunocytochemical staining suggests that Ub exists in the mid-portion of neuropil threads (the largest source of PHF in the AD brain), whereas both ends of the threads are reactive for only tau, not Ub (Iwatsubo et al. 1992). This finding suggests that neuropil threads may extend at both ends: Tau may aggregate and deposit first, followed by its ubiquitination.
Ubiquitination of Other Neuropathological Inclusions
Following the discoveries about Ub and PHFs reviewed above, a variety of types of Ub–protein conjugates were detected in other neurodegenerative diseases. Although such findings were initially based solely on immunocytochemical evidence, we and others postulated that ubiquitination may be widely involved in the degradation of cytoplasmic proteins accumulating during aging and in a number of neurodegenerative diseases. Therefore, ubiquitinated inclusions may represent a failure of degradation by the Ub–proteasome system of neurons or glia. Early on, it was thought that Ub was associated with intermediate (10-nm) filaments in cells, because many inclusions seemed to consist of “altered” intermediate filaments (Lowe et al. 1988).
With the knowledge that DF2 recognizes Ub, we investigated whether DF2 stains other types of known intracellular inclusions. In the cortex, DF2 revealed fine dot-like stainings that had not been described before, and larger dot-like stainings were seen in the white matter, possibly derived from multivesicular and lysosome-related bodies. In addition, DF2 intensely labeled the classical granulovacuolar changes in the hippocampus in AD and other neurodegenerative disorders (Fig. 1, inset). We soon found that DF2 strongly stained cortical and brain stem Lewy bodies in brain sections from “diffuse Lewy body disease” (Kuzuhara et al. 1988), as originally described by Kenji Kosaka (1978), who proposed that some elderly subjects dying with dementia had many cortical Lewy bodies. In this dementia, Lewy bodies are abundant in cortical neurons, especially in the cingulate gyrus, in addition to their presence in the substantia nigra and locus ceruleus, their prototypical loci in Parkinson’s disease. The eosinophilic round bodies with clear margins in brainstem neurons and the somewhat indistinct, smaller round bodies in cortical neurons are sometimes difficult to recognize with conventional hematoxylin–eosin stains, but anti-Ub antibodies such as DF2 labeled them well. Furthermore, DF2 revealed a possible evolution of Lewy bodies: Diffuse cytoplasmic accumulation of Ub-positive material might be followed by its gradual coalescence into an amorphous perinuclear inclusion (Kuzuhara et al. 1988). DF2 and other Ub antibodies also labeled elongated neurites (now known as Lewy neurites) in and near the sites of Lewy bodies. Because Lewy bodies did not stain for tau, the target protein of ubiquitination had to be distinct. In 1997, α-synuclein was shown to be the principal component of Lewy bodies (Spillantini et al. 1997). The accumulated α-synuclein was then shown to be ubiquitinated (Hasegawa et al. 2002).
The DF2 immunoreactivity of Lewy bodies led us to search for similar DF2-positive inclusions, and we found that Lewy-like bodies in motor neurons in amyotrophic lateral sclerosis (ALS; Murayama et al. 1990a) and Pick bodies in Pick's disease (Murayama et al. 1990b) were strongly reactive; the latter stained also for tau, whereas the former stained neither for tau nor α-synuclein. In this regard, the entity of frontotemporal lobar degeneration (FTLD) with Ub-positive/tau-negative inclusions was described later, and TDP43 was identified as the ubiquitinated protein in both this disorder and ALS (Neuman et al. 2006). It is noteworthy that the specific protein targets of ubiqutination that accumulate in insoluble deposits in distinct disorders have apparent major roles in the respective neurodegenerative processes: In AD, tau is ubiquitinated, in Parkinson's disease and dementia with Lewy bodies, it is α-synuclein, and in ALS and FTLD-U, it is TDP-43.
Significance of Ubiquitinated Neurofibrillary Tangles
The discovery of tau ubiqutination in PHFs suggested that living (but presumably injured) neurons may mount an attempt to remove the abnormal inclusions (NFTs) by Ub tagging, although this apparently fails to efficiently remove the tangles. The UPS was believed to be involved principally in the turnover of short-lived proteins (cytosolic, nuclear, and endoplasmic reticulum [ER]), and especially tightly regulated transcription factors; for this process, the proteolytic system requires adenosine triphosphate (ATP). There was initially a view that long-lived proteins, including tau, may be degraded principally by lysosomes, the other major protein degradation machinery of the cell.
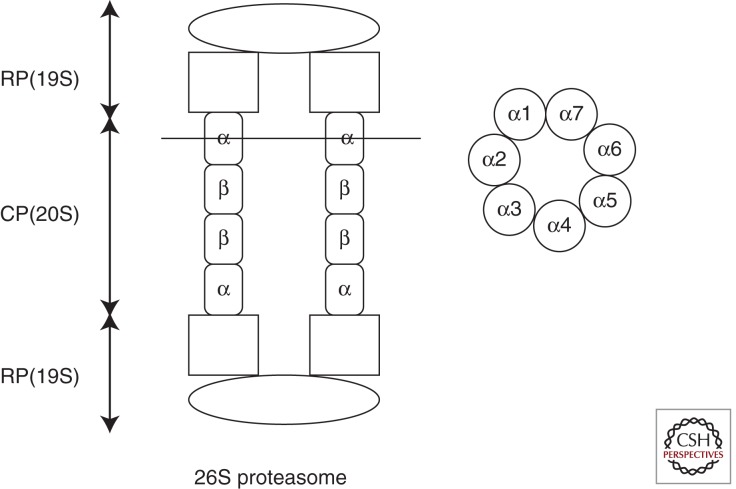
Ubiquitination occurs in three steps. The first step is activation of the carboxyl terminus of Ub by an E1 protein (i.e., a Ub-activating enzyme), which consumes ATP. Second, there is conjugation of the ATP-activated Ub to an E2 protein (i.e., a Ub-conjugating enzyme). Third, there is ligation of Ub with an ε-amino group of lysine in the target protein by an E3 ligase. E3 ligase binds both the target protein and the E2–Ub complex; thousands of substrate-specific E3s ensure selective protein tagging and degradation. An E4 enzyme catalyzes the polyubiquitination of the target substrate that is bound to the E2–E3 complexes. Proteins tagged with chains of four or more K48-linked multiubiquitins provide the strongest signal for degradation by the 26S proteasome, because a chain of at least four Ub moieties is required for substrate recognition by the 26S proteasome complex. Ubiquitinated target proteins bind to the 19S cap (RP), which has a Ub binding site and ATPase activity, and this leads to cleavage of Ub moieties from the target by deubiquitinating enzymes, unfolds the polypeptides and sends them to the narrow channel of the 20S core particle. The narrow canal of 20S proteasome (∼700 kDa) consists of heptameric stacks of four rings, with the inner two being composed of β subunits and the outer two of α subunits (Gallastegui and Groll 2010). Thus, the general structure of the 20S core particle is α1–7β1–7β1–7α1–7 (Fig. 4). These subunits have activities as trypsin-like, chymotrypsin-like, and peptidyl–glutaminyl hydrolyzing activities. The catalytic portions face the interior surface; during its passage, the target protein is processed to short peptides, mostly between six and 10 amino acids. Deubiquitinating activity attached to the 19S cap is essential for complete degradation; if this is blocked, degradation is inhibited, and ubiquitinated substrates accumulate in the cytoplasm.
Figure 4.
Schematic illustration of 26S proteasome. It consists of 19S regulatory particle (RP) and 20S core particle (CP). The narrow canal of CP consists of heptameric stacks of four rings, with the inner two being composed of β subunits and the outer two of α subunits. The image on the right shows a cross section at an indicated line (left). These subunits have trypsin-like, chymotrypsin-like, and peptidyl glutaminyl hydrolyzing activities. (Modified from Gallastegui and Groll 2010.)
The 26S proteasome is primarily a cytosolic enzyme, but it is also found in ER membranes and localized to nuclei. The former activity is known as ER-associated degradation. The latter (nuclear) activity is thought to be involved in the development of the ubiquitinated nuclear inclusions found in Huntington's disease and some types of spinocerebellar ataxia.
The question arises whether a fundamental impairment of proteasome function is involved in the formation of intracellular aggregates, in particular the NFT of AD. AD is the most common of numerous age-associated brain diseases, and the activity of brain proteasomes appears to decline with age (Keller et al. 2002). PHF have been associated with inhibition of the activity of the proteasome in a brain region–specific manner (Keller et al. 2000). That is, in areas where NFTs formed abundantly, including hippocampus and parahippocampal gyrus and superior and middle temporal gyri, proteasome activity (as assessed by chymortrypsin-like and postglutamyl peptidases) appeared to be most affected, whereas occipital gyri and cerebellum, which often have few or no NFTs, were least affected (Keller et al. 2000). Despite the decreased proteasomal enzymatic activity observed in AD brains, the levels of the α and β subunits were not decreased. This suggests that posttranslational modifications could cause the decreases in the activities of proteasome. Another study of AD brain tissues showed that hyperphosphorylated tau was bound to the proteasome, presumably to the 19S cap portion, and the more tau that was bound, the more that proteasomes appeared to be inhibited (Keck et al. 2003). In vitro experiments further showed that aggregated (recombinant) tau—but not nonaggregated (monomeric) tau—can inhibit the proteasome activity. The hyperphosphorylation of tau may not be related to suppression of proteasomes. Based on these various findings, it has been speculated that small aggregates of PHFs may bind to the cap portion of the 26S proteasome and inhibit its activity. This hypothesis is consistent with related observations made in other cell model systems (Bence et al. 2001). Taken together, the available evidence suggests that PHF-bearing neurons have defective activity of some of their proteasomes, which may contribute to the neurodegeneration. On the other hand, it seems unlikely that the decreasing activity of the proteasome leads to NFT formation. Aggregate formation itself may be a protective event in AD neurons and help extend their life spans, as compared with neurons not bearing NFTs (Gomez-Isla et al. 1997).
The currently prevailing view of the temporal involvement of Ub in PHF evolution is that the aggregation of hyperphosphorylated tau is followed by ubiquitination. This is consistent with previous immunocytochemical observations (Bancher et al. 1991) and also with confocal microscopy reconstructions, suggesting that, in the neuropil threads of AD cortex, both growing ends of the threads contain full-length tau, whereas Ub is present at the mid portion, where the amino-terminal region of tau may have been processed (Iwatsubo et al. 1992). Similarly, in our biochemical studies, ubiquitinated tau was undetectable in the soluble fraction of cortex (but see Cripps et al. 2006), and only a portion of amino-terminally processed tau in the insoluble fraction was ubiquitinated (Morishima-Kawashima et al. 1993). The question remains why only a minority of the ubiquitin tagged to the processed tau in PHF is in the form of polyubiquitin chains, and what the significance is of the monoubiquitination that appears to represent the majority of ubiquitin found in PHFs (Morishima-Kawashima et al. 1993).
The Ubiquitin–Proteasome System in AD
Since ubiquitin immunoreactivity has been documented in a range of inclusion bodies, a role of the UPS in the pathogenesis of a number of neurodegenerative diseases, especially in AD, has been considered. Several molecules that are relevant to the UPS have been shown to be associated with AD. Let us first discuss UBB+1, a mutant form of ubiquitin extending 19 amino acids at its carboxyl terminus which is generated owing to dinucleotide deletions of the ubiquitin mRNA through “molecular misreading” (van Leeuwen et al. 1998). UBB+1 protein accumulates in brains affected by AD and other diseases such as Pick's disease and Huntington's disease (Fischer et al. 2003). Because of the absence of the carboxy-terminal Gly-76, UBB+1 cannot ubiquitinate other proteins, but it is itself ubiquitinated efficiently. The resulting polyubiquitinated UBB+1 cannot be degraded by proteasomes and impairs the UPS (Lam et al. 2000), which may induce neurotoxicity. Indeed, transgenic mice expressing UBB+1 have an impaired UPS and show contextual memory deficits in both water maze and fear conditioning paradigms, without specific neuropathological findings (Fischer et al. 2009). Further, UBB+1 can enhance the susceptibility of yeast to the toxicity of protein aggregates (Tank et al. 2009).
