Abstract
Autophagy is a conserved constitutive cellular process, responsible for the degradation of dysfunctional proteins and organelles. Autophagy plays a role in many diseases such as neurodegeneration and cancer; however, to date, conventional autophagy detection techniques are not suitable for clinical samples. We have developed a high throughput, statistically robust technique that quantitates autophagy in primary human leukocytes using the Image stream, an imaging flow cytometer. We validate this method on cell lines and primary cells knocked down for essential autophagy genes. Also, using this method we show that T cells have higher autophagic activity than B cells. Furthermore our results indicate that healthy primary senescent CD8+ T cells have decreased autophagic levels correlating with increased DNA damage, which may explain features of the senescent immune system and its declining function with age. This technique will allow us, for the first time, to measure autophagy levels in diseases with a known link to autophagy, while also determining the contribution of autophagy to the efficacy of drugs.
Keywords: ImageStream, PBMCs, T cells, autophagy detection, immunosenescence
Introduction
Macroautophagy (hereafter referred to as autophagy) is a dynamic housekeeping mechanism involved in cellular homeostasis. One of its functions is the degradation of cellular proteins and organelles via the lysosomal pathway. Autophagy can be induced by nutrient starvation, stress, hypoxia, reactive oxygen species (ROS) and toll-like receptor agonists. It is known to be involved in inflammation, aging, cancer, neurodegeneration, cardiac myopathies and infections.1,2
The autophagic machinery and the key molecular players (Atg genes) have been well characterized in yeast. The process starts with the formation of a double-membraned phagophore, which encircles cellular components, such as misfolded proteins, damaged organelles or recyclable cytoplasmic constituents, and forms an autophagosome. Autophagosomes then fuse with lysosomes or endosomes to form autolysosomes (also called amphisomes), where the intralumenal components are degraded by acid hydrolases. The inner and outer membranes of autophagosomes are festooned with microtubule-associated protein 1 light chain 3 (LC3) protein, the mammalian homolog of yeast Atg8/Apg8/Aut7 (LC3-I), which is post-translationally modified by a ubiquitin-like system to a lipidated form (LC3-II). From the inner membranes of autophagosomes, LC3-II is sequestered into autolysosomes before being recycled back into use.
The rate of synthesis, conversion (LC3-I to LC3-II) and turnover of LC3 are universally used to study the autophagic process in cells.1 In recent years several assays have been developed, typically in various cell lines, based on the turnover or number of punctate endogenous LC3 or GFP-LC3 structures per cell.3 Autophagy can be detected by counting LC3 puncta using confocal microscopy, by measuring lysosomal content, by LC3 western blots or by detection of double-membrane vesicles using transmission electron microscopy. These assays are not ideal to study mixed populations of primary cells like peripheral lymphocytes. Although there are flow cytometry-based autophagy assays reported, these were developed on fluorescent reporter constructs like GFP-LC3, again limiting them to use with cell lines.4,5 Here we present a novel autophagy assay based on imaging flow cytometry (ImageStream), which can be used on a mixed population of primary cells, takes the autophagic flux into account, is nonsubjective, quantitative and statistically robust.
Replicative senescence is characterized by cell cycle arrest that limits the proliferation of damaged cells and is a normal feature of every somatic cell. In the immune system, senescence is associated with a dramatic reduction in responsiveness and functional dysregulation. Although both the innate and adaptive immune system are affected by aging, the adaptive immune system, in particular T lymphocytes, are most susceptible to the deleterious effects of aging. Among T cells it has been studied primarily in CD8+ rather than CD4+ T cells during aging and chronic infections.6
Autophagy is known to decrease with age, and it is thought that this decline contributes to the process of senescence. However, while the age-related decline in chaperone-mediated autophagy has been documented in mammalian cells,7 most of the studies on autophagy have been performed in model organisms such as Drosophila, C.elegans and yeast.8 In particular, human immune cells have not been studied, presumably because of the lack of a suitable autophagy detection technique.
To address the impact of aging on autophagy, we applied this assay to peripheral blood mononuclear cells (PBMCs) from young (< 28 y) and old (> 60 y) healthy donors and show a significant decrease in both basal and starvation-induced autophagy levels in old donors. Additionally, we found that B cells have significantly lower basal levels of autophagy than T cells. We characterized senescent and nonsenescent T cells on the basis of known markers, i.e., CD57, CD28, Fas (CD95, apoptosis related molecule)9 and γH2AX (a marker for DNA double-strand breaks).10,11 We show that the senescent cells have low levels of autophagy and are incapable of inducing autophagy upon starvation, moreover that low levels of autophagy are correlated with increased DNA damage.
Results
Validation of ImageStream-based autophagy assay
It is well documented that both the relocalization of LC3 puncta to autophagosomal membranes and an increase in lysosomal content are hallmarks of autophagy.1 The detection of autophagy should ideally measure both of these hallmarks, and also include the delivery of LC3 to the lysosomes (i.e., the formation of autolysosomes). This delivery is reflected by the colocalization of LC3 and lysosomal markers. Both the fluorescence intensity of endogenous LC3 and a lysosomal marker, along with their colocalization index (= bright detail similarity, BDS) can be detected by the novel ImageStream technology as depicted in Figure S1.
ImageStream is a benchtop multispectral imaging flow cytometer designed for the acquisition of six channels of cellular imagery. It combines the per-cell information content provided by standard microscopy with the statistical significance afforded by the large sample sizes common to standard flow cytometry. With this system, fluorescence intensity measurements are acquired as with a conventional flow cytometer; however, it takes advantage of the system’s imaging abilities to locate and quantitate the distribution of signals on or within cells. This system is run on Inspire software and the data analysis is done using Ideas software (see the details under the methods section).
BDS is a feature that calculates the degree of overlapping pixel intensities taken from different channels of fluorescent imagery.12 BDS is the log-transformed Pearson’s correlation coefficient that is non-mean normalized and is applied to the open residue image. For this feature to be accurate, it is essential to gate on cells that are bright for both fluorescent markers of interest. As in flow cytometry, BDS gating was determined for each experiment; BDS cut-off was defined by a negative control generated from the appropriate cell type and its gating strategy. We used either mean BDS or % cells gated with BDShi (both show similar trends) on cells double positive for the lysosomal marker and LC3 as the read-out for autophagy levels, depending on the different cell types. To detect the autophagy, a continuous degradative process, the autophagic flux has to be arrested with lysosomal protease inhibitors. The addition of membrane-permeable cathepsin inhibitors E64d and Pepstatin A (PepA) resulted in accumulation of autolysosomes.1,3 Addition of lysosomal inhibitors also controls for lysosomal defects.