Ubiquitin carboxy-terminal hydrolase L1 (UCH-L1), which was originally identified as a deubiqutinating enzyme, has multiple functions, including as a ubiquitin ligase and a stabilizer of monoubiquitinated proteins (Setsuie and Wada 2007). Although UCH-L1 is genetically associated with Parkinson's disease (i.e., it is the PARK5 gene; Belin and Westerlund 2008), it has also been implicated in the pathogenesis of AD. A proteomic analysis has shown down-regulation and oxidative modification of UCH-L1 in the AD brain, and the levels of soluble UCH-L1 were inversely proportional to the number of NFTs (Choi et al. 2004). Moreover, administration of UCH-L1 can reverse the amyloid β-protein–induced synaptic dysfunction and memory loss in transgenic mice overexpressing APP and PS1 (Gong et al. 2006).
Next, ubiquilin-1 has been reported to be genetically linked to AD. Ubiquilin-1 interacts with both proteasomes and ubiquitinated proteins and regulates the proteasomal degradation of various proteins, including presenilin 1 (Haapasalo et al. 2010). An intronic polymorphism involving alternative splicing of exon 8 in the ubiquilin 1 gene (UBQLN1), which is genetically located near a well-established linkage peak for AD on chromosome 9q22, has been associated with increased risk for late-onset AD (Bertram et al. 2005). However, this finding has not been replicated by others (Smemo et al. 2006).
The earliest symptoms of AD are believed to be due to synaptic dysfunction, and in this context, numerous studies have established a significant role of the UPS in the regulation of synaptic plasticity. Although ubiquitin tagging was originally identified as a signal for protein destruction by the proteasome system, ubiquitination is now known to be a posttranslational modification in which ubiquitins can serve as signals for protein localization and sorting in various physiological phenomena such as signal transduction, the cell cycle, transcription, DNA repair, and endocytosis. In particular, the critical role of the UPS in synaptic plasticity, learning and memory formation needs to be emphasized (Bingol and Sheng 2011). The role of the UPS in synaptic plasticity was first highlighted in Aplysia. During the induction of long-term facilitation in the snail, the regulatory subunit of cAMP-dependent protein kinase (PKA) is ubiquitinated and degraded by the proteasome, generating persistently activated PKA (Hegde et al. 1993). Activated PKA induces transcription of ApUCH (UCH-L1 in mammals), a deubiquitinating enzyzme, which has been found to be critical for the induction of long-term facilitation (Hegde et al. 1997). It is now known that the UPS also regulates turnover of neurotransmitter receptors, protein kinases, synaptic proteins, transcription factors, and other molecules critical for proper synaptic plasticity, thus controlling synaptic strength and connections during brain development in mammals (Bingol and Sheng 2011). The UPS is also critically involved in learning and memory. Bilateral injection of lactacystin, a specific proteasome inhibitor, into the CA1 region of the rat hippocampus blocks long-term memory formation (Lopez-Salon et al. 2001). This and numerous related findings suggest that degradation of certain critical proteins by the UPS is required during long-term memory formation. One of these proteins is arc, a negative regulator of synaptic strength that promotes the internalization of AMPA receptors and is degraded via the E3 ligase, UBE3A (Greer et al. 2010).
Synaptic loss has long been documented in AD brain (Gonatas et al. 1967) and, as expected, is strongly correlated with the degree of cognitive impairment (Terry et al. 1991). In transgenic mouse models of AD, synaptic deficits have been detected prior to the formation of amyloid plaques (Hsia et al. 1999). Among the various molecular species of Aβ present in the brain, soluble oligomeric forms of Aβ are arguably the most plausible candidates to impair synaptic function (reviewed in Walsh and Selkoe 2004). Soluble Aβ oligomers inhibit hippocampal long-term potentiation and alter memory and learning performance. They also facilitate long-term depression by, among other effects, disrupting synaptic glutamate uptake (Li et al. 2009). Recently, it has been shown that soluble Aβ oligomers isolated from AD cortex can induce tau hyperphosphorylation at AD-relevant epitopes and subsequent neuritic degeneration (Jin et al. 2011). Beyond an age-related reduction (Keller et al. 2002), proteasome activities decrease in AD in a brain region–specific manner, particularly in hippocampus, parahippocampal gyrus, superior and middle temporal gyri, and the inferior parietal lobule (Keller et al. 2000), areas that are especially critical for long-term memory formation. Moreover, soluble Aβ oligomers themselves can inhibit proteasomal activity (Tseng et al. 2008). Thus, it is possible that additive effects of Aβ oligomers and reduced proteasome activities in AD may accelerate synaptic dysfunction.
It is still unsettled how the degradation of tau is regulated under physiological conditions. It has been reported that tau is degraded by several major cellular degradation systems, including calpain, caspases, lysosomes, and proteasomes. As regards the proteasome, both the ATP-dependent 26S proteasome and the ATP-independent 20S proteasome have been reported to degrade normal, soluble tau (Cardozo et al. 2002; Zhang et al. 2005). However, administration of inhibitors of the proteasome has provided conflicting results: Treatment with lactacystin either did or did not suppress the degradation of tau (David et al. 2002; Brown et al. 2005a). On the other hand, it has been reported that the E3 ligase CHIP (carboxyl terminus of the Hsc70-interacting protein) binds to tau and is involved in the degradation of abnormal forms of tau, including insoluble tau and hyperphosphorylated tau, coordinately with Hsp70 (Petrucelli et al. 2004; Dickey et al. 2006). As described above, it has also been found that the 20S proteasome interacts with tau aggregates (Keck et al. 2003). Collectively, such findings suggest that the UPS may be implicated more in the degradation of abnormal forms of tau than of normal, soluble tau.
As in the case of tau, an impairment of proteasome activity by protein aggregates is observed for other proteins, such as polyglutamine-expanded proteins, the prion protein, and Aβ (Bence et al. 2001). Notably, immunotherapy against Aβ in the 3xTg-AD mice, which reduces Aβ oligomers, reverses the proteasome deficits of these transgenic mice (Tseng et al. 2008). Thus, the accumulation of tau and of Aβ, forming the two major protein lesions of AD, impairs proteasome activity in vivo.
Despite the substantial indirect evidence just reviewed that associates UPS dysfunction with key features of AD, it remains unclear whether aberration in the UPS plays a causative or only a secondary role. Besides its key role in the UPS, ubiquitin is now known to function as a signal tag in the lysosome–autophagy system (see below). Indeed, many recent studies suggest the involvement of autophagy in the pathogenesis of AD. For example, immunocytochemistry showing the presence of K63-linked polyubiquitin in a fraction of the NFTs in AD cortex (Paine et al. 2009) suggests an active involvement of autophagy in the mechanism of AD.
PART II: THE AUTOPHAGY AND THE ENDOSOMAL–LYSOSOMAL SYSTEM IN AD
The Lysosomal Network
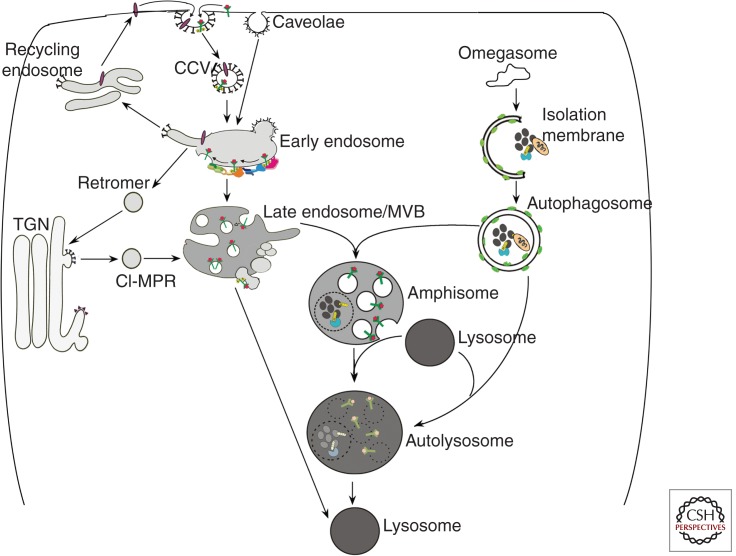
The endocytic and autophagic pathways share in common the role of delivering unneeded cellular materials to lysosomes for degradation and recycling to provide energy and new synthesis. Vesicular compartments in these pathways actively cross-talk and have common points of regulation (Simonsen et al. 2009; Settembre et al. 2011). Under pathological conditions, a deficit in one pathway often impedes functioning of the other. Therefore, it is reasonable to view these two pathways, which intersect at the lysosome, as comprising a lysosomal network (Fig. 5).
Figure 5.
Schematic illustrating the endocytic and the autophagic pathways to the lysosome. See the text for details.
Although key to the survival of all cells, endocytosis supports unique neuronal functions, including aspects of synaptic transmission and plasticity underlying memory and learning. Also, extracellular signals (e.g., growth factors) activate surface receptor complexes that, upon internalization, undergo retrograde axonal transport within “signaling endosomes” to promote neuronal survival and differentiation (Delcroix et al. 2003). That these vital processes are initiated in synaptic terminals at great distances from the perikaryon renders neurons particularly vulnerable to impairments of endocytic function and vesicular transport.
During the initiation of endocytosis, the invagination of plasma membranes into vesicles is mediated most often by the clathrin/adaptor protein complex, but also via caveolae or bulk macropinocytosis (Lim et al. 2011). Cargoes are delivered to early sorting endosomes through fusion mediated by the small GTPase Rab5 and additional Rab5 effector proteins (Fig. 5). Many cell surface proteins and lipids are returned to the plasma membrane via recycling endosomes, whereas other components are delivered to the Golgi by the retromer complex (Kelly et al. 2011). Still other cargoes reach late endosomes when Rab7 and its effectors replace Rab5 and initiate further endosomal maturation (Poteryaev et al. 2010). An MVB is then created by inward budding of the surface membrane (Piper et al. 2001), thereby sorting cargoes into intraluminal vesicles. Ubiquitin, the crucial signal for efficient sorting of proteins into the MVB (Babst et al. 1997), initiates this process, which is mediated by a group of ESCRT complexes (endosomal sorting complex required for transport; Hurley 2010). Acid hydrolases, including cathepsins, are delivered from the trans-Golgi network (TGN) to MVBs/late endosomes by either of two mannose-6-phosphate receptors: cation-dependent 46 kDa MPR (CD-MPR) and cation-independent 215 kDa MPR (CI-MPR; Mullins et al. 2001). Proteolysis begins in late endosomes/MVBs but accelerates upon fusion with lysosomes, where cathepsins are activated by the highly acidic (pH 4.5–5.5) intraluminal environment. The acidic pH is achieved mainly through the ATP-dependent proton pump, vacuolar ATPase (Nishi et al. 2002), but is also modulated by a group of chloride, sodium, calcium, and potassium transporters (Pillay et al. 2002).
Autophagy is the cell’s principal degradative pathway for eliminating unwanted organelles and long-lived proteins and for clearing damaged, aggregated, or obsolete proteins (Wong et al. 2010). Autophagy refers to at least three processes by which intracellular constituents reach the lumen of MVBs or lysosomes for degradation: chaperone-mediated autophagy (CMA), microautophagy, and macroautophagy (Cuervo 2004; Levine et al. 2011). In CMA, cytosolic proteins containing a KFERQ motif (including proteins pathogenic in some neurodegenerative diseases) are selectively targeted by certain chaperones to the lysosomal lumen for degradation (Arias et al. 2011). Microautophagy involves the nonselective entry of small quantities of cytoplasm into lysosomes or late endosomes/MVBs when the limiting membranes of these compartments invaginate and pinch off small vesicles for digestion within the lumen (Sahu et al. 2011). Finally, macroautophagy mediates bulk or targeted degradation of cytoplasmic constituents. During macroautophagy, an elongated “isolation” membrane created from a preautophagosomal structure sequesters a region of cytoplasm to form a double-membrane-limited autophagosome (Fig. 5). Two ubiquitin-like protein conjugation pathways are known to coordinate this process (Ohsumi 2001). Sequestered material within autophagosomes is digested when lysosomes or late endosomes fuse with the outer membrane of the autophagosome (Gordon et al. 1988). Amphisomes formed by the fusion of autophagosomes with early endosomes or MVBs/late endosomes are especially important in neurons, where a considerable proportion of endocytosed cargo is directed to the autophagic pathway prior to being degraded by lysosomes (Larsen et al. 2002). This is especially true in axons (Lee et al. 2011). Induction of autophagy is generally controlled by the mTOR kinase (mammalian Target of Rapamycin), which is regulated by growth factors (especially insulin) and nutrient levels. Autophagy is constitutively active in neurons and is nonselective under nutrient deprivation conditions but may be selective when damage uncovers a molecular target on an organelle such as a mitochondrion (Youle et al. 2011), which initiates signaling and a ubiquitin-ligase-mediated chain of events that triggers selective sequestration of the organelle (Dikic et al. 2010; Weidberg et al. 2011).
Molecular Pathology of the Lysosomal Network
A continuum of pathological changes of the lysosomal network unfolds in neurons as Alzheimer disease progresses, including dysregulation of endocytosis, increased lysosome biogenesis and, later, progressive failure of lysosomal clearance mechanisms (Fig. 6; Nixon et al. 2006). These changes can be driven by known AD genetic and environmental risk factors, as discussed later. Endosome anomalies are the earliest specific pathology reported in AD brain tissue. Neuronal endosome enlargement, which is not characteristically observed in other major neurodegenerative diseases, develops in pyramidal neurons of the neocortex at a stage when plaques and tangles are restricted only to the hippocampus (Braak stage 2) and not in brains of similarly aged individuals free of AD-like hippocampal pathology (Cataldo et al. 1997, 2000). Similar endosomal anomalies develop gradually in Down syndrome brain, beginning decades before the appearance of classical AD pathology (Cataldo et al. 2000). Genes related to endocytosis, such as Rab5, Rab7, and Rab4, are among the earliest groups to show up-regulated transcription in AD (Ginsberg et al. 2010), and their corresponding proteins are abnormally recruited to endosomes, where they promote fusion and abnormal enlargement of early and late endosomes (Cataldo et al. 1997, 2008). This pattern is not seen in normal aging brain and is specific for AD among aging-related neurodegenerative diseases that have been studied (Cataldo et al. 2000).
Figure 6.
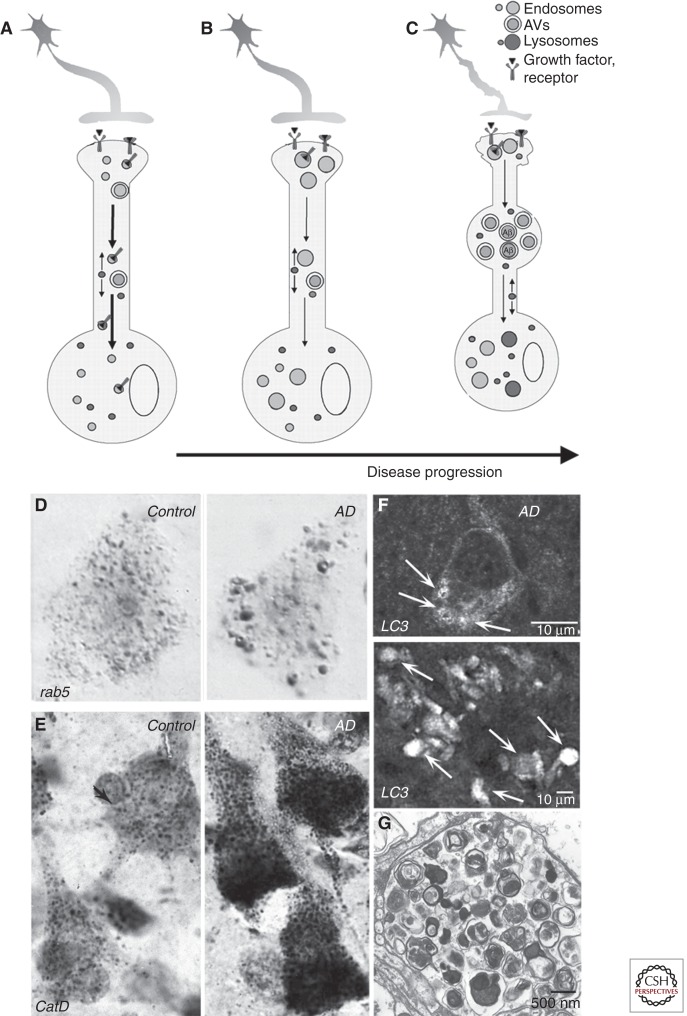
Progression of pathological changes in the lysosomal network in Alzheimer disease (AD). A normal neuron is depicted in A. At the earliest stages of AD (B), accelerated endocytosis, mediated in part by βCTF and/or by pathological activation of rab5, causes endosome enlargement (D; rab5 immunocytochemistry), defective retrograde transport of endosomes and their neurotrophin cargoes, proapoptotic pathway activation, and cholinergic neurodegeneration. Initial up-regulation of lysosome biogenesis (E; cathepsin D immunocytochemistry) is followed by progressive failure of lysosomal proteolysis (C), which impedes autophagy and the axonal transport of late endosomes/MVBs and autophagy-related vesicular compartments. These compartments, containing incompletely digested protein substrates, selectively accumulate within neurons and especially dystrophic neurites (C), as depicted in F (LC3 immunocytochemistry) and G (ultrastructure of AVs in a dystrophic neurite). Failure of lysosomal proteolysis and autophagy in AD is caused by PS1 mutations in early-onset FAD and is also promoted by normal aging, oxidative stress, ApoE ε4, intracellular Aβ, and other AD-related genetic and environmental risk factors.
As neuronal endosomes begin to enlarge, lysosomal biogenesis rises, as evidenced by lysosome proliferation and increased expression of acid hydrolases and other lysosome-related proteins (Nixon et al. 2006). Lysosome proliferation is followed by progressive enlargement and distortion of these compartments. Cathepsin-positive vacuoles massively accumulate within swellings of dystrophic neurites throughout the neuropil and in senile plaques as they develop (Cataldo et al. 1990, 1991). Ultrastructural investigations of AD brain biopsies have revealed that the most prevalent organelles by far within dystrophic swellings are autophagic vacuoles (AVs), representing all stages of the macroautophagic process (Nixon et al. 2005), including autophagosomes. This implies that the progression of autophagy is delayed or impaired, because AVs are relatively rare in normal brain. AV accumulations are not specific to the degenerative phenomena of AD; however, in AD brain, the extensive numbers of dystrophic neurites (Masliah et al. 1993; Schmidt et al. 1994), their characteristic marked distension, and the fact that they are predominantly filled with AVs distinguish the pattern and magnitude of this pathology from that of other aging-related neurodegenerative diseases (Benzing et al. 1993). In non-AD neurodegenerative disorders, many types of vesicles and cytoskeletal elements are relatively abundant, suggesting general axonal transport failure distinct from the selective transport deficits seen in AD (Lee et al. 2011a,b).
The profuse and selective accumulation of AVs in neurons in AD reflects a defect in the clearance of AVs by lysosomes rather than an abnormally elevated induction of autophagy. The accumulating AVs are mainly electron-dense autolysosomes, indicating that autophagosome–lysosome fusion is relatively competent. Similar autophagy pathology is observed when lysosomal proteolysis is inhibited (Ivy et al. 1984; Koike et al. 2005; Yang et al. 2008). That presenilin 1 mutations, which are a cause of early-onset familial AD, impede lysosome proteolysis and accelerate neuritic dystrophy also supports a primary role for failure of proteolytic clearance (Lee et al. 2010). Increased autophagy induction in AD could, however, compound the defect in AV clearance in AD. Transcription of many lysosomal genes and factors promoting autophagy is up-regulated, whereas negative regulators of autophagy are down-regulated (Lipinski et al. 2010), although not all studies support enhanced autophagy induction in AD (Pickford et al. 2008; Nixon et al. 2011).
AD Genetics Implicates the Lysosomal Network in Pathogenesis
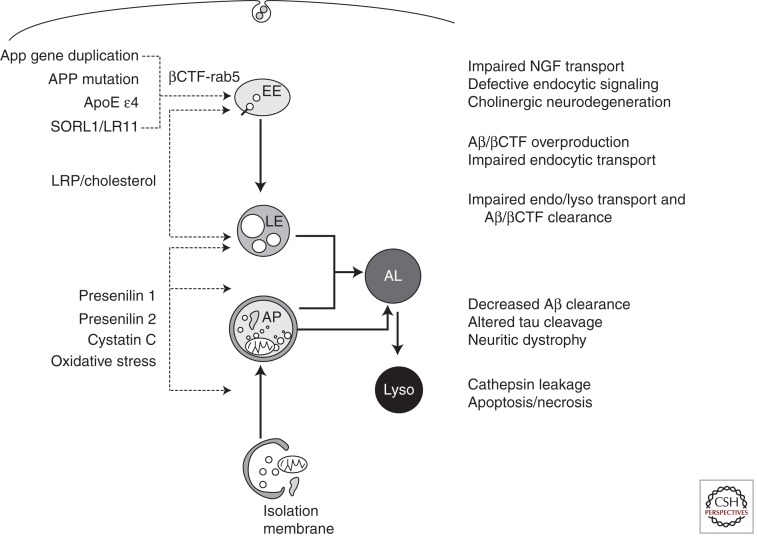
Genetic factors that represent the strongest support for the amyloid/Aβ cascade hypothesis also act directly through Aβ-independent mechanisms to dysregulate the lysosomal network and contribute not only to Aβ accumulation but also to other critical aspects of AD pathogenesis (Fig. 7). Beyond its role as a component of γ-secretase, Presenilin 1 (PS1) is required for lysosome acidification, which is needed to activate cathepsins and other hydrolases that carry out digestion during autophagy (Lee et al. 2010). In the absence of PS1, the V0a1 subunit of v-ATPase is not N-glycosylated in the ER and is degraded before sufficient amounts can be delivered to autolysosomes/lysosomes to support lysosomal acidification. This function of PS1 is independent of the aspartyl protease activity of the molecule because proteolytically inactive PS1 mutated at one or both catalytic aspartates is able to restore normal lysosome acidification in PS1-deleted cells (Lee et al. 2010). Importantly, PS1 mutations also inhibit this process in fibroblasts from patients with familial AD (Lee et al. 2010), providing a basis for defective autophagy and the potentiation of autophagic–lysosomal and amyloid pathology and accelerated neuronal cell death observed in PS-FAD (Cataldo et al. 2004).
Figure 7.
Dysfunction of autophagic and endocytic pathways to lysosomes driven by relevant genes and other risk factors in Alzheimer disease (left side of the diagram) causes or promotes pathophysiology critical to the development and progression of Alzheimer disease (right side of the diagram). See the text for further details.
An additional copy of the App gene (genomic duplication) is sufficient to cause another form of early-onset FAD (Rovelet-Lecrux et al. 2006; Sleegers et al. 2006). App promoter polymorphisms that increase APP expression are also associated with early-onset AD (Athan et al. 2002). These findings support a longstanding hypothesis that the App gene on the trisomic copy of human chromosome 21 (HSA21) in Down syndrome (DS) is principally responsible for the invariant early development of AD in DS individuals (Margallo-Lana et al. 2004). In DS, the extra copy of App causes endocytic up-regulation and endosome pathology similar to that seen at the earliest stages of sporadic AD, but beginning even earlier. Recently, these effects of increased App dosage were shown to be mediated specifically by the β-cleaved carboxy-terminal fragment of APP, βCTF (Jiang et al. 2010), which binds to a complex of signaling molecules on endosomes that pathologically activates rab5 (S Kim and RA Nixon, unpubl.). It is this abnormal rab5 activation that causes protein and lipid accumulation in endosomes, slowed lysosomal degradation of endocytic cargoes, endosome swelling (Cataldo et al. 2008), and disrupted retrograde transport of endosomes (S Kim and RA Nixon, unpubl.). Interactions between FAD-mutant forms of APP and APP binding protein (APP-BP1) on endosomes also initiate pathological rab5 activation, which was shown to promote a neuronal apoptosis cascade (Laifenfeld et al. 2007).