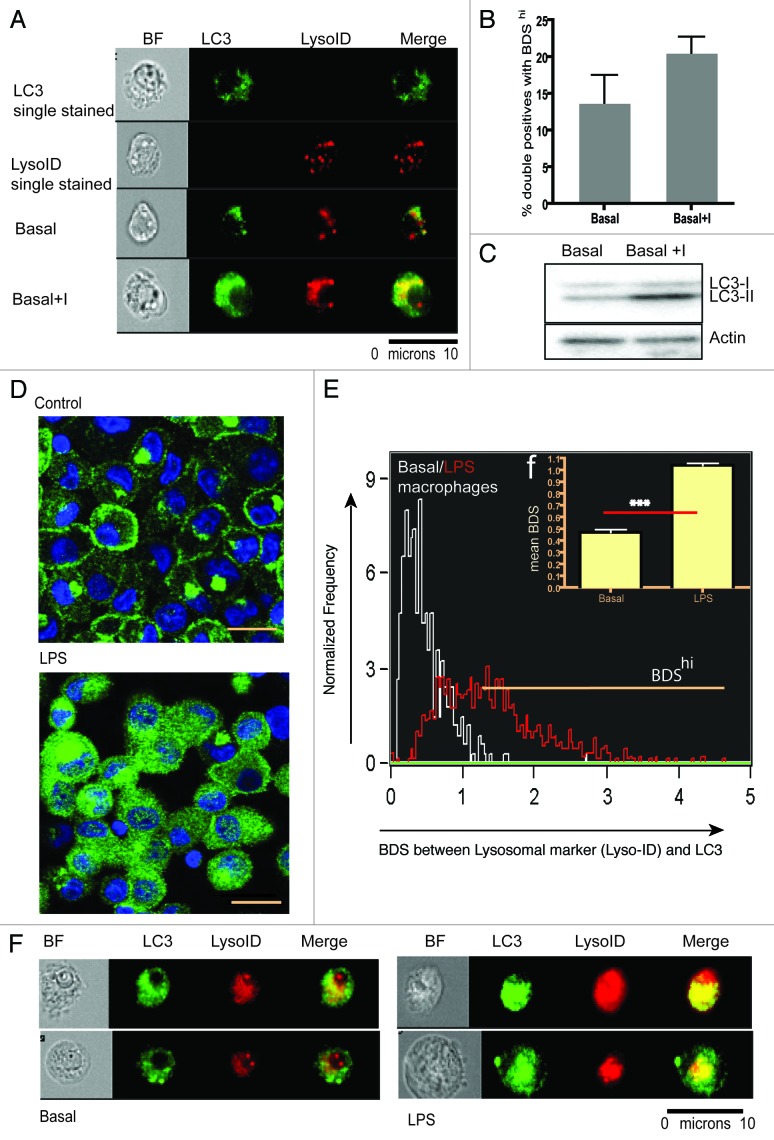
First, we compared this novel technique to the classical detection technique of western blotting using the HEK293 cell line and detection of endogenous LC3. As expected, western blotting of LC3-I and II indicate that basal levels of autophagy can be measured by lipidation of LC3-I to LC3-II when turnover is inhibited by lysosomal inhibitors E64d/PepA (Fig. 1C). Another aliquot of the same HEK293 cells was also analyzed by ImageStream. In Figure 1A we show examples of cells that were either single-stained for LC3 or lysosomal marker (Lyso-ID) or double-stained for these markers under basal and induced conditions. The number of cells positive for both markers (double positive) increased marginally when lysosomal turnover is inhibited. However, within the double positive population, the BDS of the two markers increased as it does in the western blot (Fig. 1B and C). The turnover in the ImageStream assay from Figure 1B (HEK cell line) is 30% compared with LC3 turnover (conversion of LC3-I to LC3-II) in the western blot of 60% (Fig. 1C). We believe that this technique does not directly assay the LC3-II turnover, rather it shows how much of LC3 is susceptible to degradation by lysosomes in a given cell population. LC3-II may increase by a certain amount and cause an increase in autophagic degradation or colocalization, as measured by our assay. But there can also be other factors such as transportation of LC3-II to lysosomes along with the rate of fusion and degradation of lysosomes that can affect colocalization of LC3 and lysosomes in a cell population, resulting in a different absolute value for a similar trend.
Figure 1. Basal autophagy levels in HEK293 cell line and monocyte-derived macrophages by conventional LC3 western blot, confocal microscopy and ImageStream. HEK293 cells were cultured in nutrient-rich medium with or without E64d/PepA. Cells were split into two aliquots, (A) representative ImageStream images of cells, either single stained (upper panels), or stained for both markers (bottom panels), without and with E64d/PepA (B) BDS histograms of gated double positive cells in basal medium or basal medium plus lysosomal inhibitors (Bar graph represents mean of triplicates ± SEM). (C) Western blot for LC3-I to LC3-II conversion for the same sample (D) Confocal images of monocyte-derived macrophages cultured in medium (upper panel) or treated with LPS (lower panel) (scale bar = 10 μm). (E) BDS overlay of monocyte-derived macrophages cultured in medium (white line) or treated with LPS (red line). (F) Mean BDS histogram on technical replicates for monocyte derived macrophages on (E) (***p < 0.0001). (F) Merged images of two representative cells/condition of monocyte-derived macrophages showing from left to right: bright field, LC3 (green), Lyso-ID (red) and a merge of LC3 and Lyso-ID.
To ascertain that this technique can also measure autophagy in primary cells, we generated a 90% pure population of monocyte-derived macrophages from healthy donors. There is ample evidence that toll-like receptor antagonists such as lipopolysaccharide (LPS) induce autophagy in dendritic cells and macrophages.13-15 We stimulated macrophages for 12 h with LPS and stained the monocyte derived macrophages for LC3 before visualizing them using confocal microscopy. An increase of LC3 puncta in LPS-treated cells is shown in Figure 1D. Although markers are present at basal conditions by ImageStream, treatment with LPS resulted in a distinct punctate bright pattern of LC3 and increased lysosomal staining, as demonstrated in the selected images of two cells per condition obtained by ImageStream showing bright field, each channel alone and the merged image (Fig. 1G). However, a more robust autophagy signal was obtained when LC3hiLysohi macrophages were analyzed for their colocalized pattern. This is represented as an overlaid BDS histogram (Basal vs. LPS) in Figure 1E and F.