Acceleration of endosome pathology is also seen in individuals who inherit the ε4 allele of APOE, a key mediator of neuronal cholesterol transport and the major genetic risk factor for late-onset AD (Cataldo et al. 2000). Moreover, among a very few additional pathological conditions associated with AD-like endosomal pathology are APP transgenic mice fed elevated dietary cholesterol and individuals with Niemann–Pick type C (NPC), a disorder of cholesterol homeostasis. High dietary LDL-cholesterol and overexpression of its receptor ApoE (particularly ApoE ε4) elevate βCTF levels (Ji et al. 2006; Cossec et al. 2010), and these levels are also elevated in NPC, DS, and AD, particularly in early-onset forms of AD caused by certain mutations of APP. The downstream consequences of βCTF-mediated rab5 activation and abnormal endosomal signaling for AD progression are many, as discussed later. Although less well characterized, a growing number of AD risk genes for late-onset AD identified through genome-wide screens and association studies have direct links to endocytic regulation (Seshadri et al. 2010; Hollingworth et al. 2011; Hu et al. 2011).
The genetics of other neurological diseases further support a close connection between disruption of lysosomal network function and selective vulnerability to neurodegeneration (Nixon et al. 2008). Lysosomal storage disorders involving primary defects in lysosome function commonly exhibit prominent neurodegenerative phenotypes, including neuritic dystrophies closely resembling the ultrastructural morphologies of dystrophic neurites in AD and, in some of these disorders, neurofibrillary tangles as well as increased amyloidogenic processing of APP and diffuse β-amyloid deposits (Ryazantsev et al. 2007; Ohmi et al. 2009; Bellettato et al. 2010). Inherited defects in the endocytic pathway are also associated with neurodegenerative disorders at a high frequency (Nixon 2004; Nixon et al. 2008; Rusten et al. 2008; Zhang 2008).
Biology of Lysosomal Network Dysregulation in AD and Its Pathological Consequences
Neuritic Dystrophy
Endocytosis and autophagy are highly active at synaptic terminals and axons and generate large numbers of endosomes and autophagosomes, representing a considerable burden of retrograde organelle trafficking to the perikaryon (Overly et al. 1996). Transport and autophagy must, therefore, be especially efficient to prevent “traffic jams.” In healthy neurons, these mechanisms are, in fact, exceptionally efficient, as evidenced by a scarcity of autophagy-related intermediates, a high autophagic clearance capacity, and the rapidity with which axonal AVs accumulate when their clearance is impeded (Nixon et al. 2011), but they are also highly vulnerable to disruption (Boland et al. 2008; Cai et al. 2010; Lee et al. 2011a).
AVs and lysosomes constitute more than 95% of the organelles in dystrophic neuritic swellings in AD and AD mouse models, implying a cargo-specific defect in axonal transport, rather than a global one. Indeed, when lysosomal proteolysis is inhibited by blocking acidification or directly inhibiting cathepsins, axonal transport of autophagy-related compartments is selectively slowed and intermittently interrupted. These organelles then accumulate selectively within axon swellings that acquire additional AD-like features, including local cytoskeletal protein hyperphosphorylation and ubiquitin immunolabeling (Lee et al. 2011a). These observations suggest how PS1 mutations, which impede lysosomal acidification (Lee et al. 2010), may markedly accelerate and amplify neuritic dystrophy in AD.
Similar neuritic dystrophy eventually develops in all forms of AD and in mouse AD models where only FAD-causing mutant APP is overexpressed. In these cases, the mechanism is not yet clear. Aβ administered directly into brains of wild-type rodents does not induce neuritic dystrophy, despite diffuse amyloid deposition (Frautschy et al. 1996, 1998), raising the possibility that other APP metabolites within neuronal compartments besides or in addition to Aβ could be important to the development of neuritic dystrophy. In this regard, elevated βCTF levels induced by APP overexpression, elevated dietary cholesterol, or overexpression of its receptor ApoE (particularly ApoE ε4) can up-regulate endocytosis and enlarge endosomes (Laifenfeld et al. 2007; Chen et al. 2010; Cossec et al. 2010), leading to impaired endosome retrograde transport (S Kim and RA Nixon, unpubl.). Accelerated endocytosis also increases protein and lipid accumulation in endosomes and slows lysosomal degradation of endocytic cargoes (Cataldo et al. 2008), leading to lysosomal instability and neurodegeneration, as discussed below.
Protein Clearance Failure and Amyloidogenesis
In APP transgenic mouse models of AD, undigested autophagy substrates including LC3-II, p62, and ubiquitinated proteins accumulate in neuronal AVs, establishing that autophagic protein turnover in lysosomes is impeded (Yang et al. 2011). This general failure to clear autophagy substrates affects clearance of various proteins relevant to AD pathogenesis, including Aβ, tau, and other factors, such as damaged mitochondria and activated caspases, that promote cell death (Yang et al. 2008).
Autophagy sequesters and digests unneeded or damaged organelles, some of which are APP-rich (Pickford et al. 2008). AVs are also enriched in APP substrates and secretases and, during autophagy, Aβ peptide is generated from APP (Yu et al. 2005), although it is subsequently degraded in lysosomes under normal circumstances (Heinrich et al. 1999; Bahr et al. 2002; Florez-McClure et al. 2007). Although less well studied as “Aβ degrading proteases” than the zinc metallopeptidase family (Guenette 2003; Eckman et al. 2005), cathepsins are considered an important route for Aβ/amyloid clearance (Mueller-Steiner et al. 2006; Nixon 2007; Butler et al. 2011) and human neurons may be particularly dependent on this mechanism (LeBlanc et al. 1999; reviewed in Saido and Leissring 2011). Chronic low-level stimulation of autophagy through peripheral administration of rapamycin or other agents (Tian et al. 2011), or enhancing lysosomal proteolysis selectively (Sun et al. 2008; Yang et al. 2011), can markedly diminish Aβ levels and amyloid load in APP transgenic mice, underscoring the importance of lysosomal clearance of Aβ.
Endocytic pathway up-regulation in AD stemming in part from pathological rab 5 activation generates higher levels of Aβ (Mathews et al. 2002; Grbovic et al. 2003) that must be cleared in part by lysosomes. Pathological rab5 activation, which in Down syndrome is dependent on βCTF generation (Jiang et al. 2010), can up-regulate endocytosis in a manner functionally equivalent to the elevated endocytosis associated with increased synaptic activity, which is considered a source of Aβ generation (Cirrito et al. 2008). An unknown amount of Aβ produced in endosomes reaches lysosomes directly or via production during autophagy (Nixon 2007), and accumulated AVs and MVBs appear to be the major intracellular reservoirs of Aβ immunoreactivity in AD brain (Takahashi et al. 2002; Yu et al. 2005). The acidic environment in lysosomes is particularly favorable for the initial stages of Aβ oligomerization (Peralvarez-Marin et al. 2008).
Tauopathy
Neurofibrillary tangles composed of tau proteins in a hyperphosphorylated state are rarely observed in abundance except in AD and a limited number of aging-related tauopathies. However, they are a prominent feature in at least two lysosomal disorders, NPC and mucopolysaccharidosis type IIB (MPS IIB; Ohmi et al. 2009), and in a mouse model of MPS IIB (Ryazantsev et al. 2007). In NPC, which arises from a defect in endosomal trafficking of cholesterol and the accumulation of unesterified cholesterol in late endosomes/lysosomes, additional features associated with AD are seen, including allele-selective influences of the APOE genotype on pathology and increased amyloidogenic processing of APP (Nixon 2004). The mechanisms explaining this connection are unclear, but tau has been found to be an autophagy substrate, at least when overexpressed (Berger et al. 2006). Incomplete charperone-mediated autophagy of tau generates fragments that aggregate and are cleared by macroautophagy (Wang et al. 2009). Moreover, autophagy preferentially degrades a caspase-cleaved fragment of tau implicated in tau neurotoxicity (Dolan et al. 2010). Consistent with these findings, rapamycin induction of autophagy reduces tau pathology in the triple transgenic AD mouse model (Caccamo et al. 2010), whereas in other models, autophagic–lysosomal dysfunction amplifies tau pathology and tau neurotoxicity (Hamano et al. 2008; Khurana et al. 2010).
Synaptic Dysfunction
Pathological Rab5 activation driving endocytic dysfunction in AD may negatively impact long-term potentiation (LTP) and long-term depression (LTD) aspects of synaptic plasticity closely associated with learning and memory (Kessels et al. 2009). LTD induction requires surface removal of AMPA receptors driven by their Rab5-dependent internalization (Brown et al. 2005b). Conversely, deletion of the neuronal rab5 GEF, rin1, reduces rab5 activation, increases LTP induction in the amygdala, and enhances fear learning and memory, most likely by increasing surface levels of AMPA receptors (Dhaka et al. 2003). Autophagy may also modulate synaptic plasticity, which involves structural remodeling of nerve terminals (Boland et al. 2006) and the trafficking and degradation of receptors and other synaptic proteins (Leil et al. 2004; Rowland et al. 2006).
Neurodegeneration
A close connection between lysosomal network dysfunction and mechanisms of neurodegeneration is well documented (McCray et al. 2008; Nixon et al. 2008; Bellettato et al. 2010; Cherra et al. 2010). Deprived of the cytoprotective functions of autophagy, neurons, which cannot dilute toxic protein buildup by cell division, are particularly vulnerable to potentially toxic mutant, oxidized and aggregated proteins and peptide fragments. An important possible outcome of the accumulation of the latter in lysosomes is increased membrane permeability and release of hydrolases into the cytoplasm, even from otherwise intact lysosomes (Kroemer et al. 2005). Cataclysmic disruption of lysosomal membranes releases hydrolases that act as both the trigger and executioner in rapid necrosis (Syntichaki et al. 2003; Kroemer et al. 2005), whereas slow release of cathepsins more likely operates through signaling pathways to trigger apoptosis (Kroemer et al. 2005).
By up-regulating endocytosis, high dietary LDL-cholesterol and overexpression of its receptor ApoE (particularly ApoE ε4) elevate βCTF levels and increase delivery of Aβ1–42 to lysosomes in cellular model systems (Ji et al. 2006; Cossec et al. 2010). Accumulation of Aβ1–42, which is less efficiently degraded than Aβ1–40 owing to its strong propensity to aggregate, causes leakage of lysosomal enzymes into the cytosol prior to other morphological signs of cellular toxicity in vitro (Yang et al. 1998; Glabe 2001). Consistent with these findings, strong overexpression of human Aβ42, but not Aβ40, in Drosophila neurons induces age-related accumulation of Aβ in autolysosomes and neurotoxicity (Ling et al. 2009). Aβ42-induced neurotoxicity is further enhanced by autophagy activation and is partially rescued by autophagy inhibition. Expression of the ApoE ε4 allele, but not ApoE ε3, in mice administered a neprilysin inhibitor increases Aβ immunoreactivity in lysosomes and causes neurodegeneration of hippocampal CA1, entorhinal, and septal neurons (Belinson et al. 2008). ApoE ε4 that trafficks to lysosomes more readily than ApoE ε3, promotes leakage of acid hydrolases, and induces apoptosis in cultured neuronal cells by forming membrane-damaging intermediates in the low-pH environment (Ji et al. 2002).
Beyond influencing Aβ generation and toxicity, defective endosome functioning plays a crucial Aβ-independent role in the failure of retrograde NGF signaling that leads to basal forebrain cholinergic neuron degeneration in the Ts65Dn mouse model of DS (Cooper et al. 2001; Delcroix et al. 2004). Proteins known to play roles in both endocytosis regulation and cell survival decisions link the endosome dysregulation seen in AD to cell death cascade signaling (Vito et al. 1996, 1999; Missotten et al. 1999; Chen et al. 2000; Gout et al. 2000).
Remediating Lysosomal Network Dysfunction as AD Therapy
Recent evidence supports the value of targeting autophagy efficiency as a possible therapeutic strategy for AD. Peripheral administration of rapamycin to strongly stimulate autophagy substantially reduces amyloid deposition and tau pathology in both APP and triple transgenic mouse models of AD pathology (Caccamo et al. 2010; Spilman et al. 2010; Tian et al. 2011). Autophagy induction also has beneficial effects in transgenic models of several aging-related neurodegenerative diseases (Garcia-Arencibia et al. 2010). Stimulating lysosomal proteolytic efficiency in the TgCRND8 APP mouse model by deleting an endogenous inhibitor of lysosomal cysteine proteases (cystatin B) rescues lysosomal pathology, eliminates abnormal autolysosomal accumulation of autophagy substrates, including Aβ, decreases Aβ and amyloid deposition, and ameliorates learning and memory deficits (Yang et al. 2011). Similar therapeutic effects, including restoration of synaptic functions, are seen in APP mouse models after deleting cystatin C (Sun et al. 2008), by overexpressing cathepsin B (Mueller-Steiner et al. 2006), or by enhancing its activity (Butler et al. 2011). Collectively, these observations support the pathogenic significance of autophagic–lysosomal dysfunction in AD and specifically the importance of deficient lysosomal proteolysis.
CONCLUSION
As postmitotic cells, neurons are especially reliant on the lysosomal network to sort and clear normal and damaged proteins and support vital signaling functions. Not surprisingly, a defective endosomal–lysosomal pathway and certain autophagy-related genes are being linked with an unusually high frequency to neurodegenerative diseases, and the mechanistic relationship to neurodegeneration is becoming increasingly well established. Driven by the same genetic and environmental risk factors implicated in the Aβ/amyloid cascade hypothesis, a continuum of highly characteristic pathological changes in the lysosomal network evolves during AD, including dysregulation of endocytosis, increased lysosome biogenesis, and progressive failure of lysosomal proteolysis and autophagy. These deficits cripple neuronal functions critical for synaptic plasticity and neuron survival, lead to the hallmark neuritic dystrophy of AD, promote accumulation of toxic proteins—including Aβ, tau, activated caspases, and other cell death-promoting proteins—and initiate multiple neurodegenerative cascades. Remediation of lysosomal network dysfunction by several different approaches has been shown to have significant ameliorative effects on pathological and cognitive deficits in animal models of AD, underscoring the pathogenic significance of this dysfunction and the promise of therapeutic strategies targeting the lysosomal network in AD and other neurodegenerative disorders.
ACKNOWLEDGMENTS
The work of the Ihara laboratory on ubiquitin and PHF was carried out from 1985 to 1993, and Dr. Ihara thanks all laboratory members from that period (Department of Clinical Physiology, Tokyo Metropolitan Institute of Gerontology; Department of Neuropathology, Faculty of Medicine, University of Tokyo). In particular, the preparation of the DF2 antibody and identification of ubiquitin in PHF was performed collaboratively with Dr Hiroshi Mori, Department of Neuroscience, Osaka City University Graduate School of Medicine. For the introduction and section on autophagy and the lysosomal system, the assistance of Nicole Piorkowski and Corrinne Peterhoff in manuscript and figure preparation is gratefully acknowledged. Research from the Nixon laboratories is supported by the National Institute on Aging.
Footnotes
Editors: Dennis J. Selkoe, Eckhard Mandelkow, and David M. Holtzman
Additional Perspectives on The Biology of Alzheimer Disease available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Anderton BH, Breinburg D, Downes MJ, Green PJ, Tomlinson BE, Ulrich J, Wood JN, Kahn J 1982. Monoclonal antibodies show that neurofibrillary tangles and neurofilaments share antigenic determinants. Nature 298: 84–86 [DOI] [PubMed] [Google Scholar]
- Arias E, Cuervo AM 2011. Chaperone-mediated autophagy in protein quality control. Curr Opin Cell Biol 23: 184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athan ES, Lee JH, Arriaga A, Mayeux RP, Tycko B 2002. Polymorphisms in the promoter of the human APP gene: Functional evaluation and allele frequencies in Alzheimer disease. Arch Neurol 59: 1793–1799 [DOI] [PubMed] [Google Scholar]
- Babst M, Sato TK, Banta LM, Emr SD 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J 16: 1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahr BA, Bendiske J 2002. The neuropathogenic contributions of lysosomal dysfunction. J Neurochem 83: 481–489 [DOI] [PubMed] [Google Scholar]
- Bancher C, Grundke-Iqbal I, Iqbal K, Fried VA, Smith HT, Wisniewski HM 1991. Abnormal phosphorylation of tau precedes ubiquitination in neurofibrillary pathology of Alzheimer disease. Brain Res 539: 11–18 [DOI] [PubMed] [Google Scholar]
- Belin AC, Westerlund M 2008. Parkinson's disease: A genetic perspective. FEBS J 275: 1377–1383 [DOI] [PubMed] [Google Scholar]
- Belinson H, Lev D, Masliah E, Michaelson DM 2008. Activation of the amyloid cascade in apolipoprotein E4 transgenic mice induces lysosomal activation and neurodegeneration resulting in marked cognitive deficits. J Neurosci 28: 4690–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellettato CM, Scarpa M 2010. Pathophysiology of neuropathic lysosomal storage disorders. J Inherit Metab Dis 33: 347–362 [DOI] [PubMed] [Google Scholar]
- Bence NF, Sampat RM, Kopito RR 2001. Impairment of the ubiquitin–proteasome system by protein aggregation. Science 292: 1552–1555 [DOI] [PubMed] [Google Scholar]
- Benzing WC, Mufson EJ, Armstrong DM 1993. Alzheimer's disease-like dystrophic neurites characteristically associated with senile plaques are not found within other neurodegenerative diseases unless amyloid β-protein deposition is present. Brain Res 606: 10–18 [DOI] [PubMed] [Google Scholar]
- Berger Z, Ravikumar B, Menzies FM, Oroz LG, Underwood BR, Pangalos MN, Schmitt I, Wullner U, Evert BO, O'Kane CJ, et al. 2006. Rapamycin alleviates toxicity of different aggregate-prone proteins. Hum Mol Genet 15: 433–442 [DOI] [PubMed] [Google Scholar]
- Berke SJ, Paulson HL 2003. Protein aggregation and the ubiquitin proteasome pathway: Gaining the UPPer hand on neurodegeneration. Curr Opin Genet Dev 13: 253–261 [DOI] [PubMed] [Google Scholar]
- Bertram L, Hiltunen M, Parkinson M, Ingelsson M, Lange C, Ramasamy K, Mullin K, Menon R, Sampson AJ, Hsiao MY, et al. 2005. Family-based association between Alzheimer's disease and variants in UBQLN1. New Engl J Med 352: 884–894 [DOI] [PubMed] [Google Scholar]
- Bingol B, Sheng M 2011. Deconstruction for reconstruction: The role of proteolysis in neural plasticity and disease. Neuron 69: 22–32 [DOI] [PubMed] [Google Scholar]
- Boland B, Nixon RA 2006. Neuronal macroautophagy: From development to degeneration. Mol Aspects Med 27: 503–519 [DOI] [PubMed] [Google Scholar]
- Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA 2008. Autophagy induction and autophagosome clearance in neurons: Relationship to autophagic pathology in Alzheimer's disease. J Neurosci 28: 6926–6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion JP, Couck AM, Passareiro E, Flament-Durand J 1985. Neurofibrillary tangles of Alzheimer's disease: An immunohistochemical study. J Submicrosc Cytol 17: 89–96 [PubMed] [Google Scholar]
- Brown MR, Bondada V, Keller JN, Thorpe J, Geddes JW 2005a. Proteasome or calpain inhibition does not alter cellular tau levels in neuroblastoma cells or primary neurons. J Alzheimer’s Dis 7: 15–24 [DOI] [PubMed] [Google Scholar]
- Brown TC, Tran IC, Backos DS, Esteban JA 2005b. NMDA receptor-dependent activation of the small GTPase Rab5 drives the removal of synaptic AMPA receptors during hippocampal LTD. Neuron 45: 81–94 [DOI] [PubMed] [Google Scholar]
- Butler D, Hwang J, Estick C, Nishiyama A, Kumar SS, Baveghems C, Young-Oxendine HB, Wisniewski ML, Charalambides A, Bahr BA 2011. Protective effects of positive lysosomal modulation in Alzheimer's disease transgenic mouse models. PLoS One 6: e20501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccamo A, Majumder S, Richardson A, Strong R, Oddo S 2010. Molecular interplay between mammalian target of rapamycin (mTOR), amyloid-β, and Tau: Effects on cognitive impairments. J Biol Chem 285: 13107–13120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Lu L, Tian JH, Zhu YB, Qiao H, Sheng ZH 2010. Snapin-regulated late endosomal transport is critical for efficient autophagy–lysosomal function in neurons. Neuron 68: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo C, Michaud C 2002. Proteasome-mediated degradation of tau proteins occurs independently of the chymotrypsin-like activity by a nonprocessive pathway. Arch Biochem Biophys 408: 103–110 [DOI] [PubMed] [Google Scholar]
- Cataldo AM, Nixon RA 1990. Enzymatically active lysosomal proteases are associated with amyloid deposits in Alzheimer brain. Proc Natl Acad Sci 87: 3861–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Paskevich PA, Kominami E, Nixon RA 1991. Lysosomal hydrolases of different classes are abnormally distributed in brains of patients with Alzheimer disease. Proc Natl Acad Sci 88: 10998–11002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Barnett JL, Pieroni C, Nixon RA 1997. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer's disease: Neuropathologic evidence for a mechanism of increased β-amyloidogenesis. J Neurosci 17: 6142–6151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Troncoso JC, Gomez-Isla T, Hyman BT, Nixon RA 2000. Endocytic pathway abnormalities precede amyloid β deposition in sporadic Alzheimer's disease and Down syndrome: Differential effects of APOE genotype and presenilin mutations. Am J Pathol 157: 277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Peterhoff CM, Schmidt SD, Terio NB, Duff K, Beard M, Mathews PM, Nixon RA 2004. Presenilin mutations in familial Alzheimer disease and transgenic mouse models accelerate neuronal lysosomal pathology. J Neuropathol Exp Neurol 63: 821–830 [DOI] [PubMed] [Google Scholar]
- Cataldo AM, Mathews PM, Boiteau AB, Hassinger LC, Peterhoff CM, Jiang Y, Mullaney K, Neve RL, Gruenberg J, Nixon RA 2008. Down syndrome fibroblast model of Alzheimer-related endosome pathology. Accelerated endocytosis promotes late endocytic defects. Am J Pathol 173: 370–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Borinstein SC, Gillis J, Sykes VW, Bogler O 2000. The glioma-associated protein SETA interacts with AIP1/Alix and ALG-2 and modulates apoptosis in astrocytes. J Biol Chem 275: 19275–19281 [DOI] [PubMed] [Google Scholar]