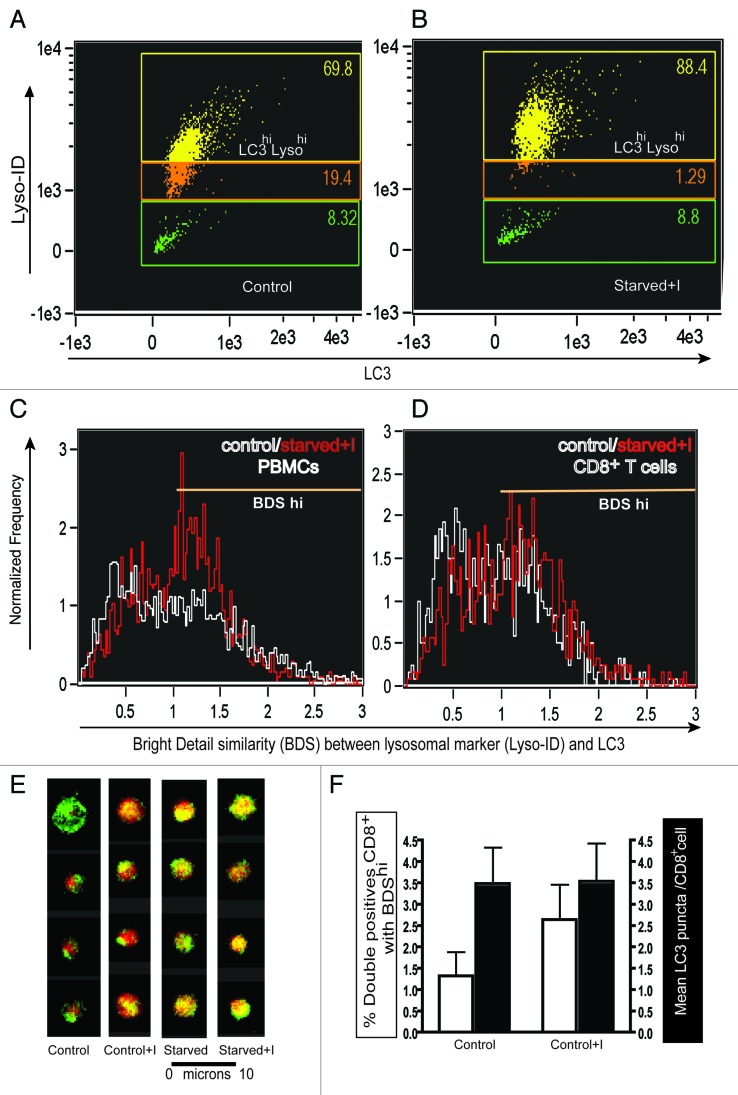
Next, we tested whether this assay works in primary blood-derived cells. We induced autophagy in PBMCs isolated from healthy donors. Cells were either cultured in medium, medium with E64d/PepA, starvation medium (HBSS) or HBSS with E64d/PepA for 2 h. Subsequently, PBMCs were stained with Lyso-ID and intracellular LC3. First, we demonstrated that the number of cells highly positive for LC3 and the lysosomal marker (double-positive population) increased upon inducing and blocking basal autophagy via lysosomal inhibitors (Fig. 2A and B). Clearly in these cells the increase of lysosomal staining was more pronounced than the increase in LC3 intensity. However, when we gated on the LC3hiLysohi (double positive) population and determined the BDS between the two probes for total PBMCs (Fig. 2C) or CD8+ T lymphocytes (Fig. 2D) cultured in medium (white line) or in starvation medium plus lysosomal inhibitors (red line), there was a clear increase in autolysosomes in the latter. In Figure 2E, a row of four representative PBMCs (out of 20,000 acquired) were chosen to show merged images for LC3 and Lyso-ID for all four conditions. Figure 2F shows a comparison between two different ways to analyze the ImageStream data: (1) double-positive cells with BDShi and (2) classical counting LC3 puncta. The former technique detects significantly increased autolysosomes when lysosomal inhibitors arrest the basal autophagic flux whereas the latter fails to do so.
Figure 2. The ImageStream-based autophagy assay detects autophagy in PBMCs. PBMCs from a healthy young donor, stained for LC3 and Lyso-ID after 2 h culture in (A) control medium (B) in starvation medium with lysosomal inhibitors E64d/PepA. BDS histograms of gated double-positive cells in control medium (white line) or starvation medium plus lysosomal inhibitors (red line) (C) in total PBMCs, (D) in CD8+ T cells. (E) Representative images of single cells merged for fluorescent signals from LC3 and Lyso-ID under control, control + inhibitors (C+I), starved or starved plus inhibitors (starved + I) conditions in total PBMCs. (F) Autophagy levels as measured by % double positives with high BDS (left Y axis, white bars) or mean LC3 puncta (right Y axis, black bars) on CD8+ T cells from healthy donors under control conditions and in the presence of E64d/PepA (mean ± s.d., n = 7).
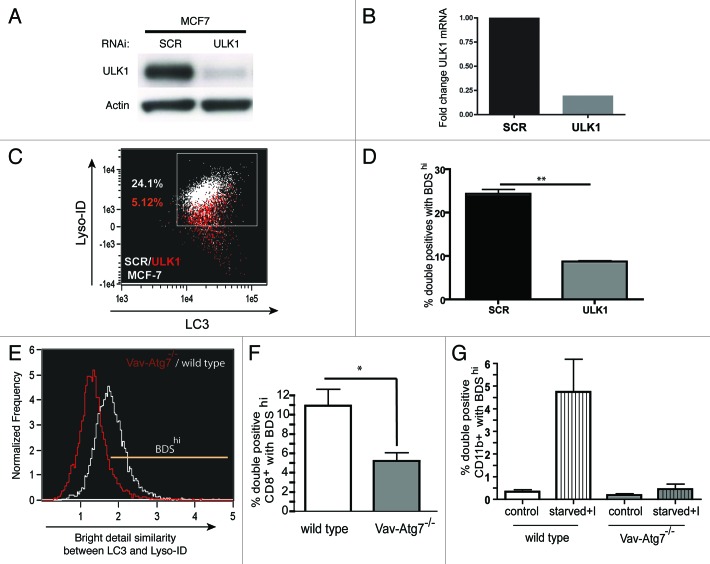
To validate the assay, we measured autophagy in cell lines and mouse splenocytes deficient in autophagy. First, we knocked down ULK1 by siRNA in two cancer cell lines, MCF7 and A431. ULK1 is the mammalian ortholog of yeast Atg1 and is a critical player at the initiating step of the autophagy cascade.16 Mammals have two Atg1 orthologs, ulk1 and ulk217 but it is thought that only ULK1 has a role in autophagy modulation.18 The knockdown of ULK1 was confirmed by western blot (Fig. 3A). The RT-qPCR showed that 80% of ULK1 mRNA expression was knocked down in MCF7 cells by ULK1 siRNA as compared with scrambled siRNA (Fig. 3B). When analyzed on ImageStream, a dot plot showing LC3 and LysoID of scrambled siRNA-treated cells (white) was overlaid with ULK siRNA-treated cells (red). Interestingly, in the case of A431 cells (data not shown), the ULK1 siRNA-treated cells showed such weak lysosomal staining that we were unable to gate on the double positive LC3+LysoID+ population required for accurate BDS analysis, compared with ULK1 siRNA-treated MCF-7 cell line (Fig. 3C). In both cell types, to a different extent, knocking down ULK1 decreased LC3, lysosomal staining along with colocalization of both the markers, whereas control cells expressed both markers highly. Figure 3D shows the quantification of BDS in MCF7 cells. It is worth noting here that, as opposed to primary cells, counting LC3 puncta among the double-positive population also yielded a good signal to noise ratio, whereas counting LC3 puncta in all cells showed no difference (data not shown).
Figure 3. Validation of assay using cells deficient for autophagy. MCF7 cells were either transfected with scrambled (SCR) or ULK1 siRNA, then processed for (A) western blot of cell lysates detected with rabbit-anti-ULK1 or (B) RT-qPCR, graph indicates fold change of ULK1mRNA in the MCF7 cell line. (C) Representative dot plot cells stained for LC3 and lysosomes, ULK1 siRNA-treated cells (red) overlaid with scrambled siRNA treated cells (white) (D) % double positives with BDShi (LC3+Lyso+ in MCF7 cell line) (mean ± SEM, n = 2). Splenocytes from Vav-Atg7−/− mice (gray) or age-matched wild-type mice (white) were stained for LC3 and lysosomes. (E) Representative BDS overlay histogram from Vav-Atg7−/− mice (gated on % total double positives) (red) or age-matched wild-type mice (white) on all splenocytes. (F) Basal autophagy in CD8+ splenocytes measured as % double positives that are BDShi (mean ± SEM, n = 3), (G) basal and induced autophagy levels in CD11b+ splenic myeloid cells (mean ± SEM, n = 2).
Second we tested this assay on murine Atg7−/− splenocytes obtained from Vav-Atg7−/− mice.19 Atg7 encodes the E1-like enzyme required for both of the ubiquitin-like conjugation systems that are essential for the complete formation of autophagosomes.20,21 Atg7-deficient CD8+ T cells obtained from spleen had low autophagy levels (BDShi in Fig. 3F), as compared with wild-type CD8+ T cells (Fig. 3F). It is also shown as a BDS overlay from representative Atg7−/− (red) and WT (white) (Fig. 3E) splenocytes. We also plotted the intensity of LC3 and Lyso-ID staining in Atg7−/− or wild-type CD11b+ myeloid splenocytes that were either cultured in full medium or starved in the presence of lysosomal inhibitors for 2 h. In CD11b+ splenocytes, constitutive levels of autophagy were reduced in the absence of Atg7; furthermore, the increase in autophagy observed in wild-type CD11b+ cells was not seen in Atg7 deficient CD11b+ splenocytes upon induction (Fig. 3G).
For all our experiments we have used a novel acidic organelle-selective dye, which has a long wavelength red emission, Lyso-ID, as a lysosomal marker. It does not photo-convert to a green emitting dye and does not have complicating metachromatic dual emission artifacts.22 It is possible, however that due to the fixation step in our staining protocol, this marker may leak into the cytoplasm. To exclude this we compared Lyso-ID with an antibody based staining (anti-LAMP1) of lysosomes in PBMCs. We observed a high degree of colocalization between the two markers (Fig. S2A) with good correlation of their intensities (Fig. S2B). To confirm that the resolution of the ImageStream allows us to distinguish between vesicles from different stages of the autophagy process, we included WIPI-1 (a mammalian Atg18 paralogue) as a marker. WIPI-1 functions downstream of ULK1 and PI3-kinase complexes but upstream of the Atg12–Atg5-Atg16L1 complex and LC3.23 Thus, WIPI-1 is thought to have a role in the formation of the phagophore but not in the complete autophagosome assembly.24 We could observe significant colocalization between LC3 and Lyso-ID (Fig. S2D and S2E) but minimum colocalization between WIPI-1 and Lyso-ID (Fig. S2C and S2E).
Taken together, autophagy can be measured when either the intensity of two autophagy markers or their level of colocalization was taken into account. First, we found that counting LC3 spots was not reliably measuring autophagy in primary cells. Second, to quantify delivery of LC3 into the lysosomes (colocalization of these markers), gating on double-positive population is necessary, as BDS performs more reliably when analyzing bright pixel intensity. We noticed that this avoids measuring artifact signals generated from the individual markers on their own.
T cells have higher levels of autophagy than B cells
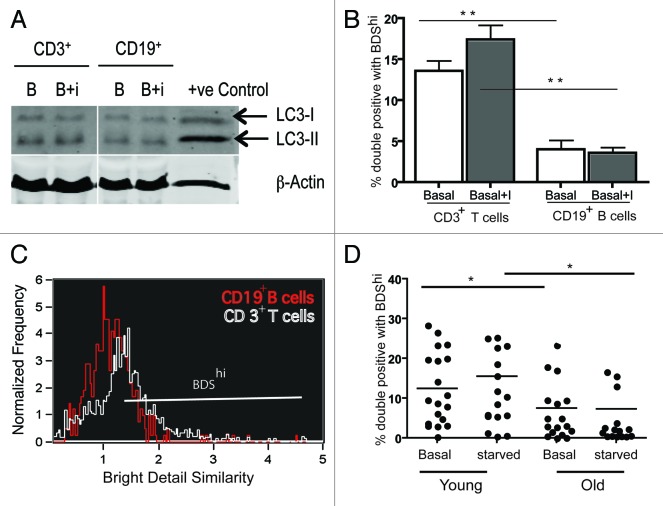
To further subdivide PBMCs, we stained for surface markers using anti-CD3, anti-CD8 and anti-CD19 monoclonal antibodies and assessed levels of autophagy. Strikingly, we found that among lymphocytes, CD8+ T cells have the highest percentage of double-positive cells at basal level (data not shown). When further stained for CD3+ (T lymphocytes) and CD19+ cells (B lymphocytes), T lymphocytes had a 10-fold higher level of autophagic activity than B cells as shown by % BDShi among double-positive cells (Fig. 4B) and an overlay of BDS histograms for CD3+ T cells (white) and CD19+ B cells (red) (Fig. 4C). We further bead sorted these populations from whole blood to do a western blot. The results (blot for a representative donor out of 5 donors, Fig. 4A) show a similar result as the ImageStream but less clear, which may be due to increased stress levels during cell sorting inducing autophagy.