- Chen X, Wagener JF, Morgan DH, Hui L, Ghribi O, Geiger JD 2010. Endolysosome mechanisms associated with Alzheimer's disease-like pathology in rabbits ingesting cholesterol-enriched diet. J Alzheimer’s Dis 22: 1289–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherra SJ 3rd, Dagda RK, Chu CT 2010. Review: Autophagy and neurodegeneration: Survival at a cost? Neuropathol Appl Neurobiol 36: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L 2004. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J Biol Chem 279: 13256–13264 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Finley D, Varshavsky A 1984. Ubiquitin dependence of selective protein degradation demonstrated in the mammalian cell cycle mutant ts85. Cell 37: 57–66 [DOI] [PubMed] [Google Scholar]
- Cirrito JR, Kang J-E, Lee J, Stewart FR, Verges DK, Silverio LM, Bu G, Mennerick S, Holtzman DM 2008. Endocytosis is required for synaptic activity-dependent release of amyloid-β in vivo. Neuron 58: 42–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JD, Salehi A, Delcroix JD, Howe CL, Belichenko PV, Chua-Couzens J, Kilbridge JF, Carlson EJ, Epstein CJ, Mobley WC 2001. Failed retrograde transport of NGF in a mouse model of Down's syndrome: Reversal of cholinergic neurodegenerative phenotypes following NGF infusion. Proc Natl Acad Sci 98: 10439–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossec JC, Simon A, Marquer C, Moldrich RX, Leterrier C, Rossier J, Duyckaerts C, Lenkei Z, Potier MC 2010. Clathrin-dependent APP endocytosis and Aβ secretion are highly sensitive to the level of plasma membrane cholesterol. Biochim Biophys Acta 1801: 846–852 [DOI] [PubMed] [Google Scholar]
- Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, Yang AJ 2006. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J Biol Chem 281: 10825–10838 [DOI] [PubMed] [Google Scholar]
- Cuervo AM 2004. Autophagy: Many paths to the same end. Mol Cell Biochem 263: 55–72 [DOI] [PubMed] [Google Scholar]
- Cuervo AM, Palmer A, Rivett AJ, Knecht E 1995. Degradation of proteasomes by lysosomes in rat liver. Eur J Biochem 227: 792–800 [DOI] [PubMed] [Google Scholar]
- David DC, Layfield R, Serpell L, Narain Y, Goedert M, Spillantini MG 2002. Proteasomal degradation of tau protein. J Neurochem 83: 176–185 [DOI] [PubMed] [Google Scholar]
- Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC 2003. NGF signaling in sensory neurons: Evidence that early endosomes carry NGF retrograde signals. Neuron 39: 69–84 [DOI] [PubMed] [Google Scholar]
- Delcroix JD, Valletta J, Wu C, Howe CL, Lai CF, Cooper JD, Belichenko PV, Salehi A, Mobley WC 2004. Trafficking the NGF signal: Implications for normal and degenerating neurons. Prog Brain Res 146: 3–23 [DOI] [PubMed] [Google Scholar]
- Dhaka A, Costa RM, Hu H, Irvin DK, Patel A, Kornblum HI, Silva AJ, O'Dell TJ, Colicelli J 2003. The RAS effector RIN1 modulates the formation of aversive memories. J Neurosci 23: 748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey CA, Yue M, Lin WL, Dickson DW, Dunmore JH, Lee WC, Zehr C, West G, Cao S, Clark AM, et al. 2006. Deletion of the ubiquitin ligase CHIP leads to the accumulation, but not the aggregation, of both endogenous phospho- and caspase-3-cleaved tau species. J Neurosci 26: 6985–6996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I, Johansen T, Kirkin V 2010. Selective autophagy in cancer development and therapy. Cancer Res 70: 3431–3434 [DOI] [PubMed] [Google Scholar]
- Dolan PJ, Johnson GV 2010. A caspase cleaved form of tau is preferentially degraded through the autophagy pathway. J Biol Chem 285: 21978–21987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckman EA, Eckman CB 2005. Aβ-degrading enzymes: Modulators of Alzheimer's disease pathogenesis and targets for therapeutic intervention. Biochem Soc Trans 33: 1101–1105 [DOI] [PubMed] [Google Scholar]
- Finley D, Ciechanover A, Varshavsky A 1984. Thermolability of ubiquitin-activating enzyme from the mammalian cell cycle mutant ts85. Cell 37: 43–55 [DOI] [PubMed] [Google Scholar]
- Fischer DF, De Vos RA, Van Dijk R, De Vrij FM, Proper EA, Sonnemans MA, Verhage MC, Sluijs JA, Hobo B, Zouambia M, et al. 2003. Disease-specific accumulation of mutant ubiquitin as a marker for proteasomal dysfunction in the brain. FASEB J 17: 2014–2024 [DOI] [PubMed] [Google Scholar]
- Fischer DF, van Dijk R, van Tijn P, Hobo B, Verhage MC, van der Schors RC, Li KW, van Minnen J, Hol EM, van Leeuwen FW 2009. Long-term proteasome dysfunction in the mouse brain by expression of aberrant ubiquitin. Neurobiol Aging 30: 847–863 [DOI] [PubMed] [Google Scholar]
- Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD 2007. Decreased insulin-receptor signaling promotes the autophagic degradation of β-amyloid peptide in C. elegans. Autophagy 3: 569–580 [DOI] [PubMed] [Google Scholar]
- Fortun J, Dunn WA Jr, Joy S, Li J, Notterpek L 2003. Emerging role for autophagy in the removal of aggresomes in Schwann cells. J Neurosci 23: 10672–10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frautschy SA, Yang F, Calderon L, Cole GM 1996. Rodent models of Alzheimer's disease: Rat A β infusion approaches to amyloid deposits. Neurobiol Aging 17: 311–321 [DOI] [PubMed] [Google Scholar]
- Frautschy SA, Horn DL, Sigel JJ, Harris-White ME, Mendoza JJ, Yang F, Saido TC, Cole GM 1998. Protease inhibitor coinfusion with amyloid β-protein results in enhanced deposition and toxicity in rat brain. J Neurosci 18: 8311–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallastegui N, Groll M 2010. The 26S proteasome: Assembly and function of a destructive machine. Trends Biochem Sci 35: 634–642 [DOI] [PubMed] [Google Scholar]
- Garcia-Arencibia M, Hochfeld WE, Toh PP, Rubinsztein DC 2010. Autophagy, a guardian against neurodegeneration. Semin Cell Dev Biol 21: 691–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Alldred MJ, Counts SE, Cataldo AM, Neve RL, Jiang Y, Wuu J, Chao MV, Mufson EJ, Nixon RA, et al. 2010. Microarray analysis of hippocampal CA1 neurons implicates early endosomal dysfunction during Alzheimer's disease progression. Biol Psychiat 68: 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glabe C 2001. Intracellular mechanisms of amyloid accumulation and pathogenesis in Alzheimer's disease. J Mol Neurosci 17: 137–145 [DOI] [PubMed] [Google Scholar]
- Goldberg AL 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899 [DOI] [PubMed] [Google Scholar]
- Gómez-Isla T, Hollister R, West H, Mui S, Growdon JH, Petersen RC, Parisi JE, Hyman BT 1997. Neuronal loss correlates with but exceeds neurofibrillary tangles in Alzheimer's disease. Ann Neurol 41: 17–24 [DOI] [PubMed] [Google Scholar]
- Gonatas NK 1967. Neocortical synapses in a presenile dementia. J Neuropathol Exp Neurol 26: 150–151 [PubMed] [Google Scholar]
- Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, Moolman D, Zhang H, Shelanski M, Arancio O 2006. Ubiquitin hydrolase Uch-L1 rescues β-amyloid-induced decreases in synaptic function and contextual memory. Cell 126: 775–788 [DOI] [PubMed] [Google Scholar]
- Gordon PB, Seglen PO 1988. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun 151: 40–47 [DOI] [PubMed] [Google Scholar]
- Gout I, Middleton G, Adu J, Ninkina NN, Drobot LB, Filonenko V, Matsuka G, Davies AM, Waterfield M, Buchman VL 2000. Negative regulation of PI 3-kinase by Ruk, a novel adaptor protein. EMBO J 19: 4015–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbovic OM, Mathews PM, Jiang Y, Schmidt SD, Dinakar R, Summers-Terio NB, Ceresa BP, Nixon RA, Cataldo AM 2003. Rab5-stimulated up-regulation of the endocytic pathway increases intracellular β-cleaved amyloid precursor protein carboxyl-terminal fragment levels and Aβ production. J Biol Chem 278: 31261–31268 [DOI] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, et al. 2010. The Angelman syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell 140: 704–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundke-Iqbal I, Iqbal K, Quinlan M, Tung YC, Zaidi MS, Wisniewski HM 1986. Microtubule-associated protein tau. A component of Alzheimer paired helical filaments. J Biol Chem 261: 6084–6089 [PubMed] [Google Scholar]
- Grune T, Shringarpure R, Sitte N, Davies K 2001. Age-related changes in protein oxidation and proteolysis in mammalian cells. J Gerontol A Biol Sci Med Sci 56: B459–B467 [DOI] [PubMed] [Google Scholar]
- Guenette SY 2003. Mechanisms of Aβ clearance and catabolism. Neuromol Med 4: 147–160 [DOI] [PubMed] [Google Scholar]
- Haapasalo A, Viswanathan J, Bertram L, Soininen H, Tanzi RE, Hiltunen M 2010. Emerging role of Alzheimer's disease-associated ubiquilin-1 in protein aggregation. Biochem Soc Trans 38: 150–155 [DOI] [PubMed] [Google Scholar]
- Haas AL, Bright PM 1985. The immunochemical detection and quantitation of intracellular ubiquitin–protein conjugates. J Biol Chem 260: 12464–12473 [PubMed] [Google Scholar]
- Hamano T, Gendron TF, Causevic E, Yen SH, Lin WL, Isidoro C, Deture M, Ko LW 2008. Autophagic–lysosomal perturbation enhances tau aggregation in transfectants with induced wild-type tau expression. Eur J Neurosci 27: 1119–30 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Morishima-Kawashima M, Takio K, Suzuki M, Titani K, Ihara Y 1992. Protein sequence and mass spectrometric analyses of tau in the Alzheimer's disease brain. J Biol Chem 267: 17047–17054 [PubMed] [Google Scholar]
- Hasegawa M, Fujiwara H, Nonaka T, Wakabayashi K, Takahashi H, Lee VM, Trojanowski JQ, Mann D, Iwatsubo T 2002. Phosphorylated α-synuclein is ubiquitinated in α-synucleinopathy lesions. J Biol Chem 277: 49071–49076 [DOI] [PubMed] [Google Scholar]
- Hegde AN, Goldberg AL, Schwartz JH 1993. Regulatory subunits of cAMP-dependent protein kinases are degraded after conjugation to ubiquitin: A molecular mechanism underlying long-term synaptic plasticity. Proc Natl Acad Sci 90: 7436–7440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Inokuchi K, Pei W, Casadio A, Ghirardi M, Chain DG, Martin KC, Kandel ER, Schwartz JH 1997. Ubiquitin C-terminal hydrolase is an immediate-early gene essential for long-term facilitation in Aplysia. Cell 89: 115–126 [DOI] [PubMed] [Google Scholar]
- Heinrich M, Wickel M, Schneider-Brachert W, Sandberg C, Gahr J, Schwandner R, Weber T, Saftig P, Peters C, Brunner J, et al. 1999. Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J 18: 5252–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Jones L, Owen MJ, Williams J 2011. Alzheimer's disease genetics: Current knowledge and future challenges. Int J Geriatr Psychiat 26: 793–802 [DOI] [PubMed] [Google Scholar]
- Hsia AY, Masliah E, McConlogue L, Yu GQ, Tatsuno G, Hu K, Kholodenko D, Malenka RC, Nicoll RA, Mucke L 1999. Plaque-independent disruption of neural circuits in Alzheimer's disease mouse models. Proc Natl Acad Sci 96: 3228–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Pickering E, Liu YC, Hall S, Fournier H, Katz E, Dechairo B, John S, Van Eerdewegh P, Soares H 2011. Meta-analysis for genome-wide association study identifies multiple variants at the BIN1 locus associated with late-onset Alzheimer's disease. PLoS One 6: e16616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH 2010. The ESCRT complexes. Crit Rev Biochem Mol Biol 45: 463–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara Y, Abraham C, Selkoe DJ 1983. Antibodies to paired helical filaments in Alzheimer's disease do not recognize normal brain proteins. Nature 304: 727–729 [DOI] [PubMed] [Google Scholar]
- Ivy GO, Schottler F, Wenzel J, Baudry M, Lynch G 1984. Inhibitors of lysosomal enzymes: Accumulation of lipofuscin-like dense bodies in the brain. Science 226: 985–987 [DOI] [PubMed] [Google Scholar]
- Iwatsubo T, Hasegawa M, Esaki Y, Ihara Y 1992. Lack of ubiquitin immunoreactivities at both ends of neuropil threads—Possible bidirectional growth of neuropil threads. Am J Pathol 140: 277–282 [PMC free article] [PubMed] [Google Scholar]
- Ji ZS, Miranda RD, Newhouse YM, Weisgraber KH, Huang Y, Mahley RW 2002. Apolipoprotein E4 potentiates amyloid β peptide-induced lysosomal leakage and apoptosis in neuronal cells. J Biol Chem 277: 21821–21828 [DOI] [PubMed] [Google Scholar]
- Ji ZS, Mullendorff K, Cheng IH, Miranda RD, Huang Y, Mahley RW 2006. Reactivity of apolipoprotein E4 and amyloid β peptide: Lysosomal stability and neurodegeneration. J Biol Chem 281: 2683–2692 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Mullaney KA, Peterhoff CM, Che S, Schmidt SD, Boyer-Boiteau A, Ginsberg SD, Cataldo AM, Mathews PM, Nixon RA 2010. Alzheimer's-related endosome dysfunction in Down syndrome is Aβ-independent but requires APP and is reversed by BACE-1 inhibition. Proc Natl Acad Sci 107: 1630–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Shepardson N, Yang T, Chen G, Walsh D, Selkoe DJ 2011. Soluble amyloid β-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc Natl Acad Sci 108: 5819–5824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck S, Nitsch R, Grune T, Ullrich O 2003. Proteasome inhibition by paired helical filament-tau in brains of patients with Alzheimer's disease. J Neurochem 85: 115–122 [DOI] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Markesbery WR 2000. Impaired proteasome function in Alzheimer's disease. J Neurochem 75: 436–439 [DOI] [PubMed] [Google Scholar]
- Keller JN, Gee J, Ding Q 2002. The proteasome in brain aging. Ageing Res Rev 1: 279–293 [DOI] [PubMed] [Google Scholar]
- Kelly BT, Owen DJ 2011. Endocytic sorting of transmembrane protein cargo. Curr Opin Cell Biol 23: 404–412 [DOI] [PubMed] [Google Scholar]
- Kessels HW, Malinow R 2009. Synaptic AMPA receptor plasticity and behavior. Neuron 61: 340–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana V, Elson-Schwab I, Fulga TA, Sharp KA, Loewen CA, Mulkearns E, Tyynela J, Scherzer CR, Feany MB 2010. Lysosomal dysfunction promotes cleavage and neurotoxicity of tau in vivo. PLoS Genet 6: e1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, Gotow T, Peters C, von Figura K, Mizushima N, et al. 2005. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Am J Pathol 167: 1713–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo J, Honda T, Mori H, Hamada Y, Miura R, Ogawara M, Ihara Y 1988. The carboxyl third of tau is tightly bound to paired helical filaments. Neuron 1: 827–834 [DOI] [PubMed] [Google Scholar]
- Korolchuk VI, Mansilla A, Menzies FM, Rubinsztein DC 2009a. Autophagy inhibition compromises degradation of ubiquitin–proteasome pathway substrates. Mol Cell 33: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk VI, Menzies FM, Rubinsztein DC 2009b. A novel link between autophagy and the ubiquitin–proteasome system. Autophagy 5: 862–863 [DOI] [PubMed] [Google Scholar]
- Kosaka K 1978. Lewy bodies in cerebral cortex, report of three cases. Acta Neuropathol 42: 127–134 [DOI] [PubMed] [Google Scholar]
- Kosik KS, Joachim CL, Selkoe DJ 1986. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical fragments in Alzheimer disease. Proc Natl Acad Sci 83: 4044–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Jaattela M 2005. Lysosomes and autophagy in cell death control. Nat Rev Cancer 5: 886–97 [DOI] [PubMed] [Google Scholar]
- Kuzuhara S, Mori H, Izumiyama N, Yoshimura M, Ihara Y 1988. Lewy bodies are ubiquitinated. A light and electron microscopic immunocytochemical study. Acta Neuropathol 75: 345–353 [DOI] [PubMed] [Google Scholar]
- Laifenfeld D, Patzek LJ, McPhie DL, Chen Y, Levites Y, Cataldo AM, Neve RL 2007. Rab5 mediates an amyloid precursor protein signaling pathway that leads to apoptosis. J Neurosci 27: 7141–7153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Pickart CM, Alban A, Landon M, Jamieson C, Ramage R, Mayer RJ, Layfield R 2000. Inhibition of the ubiquitin–proteasome system in Alzheimer's disease. Proc Natl Acad Sci 97: 9902–9906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen KE, Sulzer D 2002. Autophagy in neurons: A review. Histol Histopathol 17: 897–908 [DOI] [PubMed] [Google Scholar]
- LeBlanc AC, Goodyer CG 1999. Role of endoplasmic reticulum, endosomal–lysosomal compartments, and microtubules in amyloid precursor protein metabolism of human neurons. J Neurochem 72: 1832–42 [DOI] [PubMed] [Google Scholar]
- Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Marinez-Vicente M, Massey AG, Sovak G, Uchiyama Y, et al. 2010. Presenilin 1 (PS1) is required for v-ATPase targeting and autolysosome acidification: PS1 mutations in Alzheimer's disease disrupt lysosomal proteolysis and autophagy. Cell 141: 1146–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Sato Y, Nixon RA 2011a. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer's-like axonal dystrophy. J Neurosci 31: 7817–7830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Saito Y, Nixon RA 2011b. Primary lysosomal dysfunction causes cargo-specific deficits of axonal transport leading to Alzheimer's-like neuritic dystrophy. Autophagy 7: in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leil TA, Chen ZW, Chang CS, Olsen RW 2004. GABAA receptor-associated protein traffics GABAA receptors to the plasma membrane in neurons. J Neurosci 24: 11429–11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ 2004. Development by self-digestion; molecular mechanisms and biological functions of autophagy. Dev Cell 6: 463–477 [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW 2011. Autophagy in immunity and inflammation. Nature 469: 323–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hong S, Shepardson NE, Walsh DM, Shankar GM, Selkoe D 2009. Soluble oligomers of amyloid β protein facilitate hippocampal long-term depression by disrupting neuronal glutamate uptake. Neuron 62: 788–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JP, Gleeson PA 2011. Macropinocytosis: An endocytic pathway for internalising large gulps. Immunol Cell Biol 10.1038/icb.2011.20 [DOI] [PubMed] [Google Scholar]
- Ling D, Song HJ, Garza D, Neufeld TP, Salvaterra PM 2009. Aβ42-induced neurodegeneration via an age-dependent autophagic–lysosomal injury in Drosophila. PLoS One 4: e4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, et al. 2010. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer's disease. Proc Natl Acad Sci 107: 14164–14169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Salon M, Alonso M, Vianna MR, Viola H, Mello e Souza T, Izquierdo I, Pasquini JM, Medina JH 2001. The ubiquitin–proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci 4: 1820–1826 [DOI] [PubMed] [Google Scholar]
- Lowe J, Blanchard A, Morrell K, Lennox G, Reynolds L, Billett M, Landon M, Mayer RJ 1988. Ubiquitin is a common factor in intermediate filament inclusion bodies of diverse type in man, including those of Parkinson's disease, Pick's disease, and Alzheimer's disease, as well as Rosenthal fibres in cerebellar astrocytomas, cytoplasmic bodies in muscle, and mallory bodies in alcoholic liver disease. J Pathol 155: 9–15 [DOI] [PubMed] [Google Scholar]
- *.Mandelkow E-M, Mandelkow E 2011. Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margallo-Lana M, Morris CM, Gibson AM, Tan AL, Kay DW, Tyrer SP, Moore BP, Ballard CG 2004. Influence of the amyloid precursor protein locus on dementia in Down syndrome. Neurology 62: 1996–1998 [DOI] [PubMed] [Google Scholar]
- Masliah E, Mallory M, Deerinck T, DeTeresa R, Lamont S, Miller A, Terry RD, Carragher B, Ellisman M 1993. Re-evaluation of the structural organization of the neuritic plaques in Alzheimer's disease. J Neuropathol Exp Neurol 52: 619–632 [DOI] [PubMed] [Google Scholar]
- Mathews PM, Guerra CB, Jiang Y, Grbovic OM, Kao BH, Schmidt SD, Dinakar R, Mercken M, Hille-Rehfeld A, Rohrer J, et al. 2002. Alzheimer's disease-related overexpression of the cation-dependent mannose 6-phosphate receptor increases Aβ secretion: Role for altered lysosomal hydrolase distribution in β-amyloidogenesis. J Biol Chem 277: 5299–5307 [DOI] [PubMed] [Google Scholar]
- McCray BA, Taylor JP 2008. The role of autophagy in age-related neurodegeneration. Neurosignals 16: 75–84 [DOI] [PubMed] [Google Scholar]
- Missotten M, Nichols A, Rieger K, Sadoul R 1999. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ 6: 124–129 [DOI] [PubMed] [Google Scholar]
- Mori H, Kondo J, Ihara Y 1987. Ubiquitin is a component of paired helical filaments in Alzheimer's disease. Science 235: 1641–1644 [DOI] [PubMed] [Google Scholar]
- Morimoto T, Ide T, Ihara Y, Tamura A, Kirino T 1996. Transient ischemia depletes free ubiquitin in the gerbil hippocampal CA1 neurons. Am J Pathol 148: 249–257 [PMC free article] [PubMed] [Google Scholar]
- Morishima-Kawashima M, Hasegawa M, Takio K, Suzuki M, Titani K, Ihara Y 1993. Ubiquitin is conjugated with amino-terminally processed tau in paired helical filaments. Neuron 10: 1151–1160 [DOI] [PubMed] [Google Scholar]
- Mueller-Steiner S, Zhou Y, Arai H, Roberson ED, Sun B, Chen J, Wang X, Yu G, Esposito L, Mucke L, et al. 2006. Antiamyloidogenic and neuroprotective functions of cathepsin B: Implications for Alzheimer's disease. Neuron 51: 703–714 [DOI] [PubMed] [Google Scholar]
- Mullins C, Bonifacino JS 2001. The molecular machinery for lysosome biogenesis. Bioessays 23: 333–343 [DOI] [PubMed] [Google Scholar]
- Murayama S, Mori H, Ihara Y, Bouldin TW, Suzuki K, Tomonaga M 1990a. Immunocytochemical and ultrastructural studies of lower motor neurons in amyotrophic lateral sclerosis. Ann Neurol 27: 137–148 [DOI] [PubMed] [Google Scholar]
- Murayama S, Mori H, Ihara Y, Tomonaga M 1990b. Immunocytochemical and ultrastructural studies of Pick's disease. Ann Neurol 27: 394–405 [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ 2009. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy 5: 706–708 [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133 [DOI] [PubMed] [Google Scholar]
- Nishi T, Forgac M 2002. The vacuolar (H+)-ATPases-nature's most versatile proton pumps. Nat Rev Mol Cell Biol 3: 94–103 [DOI] [PubMed] [Google Scholar]
- Nixon RA 2004. Niemann–Pick Type C disease and Alzheimer's disease: The APP-endosome connection fattens up. Am J Pathol 164: 757–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA 2007. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci 120: 4081–4091 [DOI] [PubMed] [Google Scholar]
- Nixon RA, Cataldo AM 2006. Lysosomal system pathways: Genes to neurodegeneration in Alzheimer's disease. J Alzheimer’s Dis 9: 277–289 [DOI] [PubMed] [Google Scholar]
- Nixon RA, Yang DS 2011. Autophagy failure in Alzheimer's disease—Locating the primary defect. Neurobiol Dis 43: 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM 2005. Extensive involvement of autophagy in Alzheimer disease: An immuno-electron microscopy study. J Neuropathol Exp Neurol 64: 113–122 [DOI] [PubMed] [Google Scholar]
- Nixon RA, Yang DS, Lee JH 2008. Neurodegenerative lysosomal disorders: A continuum from development to late age. Autophagy 4: 590–599 [DOI] [PubMed] [Google Scholar]