Figure 4. Autophagy levels in human peripheral blood lymphocytes. PBMCs from healthy donors were sorted for CD3+ and CD19+ cells and the whole cell lysates were run on 15% SDS gel or stained for LC3 and the lysosomal marker Lyso-ID after culturing them for 2 h in normal media or with inhibitors E64d/PepA, samples were run on ImageStream, 20,000 cells were acquired/treatment. Single and in focus cells were gated for CD3+, CD19+ population. They were gated for LC3hiLysohi population and these cells were further gated on BDShi (A) Representative western blot of CD3+, CD19+ cells under basal and basal+I condition out of 5 donors. (B) Basal and basal+I levels of % double positives with BDShi from three healthy donors gated on CD3+ and CD19+ populations (mean ± SEM, n = 3, **p = 0.0079) (C) Representative BDS overlay histogram under basal condition from CD3+ (white) and CD19+ (red) populations. (D) Autophagy levels (% total double positives with BDShi) in all PBMCs from young (< 28 y, n = 15, four donors were repeated) and old (> 56 y, n = 12, six donors were repeated) donors under control and induced (starved for 2 h) conditions (mean ± SEM, *p < 0.0403 (young and old controls), *p < 0.0186 (young and old starved PBMCs), Mann-Whitney test). (D) were pooled from several experiments.
Autophagy levels decrease with age in CD8+ T cells
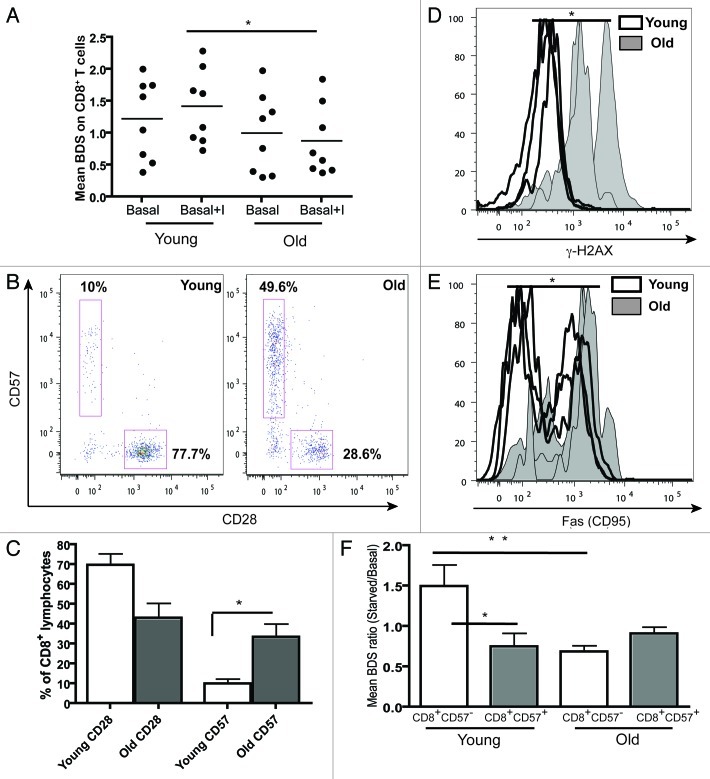
Next we sought to determine if decreased autophagy levels could be detected in senescent cells. As the highest autophagy levels were found in CD8+ T cells and CD8+ cells are best studied for senescence, we measured the autophagy levels of CD8+ T cells from young (< 28 y) and old (> 60 y) healthy donors. Despite varying rates of autophagy levels between donors, we saw a significant decrease of autophagy levels in PBMCs from old healthy donors, both for basal and induced levels of autophagy (Fig. 4D). A similar increase was observed when gated on CD8+T cells (Fig. 5A).
Figure 5. Aging and replicative senescence markers in human T lymphocytes. PBMCs from healthy young (< 28 y) and old (> 56 y) donors were stained for CD28 and CD57 and run on LSR II flow cytometer. (A) Autophagy levels (Mean BDS) in CD8+ T cells from young (< 28 y, n = 8) and old (> 56 y, n = 8) donors under basal and basal+I (for 2 h) conditions (mean ± SEM, *p < 0.0499 between young and old basal+I). (B) Representative dot plots from a young and an old donor showing percentages of CD28 and CD57 cells gated on CD8+ T cells. (C) Bar graph showing % of CD8+ lymphocytes with CD28 and CD57 markers in four young and old donors (mean ± SEM, p = 0.0571 for CD8 CD28 population and *p = 0.0286 for CD8 CD57 population). (D) Overlaid histogram of γH2AX (DNA double-strand break) levels of CD8+ lymphocytes from three young and old donors gated on CD28+CD57- population (geometric mean ± SEM, *p = 0.0286). (E) Overlaid histogram of FAS (CD95) levels of CD8+ lymphocytes from four young and old donors gated on CD28+CD57- population (geometric mean ± SEM, *p = 0.0286). PBMCs from four healthy young and old donors were cultured under control and starved conditions for 2 h and stained for CD8, CD57, LC3 and Lyso-ID. (F) Colocalization of LC3 and lysosomal marker in CD8+CD57+/− cells, expressed as mean BDS ratio between starved and basal treatments (mean ± SEM, n = 5 (young donors), n = 8 (old donors), **p = 0.0049, *p = 0.035).
Aging is strongly associated with the loss of the CD28 marker and the gain of CD57 on CD8+ cells (Fig. 5B and C). Furthermore it has been shown that expression of CD57 marks replicative senescence in CD8+ T cells.25 Additionally we characterized aging CD8+ T cells for other senescent markers. As expected the expression of both γH2AX and Fas increased with age on CD8+ lymphocytes (by flow cytometry in Fig. 5D and E, and also by ImageStream, showing γH2AX foci in CD8+ lymphocytes from young and old donors in Fig. 6D). Thus, we further gated individual donor populations into young and old CD8+ T cells using CD28 and CD57. Whereas in nonsenescent T cells (CD8+57−) basal autophagy levels were high and can be induced slightly, senescent T cells (CD8+57+) from young donors displayed low basal autophagy levels. In contrast, nonsenescent as well as senescent T cells from old donors displayed low autophagic activity and starvation fails to increase this significantly (Fig. 5F). A similar trend was observed with inhibitors (data not shown).
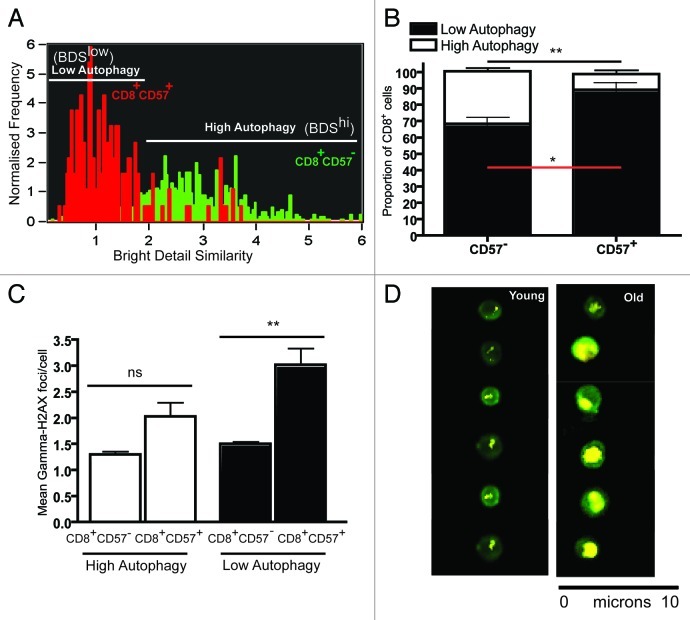
Figure 6. Levels of γH2AX foci in senescent T cells with low autophagy. PBMCs from three healthy donors were bead sorted for CD8+ T cells and stained for CD57 γH2AX, LC3 and Lyso-ID and run on ImageStream. (A) Representative BDS overlay histogram for CD8+CD57+ (red) for CD8+CD57- (green) depicts the low and high autophagy gates. (B) Bar graph showing proportion of CD8+CD57+/− lymphocytes with low and high autophagy in three healthy donors (mean ± SEM, **p = 0.0029, *p = 0.0273). (C) Bar graph showing mean γH2AX foci/cell in low and high autophagy CD8+CD57+, CD8+CD57- populations (geometric mean ± SEM, **p = 0.0057 and **p = 0.0087, respectively). (D) Representative ImageStream images of γH2AX foci in CD8+ lymphocytes from young and old cells.
Further, we bead-sorted CD8+ T cells from three healthy young donors and stained them for CD57, γH2AX, LC3 and Lyso-ID. Figure 6A shows the BDS gating strategy, BDShi (high autophagy) and BDSlow (low autophagy). It is very clear that the majority of the CD8+CD57+ T cells (red) display low autophagy levels, whereas the CD8+CD57- T cells (green) show high autophagy in this donor. Based on our results from three donors, we have plotted this in Figure 6B. Interestingly the population that has the highest mean γH2AX foci/cell is the CD57+ CD8+ cells with low autophagy levels (Fig. 6C). This further strengthens our argument that senescent T cells have low autophagy levels and increased DNA damage.
Discussion
With this novel imaging flow cytometry-based autophagy assay we have captured the dynamic autophagy process in primary cells. This assay is set up to allow the detection of autophagy in complex clinical samples, such as blood, which until now has not been possible in a statistically robust, high-throughput manner.
Mammalian autophagy detection is mostly restricted to cell lines stably expressing fluorescent proteins like GFP-LC3. Autophagy assays based on GFP-LC3 puncta count or intensity just measure the increase in autophagosomes (an intermediate step), which may represent a block in the later stages of the autophagic flux and is not sufficient to determine the overall flux.1,3 Furthermore this approach is subjective, may lead to false results due to GFP-LC3 aggregates, and importantly is not applicable to primary cells. Delivery of autophagosomes carrying autophagic substrates to lysosomes (autolysosome formation) and their subsequent degradation delineates a dynamic downstream step of the autophagic pathway and this is a more reliable indicator of the autophagy process and function under any physiological or pathophysiological condition.3 Many studies have indicated that it is difficult to catch the autophagic flux without inhibition of lysosomal hydrolases.1 E64d and Pepstatin A, membrane-permeable inhibitors of cathepsins, can perform this function and therefore enable us to examine the lysosomal turnover of intra-autophagosomal LC3-II.
This new assay measures how much of LC3 is immediately susceptible to degradation by lysosomes; in other words it is the turnover of intra-autolysosomal LC3-II measured by quantifying the cells that are double positive for LC3 and lysosomal markers and showing colocalization between these markers, both in cell lines and in subsets of primary immune cells (defined using surface markers). The addition of inhibitors controls for lysosomal defects and indicates how much autophagosomal delivery to lysosomes has taken place during the time the inhibitors were applied. This assay may give insight into a population’s autophagic flux (as we are measuring the delivery of cargo to lysosomes) if combined with evidence of subsequent degradation and recycling of cargo. The assay is high-throughput and automated, thus avoiding the subjective bias associated with some existing assays. That said, our results correlate well with LC3 immunoblot and confocal results. The ‘turnover’ in the ImageStream assay compared with LC3 turnover (measured by levels of LC3-I vs. LC3-II) in western blot is clearly similar in cell lines, and the slight differences between these two assays may be due to the different read-outs—one is measuring LC3 turnover while the other looks at the number of cells with high autolysosome throughput. Based on our results from ULK1 knockdown in MCF-7 cell lines and from Atg7−/− mouse splenocytes, we conclude that it is the lysosomal turnover of LC3 that best reflects the autophagic flux, not LC3 alone. An earlier study used multispectral imaging flow cytometry to count GFP puncta in plasmacytoid dendritic cells from LC3-GFP transgenic mice in response to VSV infection. Although GFP puncta were detected, the authors found no increase in response to VSV, possibly because LC3 alone may not accurately reflect autophagy levels.26
We should also mention that this autophagy detection assay gave us more reproducible results while working with cell lines or EBV transformed primary cell lines (data not shown) compared with primary cells from healthy donors. We came across a large variation in autophagy levels especially on induction with starvation, in human donors, which may be due to diurnal rhythm of each donor27 or differences in daily routines. ImageStream’s sensitivity, its resolution (40× camera) and acquisition time (100 cells/sec) may be some of the confounding issues; the improved newer version of ImageStream now available may overcome these hurdles. In summary we show this assay to be objective, quantitative and statistically robust, as compared with existing autophagy assays, and believe it will allow examination of autophagy in novel settings.
This unique imaging flow cytometry-based autophagy assay serves as a new tool to investigate autophagy levels in different subpopulations of blood-derived peripheral cells. The peripheral pool of mature T lymphocytes is tightly regulated in the adaptive immune system. Autophagosomes and autolysosomes have already been reported in freshly isolated mouse T cells and cultured human T lymphocytes.28-30 Also, multiple murine studies have observed that survival of the peripheral T cell compartment is dramatically reduced in the absence of autophagy.19,28 In contrast to mature T cells, mature peripheral B cells do not require autophagy for survival, as demonstrated in Atg5−/− mice31,32 and do not have detectable levels of basal autophagy. This had not been investigated quantitatively in human lymphocytes. Our assay showed similar and significant results with human peripheral T cells (CD3+) having both higher basal and induced levels of autophagy when compared with peripheral B cells (CD19+). At present we cannot explain the different levels of autophagy in these subsets of lymphocytes, both known to be long lived.