- Nukina N, Ihara Y 1986. One of the antigenic determinants of paired helical filaments is related to tau protein. J Biochem 99: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Nukina N, Kosik KS, Selkoe DJ 1987. Recognition of Alzheimer paired helical filaments by monoclonal neurofilament antibodies is due to crossreaction with tau protein. Proc Natl Acad Sci 84: 3415–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmi K, Kudo LC, Ryazantsev S, Zhao HZ, Karsten SL, Neufeld EF 2009. Sanfilippo syndrome type B, a lysosomal storage disease, is also a tauopathy. Proc Natl Acad Sci 106: 8332–8337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y 2001. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat Rev Mol Cell Biol 2: 211–216 [DOI] [PubMed] [Google Scholar]
- Overly CC, Hollenbeck PJ 1996. Dynamic organization of endocytic pathways in axons of cultured sympathetic neurons. J Neurosci 16: 6056–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine S, Bedford L, Thorpe JR, Mayer RJ, Cavey JR, Bajaj N, Sheppard PW, Lowe J, Layfield R 2009. Immunoreactivity to Lys63-linked polyubiquitin is a feature of neurodegeneration. Neurosci Lett 460: 205–208 [DOI] [PubMed] [Google Scholar]
- Peralvarez-Marin A, Barth A, Graslund A 2008. Time-resolved infrared spectroscopy of pH-induced aggregation of the Alzheimer Aβ1-28 peptide. J Mol Biol 379: 589–596 [DOI] [PubMed] [Google Scholar]
- Petrucelli L, Dickson D, Kehoe K, Taylor J, Snyder H, Grover A, De Lucia M, McGowan E, Lewis J, Prihar G, et al. 2004. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet 13: 703–714 [DOI] [PubMed] [Google Scholar]
- Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, et al. 2008. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid β accumulation in mice. J Clin Invest 118: 2190–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pike JW, Donaldson CA, Marion SL, Haussler MR 1982. Development of hybridomas secreting monoclonal antibodies to the chicken intestinal 1 α,25-dihydroxyvitamin D3 receptor. Proc Natl Acad Sci 79: 7719–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillay CS, Elliott E, Dennison C 2002. Endolysosomal proteolysis and its regulation. Biochem J 363: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Luzio JP 2001. Late endosomes: Sorting and partitioning in multivesicular bodies. Traffic 2: 612–621 [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A 2010. Identification of the switch in early-to-late endosome transition. Cell 141: 497–508 [DOI] [PubMed] [Google Scholar]
- Rovelet-Lecrux A, Hannequin D, Raux G, Le Meur N, Laquerriere A, Vital A, Dumanchin C, Feuillette S, Brice A, Vercelletto M, et al. 2006. APP locus duplication causes autosomal dominant early-onset Alzheimer disease with cerebral amyloid angiopathy. Nat Genet 38: 24–26 [DOI] [PubMed] [Google Scholar]
- Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA 2006. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci 26: 1711–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy AK, Oh T, Rivera O, Mubiru J, Song CS, Chatterjee B 2002. Impacts of transcriptional regulation on aging and senescence. Ageing Res Rev 1: 367–380 [DOI] [PubMed] [Google Scholar]
- Rusten TE, Simonsen A 2008. ESCRT functions in autophagy and associated disease. Cell Cycle 7: 1166–1172 [DOI] [PubMed] [Google Scholar]
- Ryazantsev S, Yu WH, Zhao HZ, Neufeld EF, Ohmi K 2007. Lysosomal accumulation of SCMAS (subunit c of mitochondrial ATP synthase) in neurons of the mouse model of mucopolysaccharidosis III B. Mol Genet Metab 90: 393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu R, Kaushik S, Clement CC, Cannizzo ES, Scharf B, Follenzi A, Potolicchio I, Nieves E, Cuervo AM, Santambrogio L 2011. Microautophagy of cytosolic proteins by late endosomes. Dev Cell 20: 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Saido T, Leissring MA 2011. Proteolytic degradation of amyloid β-protein. Cold Spring Harb Perspect [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt ML, DiDario AG, Lee VM, Trojanowski JQ 1994. An extensive network of PHF tau-rich dystrophic neurites permeates neocortex and nearly all neuritic and diffuse amyloid plaques in Alzheimer disease. FEBS Lett 344: 69–73 [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, et al. 2010. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303: 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setsuie R, Wada K 2007. The functions of UCH-L1 and its relation to neurodegenerative diseases. Neurochem Int 51: 105–111 [DOI] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science 332: 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA 2009. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 186: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, Del-Favero J, Cruts M, van Duijn CM, Van Broeckhoven C 2006. APP duplication is sufficient to cause early onset Alzheimer's dementia with cerebral amyloid angiopathy. Brain 129: 2977–2983 [DOI] [PubMed] [Google Scholar]
- Smemo S, Nowotny P, Hinrichs AL, Kauwe JS, Cherny S, Erickson K, Myers AJ, Kaleem M, Marlowe L, Gibson AM, et al. 2006. Ubiquilin 1 polymorphisms are not associated with late-onset Alzheimer's disease. Ann Neurol 59: 21–26 [DOI] [PubMed] [Google Scholar]
- Sohal RS, Mockett RJ, Orr WC 2002. Mechanisms of aging: An appraisal of the oxidative stress hypothesis. Free Radic Biol Med 33: 575–586 [DOI] [PubMed] [Google Scholar]
- Soto C 2003. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat Rev Neurosci 4: 49–60 [DOI] [PubMed] [Google Scholar]
- Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M 1997. Alpha-synuclein in Lewy bodies. Nature 388: 839–840 [DOI] [PubMed] [Google Scholar]
- Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, Richardson A, Strong R, Galvan V 2010. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-β levels in a mouse model of Alzheimer's disease. PLoS One 5: e9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Zhou Y, Halabisky B, Lo I, Cho SH, Mueller-Steiner S, Devidze N, Wang X, Grubb A, Gan L 2008. Cystatin C-cathepsin B axis regulates amyloid β levels and associated neuronal deficits in an animal model of Alzheimer's disease. Neuron 60: 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Tavernarakis N 2003. The biochemistry of neuronal necrosis: Rogue biology? Nat Rev Neurosci 4: 672–684 [DOI] [PubMed] [Google Scholar]
- Takahashi RH, Milner TA, Li F, Nam EE, Edgar MA, Yamaguchi H, Beal MF, Xu H, Greengard P, Gouras GK 2002. Intraneuronal Alzheimer aβ42 accumulates in multivesicular bodies and is associated with synaptic pathology. Am J Pathol 161: 1869–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tank EM, True HL 2009. Disease-associated mutant ubiquitin causes proteasomal impairment and enhances the toxicity of protein aggregates. PLoS Genet 5: e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terman A 2001. Garbage catastrophe theory of aging: Imperfect removal of oxidative damage? Redox Rep 6: 15–26 [DOI] [PubMed] [Google Scholar]
- Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R 1991. Physical basis of cognitive alterations in Alzheimer's disease: Synapse loss is the major correlate of cognitive impairment. Ann Neurol 30: 572–580 [DOI] [PubMed] [Google Scholar]
- Tian Y, Bustos V, Flajolet M, Greengard P 2011. A small-molecule enhancer of autophagy decreases levels of A{β} and APP-CTF via Atg5-dependent autophagy pathway. FASEB J 25: 1934–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP 2010. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 6: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troen BR 2003. The biology of aging. Mt Sinai J Med 70: 3–22 [PubMed] [Google Scholar]
- Tseng BP, Green KN, Chan JL, Blurton-Jones M, LaFerla FM 2008. Aβ inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol Aging 29: 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen FW, de Kleijn DP, van den Hurk HH, Neubauer A, Sonnemans MA, et al. 1998. Frameshift mutants of β amyloid precursor protein and ubiquitin-B in Alzheimer's and Down patients. Science 279: 242–247 [DOI] [PubMed] [Google Scholar]
- Vito P, Lacana E, D'Adamio L 1996. Interfering with apoptosis: Ca2+-binding protein ALG-2 and Alzheimer's disease gene ALG-3. Science 271: 521–525 [DOI] [PubMed] [Google Scholar]
- Vito P, Pellegrini L, Guiet C, D'Adamio L 1999. Cloning of AIP1, a novel protein that associates with the apoptosis-linked gene ALG-2 in a Ca2+-dependent reaction. J Biol Chem 274: 1533–1540 [DOI] [PubMed] [Google Scholar]
- Walsh DM, Selkoe DJ 2004. Deciphering the molecular basis of memory failure in Alzheimer's disease. Neuron 44: 181–189 [DOI] [PubMed] [Google Scholar]
- Wang Y, Martinez-Vicente M, Kruger U, Kaushik S, Wong E, Mandelkow EM, Cuervo AM, Mandelkow E 2009. Tau fragmentation, aggregation and clearance: The dual role of lysosomal processing. Hum Mol Genet 18: 4153–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z 2011. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 80: 125–156 [DOI] [PubMed] [Google Scholar]
- Wischik CM, Crowther RA, Stewart M, Roth M 1985. Subunit structure of paired helical filaments in Alzheimer's disease. J Cell Biol 100: 1905–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E, Cuervo AM 2010. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci 13: 805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Mirra SS, Pollock NJ, Binder LI 1986. Neurofibrillary tangles of Alzheimer disease share antigenic determinants with the axonal microtubule-associated protein tau (tau). Proc Natl Acad Sci 83: 4040–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Peng J 2006. Dissecting the ubiquitin pathway by mass spectometry. Biochim Biophys Acta 1764: 1940–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Peng J 2008. Characterization of polyubiquitin chain structure by middle-down mass spectrometry. Anal Chem 80: 3438–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AJ, Chandswangbhuvana D, Margol L, Glabe CG 1998. Loss of endosomal/lysosomal membrane impermeability is an early event in amyloid Aβ1–42 pathogenesis. J Neurosci Res 52: 691–698 [DOI] [PubMed] [Google Scholar]
- Yang DS, Kumar A, Stavrides P, Peterson J, Peterhoff CM, Pawlik M, Levy E, Cataldo AM, Nixon RA 2008. Neuronal apoptosis and autophagy cross talk in aging PS/APP mice, a model of Alzheimer's disease. Am J Pathol 173: 665–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DS, Stavrides P, Mohan PS, Kaushik S, Kumar A, Ohno M, Schmidt SD, Wesson D, Bandyopadhyay U, Jiang Y, et al. 2011. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer's disease ameliorates amyloid pathologies and memory deficits. Brain 134: 258–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii SR, Kishi C, Ishihara N, Mizushima N 2011. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem 286: 19630–19640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP 2011. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12: 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee J-H, Mohan PS, Mercken M, Farmery MR, Tjernberg LO, et al. 2005. Macroautophagy—A novel β-amyloid peptide-generating pathway activated in Alzheimer's disease. J Cell Biol 171: 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M 2008. Endocytic mechanisms and drug discovery in neurodegenerative diseases. Front Biosci 13: 6086–6105 [DOI] [PubMed] [Google Scholar]
- Zhang JY, Liu SJ, Li HL, Wang JZ 2005. Microtubule-associated protein tau is a substrate of ATP/Mg2+-dependent proteasome protease system. J Neural Transm 112: 547–555 [DOI] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL 2007. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab 6: 472–483 [DOI] [PubMed] [Google Scholar]