The frequency of CD57 expression on CD8+T cells is highly associated with the inability of those CD8+ T cells to proliferate.25 Our results show that the nonsenescent (young) CD8+CD57- cells have high basal and induced autophagy levels compared with the senescent (old) CD8+57+ T cells. Though Gerland et al.29 have reported an increase in autophagic inclusions during in vitro aging experiments on CD8+ T cells and a decrease after PHA stimulation, this may be due to the fact that they used monodansylcadaverine (MDC), a specific in vivo marker for autophagic vacuoles,33 as a readout for autophagy. In the absence of inhibitors, this may lead to inaccurate results as it only shows a snapshot of upstream autophagy pathway at a steady-state point. An overall decrease in chaperone-mediated autophagy has also been reported in aging rat liver cells.7 The observed decrease in autophagy levels (low mean BDS of colocalized double positive cells) in CD8+57+ T cells may be due to reduced fusion of lysosomes with autophagic vacuoles, altered trafficking of LC3+ autophagosomes to lysosomes due to presence of aging pigment lipofuscin, or age related change in intralysosomal pH.34 Altered lysosomal membrane dynamics have been observed with aging and it has been suggested that lysosomal enzymes are in short supply during aging.35
As such, our finding of reduced autophagosome/lysosomal colocalization reflects reduced degradation through the complete autophagic pathway (from cargo engulfment to fusion/degradation in lysosomes) when combined with lysosomal inhibitors. The presence of inhibitors in the assay eliminates the possibility of decreased lysosomal degradative capacity as an explanation for the reduction in colocalization. Our findings of reduced autophagy (either at the induction level or any step before autolysosome formation) contributes to the overall cellular aging hallmarks, and to the notion that this may be due to lower autophagic capacity in aging cells. Second, the fact that we have often seen increased lysosomal and LC3 content (e.g., in Atg7−/− cells, data not shown) without concomitant increase of colocalization of the two markers indicates further that altered lysosomal content does not necessarily reflect altered autophagy levels. The cause of the reduced autophagy levels in T cells during aging needs to be further investigated.
Furthermore we show that the senescent CD8+57+ T cells with low autophagy levels have high Fas (CD95) expression, a cell death marker, and high levels of DNA double-strand breaks (as assessed by γH2AX foci and intensity), with the opposite true in cells with high autophagy. As its absence in immune cells correlates with established signs of aging, this leads us to conclude that autophagy may have a major role to play in promoting longevity. We are further investigating this in human blood cells and mouse models by relating this decline in autophagy levels with telomere length. Indeed CD8+CD28− and CD8+CD57+ have been reported to have shorter telomeres than CD8+CD28+ and CD8+CD57− T cells.25,36 One theory is that the cellular damage in aging cells results from dysfunctional mitochondrial and their production of high levels of reactive oxygen species (ROS). Our group and others have shown that the absence of key autophagy genes can lead to increases in damaged mitochondria and ROS in T lymphocytes and other hematopoietic cells19,37,38 linking mitochondrial oxidative stress with autophagy.39
Finally, as mentioned above, we believe that this is a unique and exciting tool to investigate autophagy levels in cell lines and primary cells. This new assay will allow the study of the autophagy process in lymphocytes and other leukocytes. Even in inherited diseases that do not affect the hematopoietic system, easily accessible blood-derived lymphocytes may accurately reflect the autophagy status of the affected tissue. Many drugs currently in use are thought to modulate autophagy levels. Here again, autophagy level in lymphocytes may serve to help to gauge the contribution of autophagy to the effectiveness of these drugs.
Materials and Methods
PBMCs
40–50 ml of blood was obtained with the approval of a local ethical committee from young (< 28 y) and old (> 57 y of age) healthy donors in heparinized tubes. PBMCs were separated using lymphoprep, 1114544 (Axis-Shield). PBMCs were counted using trypan blue on a hemocytometer. They were either used immediately or from frozen, initial experiments showed similar results for both.
Cell lines
MCF7 is a breast cancer cell line, while A431 is derived from human vulva carcinoma. All cells were cultured in nutrient-rich (full) medium, i.e., RPMI1640 (Sigma, R8758), complemented with 10%FCS, L-glutamate and penicillin/streptomycin.
Monocyte-derived macrophages
CD14+ monocytes were separated from Buffy coats using MACS CD14 microbeads (Miltenyi Biotec, 12-000-305). CD14+ monocytes were plated in full medium. Purity was checked by staining with mouse anti-human CD14+ antibody conjugated to Pacific blue on LSRII (Invitrogen, MHCD1428) flow cytometer. Sterile Lumox dish 50 (Greiner Bio-One, 08121468) was used for culturing monocytes. Recombinant human GM-CSF (PeproTech, 300-03) and M-CSF (PeproTech, 300-25) were added on days 0 and 3 at 20 ng/ml each.
Preparation of murine splenocytes
Spleens were obtained from 6- to 9-week old Vav-Atg7−/− mice. The Vav-Atg7−/− were of Atg7fl/fl Vav-iCre+/− genotype,19 and the wild-type mice were littermate controls of Atg7fl/fl Vav-Cre−/− genotype. All animal experiments were approved by the local ethical review committee and performed under a home office license. Single-cell suspensions were obtained by squeezing spleen through 70 μm filter.
Induction of autophagy
Conditions were carefully established for each cell type. Conditions of culture of PBMCs included (1) full medium (2) full medium with E64d (Sigma, E8640) and Pepstatin A (Sigma, P5318) 10 µg/ml each, (3) HBSS (Invitrogen, 14060) (4) HBSS with E64d and Pepstatin A for 2 h was determined to be optimal time point for autophagy detection by ImageStream in primary cells. In human blood monocyte-derived macrophages, autophagy was induced by adding 10 ng/mL of LPS (Sigma, L1887) for 12 h.
Staining and antibodies
ImageStream and flow cytometry experiments were performed on ImageStream 100 (Amnis) and LSR 2 (Becton Dickinson), respectively. Single cell suspensions from murine splenocytes were stained with anti-mouse CD11b PE (M1/70) (BioLegend, 101208). Human monocyte-derived macrophages were stained with anti-CD14 -Pacific blue (Invitrogen, MHCD1428) and human PBMCs were surface stained with the following antibodies: anti-human CD8-Pacific blue (Molecular Probes, Invitrogen, MHCD0828), eFluor® 450 conjugated anti-CD19 (eBioscience, 48-0193-82) FITC conjugated anti-CD3, (PharMingen, 555332), PE conjugated anti-CD3 (MEM-57) (Thermo-Scientific, MA1-19624), alexa flour® 647 conjugated anti-CD57 (HCD57) (BioLegend, 322308), FITC conjugated anti-CD57 (TB01), FITC conjugated anti-CD28 (PharMingen, 550973, 555726, respectively), PE conjugated anti-CD28 (CD28.2), FITC conjugated anti-γH2AX (Ser139) (2F3) and biotin conjugated anti-CD95 (Fas/APO-1) (DX2) (eBioscience, 12-0289-42, 11-0577-42, 13-0959-82, respectively) followed by streptavidin, R-PE conjugate (Molecular Probes, Invitrogen, S-866).
For LC3, WIPI-1 and LAMP-1 staining, cells were first stained with the lysosomal marker Lyso-ID (Enzo Life Sciences, ENZ-51005-500), then surface stained and fixed/permeabilized (eBioscience Fixation and Permeabilization kit, 00-8222-49, 008333-56) and stained with either mouse monoclonal antibody to LC3 (5F10) (Nanotools, 0231-100/LC3-5F10) or with rabbit anti mouse LC3 serum (kind gift from Christian Münz, 1:1000) mouse monoclonal IgG2bWIPI-1 (38-W) (Santa Cruz Biotechnology, sc-100901) and PE conjugated anti-human CD107a (LAMP-1) (Biolegend, 328608), followed by staining with alexa flour 488® goat anti-rabbit IgG (H+L) 2 mg/ml (Invitrogen, A-11008), alexa flour® 546 goat anti-mouse IgG (H+L) (Invitrogen, A-11030), or alexa flour® 546 goat anti-rabbit (Invitrogen, A-11035). All antibodies were titrated for both ImageStream and LSR2.
siRNA transfection and western blot
RNAi sequences to ULK1 (sense strands written only) GCACAGAGACCGTGGGCAA, CCACGCAGGTGCAGAACTA,CGGAGAGCCTGCAGGAGAA, AGCAAGAGCACACGGAGA. Both specific and scrambled pool of siRNAs were provided by Eurogentec. A pool of all four siRNA duplexes was prepared and reverse transfected into cell lines at 20 nM using Qiagen’s HiPerfect, as per the manufacturer’s recommendations at a cell density of 500,000 cells in 100 mm plates. Forty-eight hours later, cells were harvested and stained. Western blot bands were detected by rabbit-anti-ULK1 (Cell Signaling, R600), 1:500 in 5% milk/TBST overnight at 4°C. Mouse anti-human actin monoclonal antibody (Sigma) was used to detect actin at a concentration of 1:2000 in 5% milk/TBST. HRP-conjugated secondary antibodies from DAKO were used at 1:10,000 in 5% milk/TBST for 1 h at room temperature. Detection was done by chemiluminesce using ECL reagent (GE Healthcare, RPN2109). RT-qPCR primers were designed using Universal Probe Library’s Assay Design Center. Sequences are as follows: forward primer TCATCTTCAGCCACGCTGT, reverse primer CACGGTGCTGGAACATCTC. The human probe #37 was obtained from Exiquon (Vedbaek). Master mix was obtained from Bioline, and cycling was done using a Rotogene RG3000 (Corbett Research). Tubulin-α-6 mRNA was used as a reference gene to normalize for differences in the amount of total RNA in each sample.
Western blot for LC3
100 µg of protein was loaded on 15% SDS-PAGE gel both for PBMC’s and HEK 293 cell lines. 786-O VHL−/− cell line treated for 24 h with 2 µM STF-62247 (Cayman Chemicals, 13084) was used as positive control for the PBMC’s experiment. Proteins separated were transferred on to nitrocellulose membrane (Bio-Rad Laboratories, 162-0115) and probed for Anti-LC3 (Clone: 51-11) (MBL, M115-3) as primary antibody, β-Actin (AC-15) (Santa Cruz Biotechnology, sc-69879) as loading control and Alexa Fluor 680 goat anti-mouse IgG (A21057, Invitrogen) was used as secondary antibody. Membranes were scanned using Odyssey Infrared Imager (LI-COR Biosciences).
ImageStream
ImageStream (IS100) is a multispectral flow cytometer combining standard microscopy with flow cytometry distributed by Cronus technology in the UK. It can acquire up to 100 cells/sec, with simultaneously acquisition of six images of each cell including bright field, scatter, and multiple fluorescent images. We used the integrated software INSPIRE to run the ImageStream. For each experiment cells (PBMCs, monocyte derived macrophages, murine splenocytes or cell lines MCF7, A431 and HEK293) were stained with respective markers and were finally suspended in 50 µl of buffer (cold PBS with 1% FCS and 0.05% sodium azide) in 0.6 ml microcentrifuge tubes. Before running the samples, the ImageStream was calibrated using speedbeads. Samples were acquired in the order of unlabeled, single color fluorescence controls (no DNA dye) and finally the experimental samples. Samples were always left on ice. The Flush/Lock/Load script was used to run individual samples. Single color controls were run to optimize the laser strength (50 mW for 405 and 100 mW for 488) to be used for the experimental samples. At least 10,000 cells/experimental sample and 2000 cells/single color control were acquired for each sample. After each sample was injected into the flow cell, the cells were allowed to form a single core stream before acquisition. Cell classifier was set on area-lower limit on bright field channel (25 for PBMCs and 50 for monocyte derived macrophages and cell lines) to avoid acquiring debris. The bright field was switched of during the acquisition of single color controls. The data files were saved as .rif files (raw files). The .rif files generated from INSPIRE were analyzed using IDEAS 4.0.735 software. Single color control files were used to create compensated files (.cif) followed by generation of data (.daf) files. To obtain single cells, the doublets or clusters of cells and debris were gated out using bright field area vs. aspect ratio feature. These single cells were now gated for in focus cells based on bright field gradient root mean square (GRMS) feature (> 300 GRMS). These in focus single cells were either analyzed for all cells or were further gated on CD8+ and CD8− populations and CD8+ were further gated on CD57+/− populations based on intensities in respective channels. These subsets were now plotted for log intensities of channel with LC3 (autophagosome marker) and Lyso-ID or Lamp1 (lysosomal marker), they were gated for LC3+Lyso+ (double positives, in some cell populations further split into LC3highLysohigh and LC3medLysomed), LC3highLysolow (more positive for LC3), LC3lowLysohigh (more positive for lysosomal marker) and LC3lowLysolow (low for both LC3 and lysosomes) populations. Autophagy levels were calculated by measuring % colocalization in double positives of total cells by plotting these LC3hiLysohi (double positives) for bright detailed similarity (BDS) between LC3 and lysosomal markers against normalized frequency of cells. BDS on double positives mostly resulted in bimodal histograms, and gating was on the brighter peak (BDShi, mostly above 1.5 or 2). In some cases where the double-positive population was absent or very small, we used % total double positives as our autophagy read-out. Initially LC3 spots were counted by creating a spot mask on CD8+ cells positive for LC3, spot to background ratio was set to 2.25 and the radii of spots was set as 1, γH2AX spots were counted in a similar way with spot to background ratio set at 5. These spot masks were then converted into a feature and were used to plot against normalized frequency of cells.
Statistical analysis
Statistical analyses were performed using Prism 4 for Mac (GraphPad Software). Error bars represent SEM and p values calculated with a two-tailed Mann–Whitney test unless stated otherwise.
Supplementary Material
Acknowledgments
We would like to thank Monika Mortensen and Daniel Puleston for providing us with mouse tissues. We would also like to thank all our donors. This study was funded by the Biomedical Research Centre Oxford (NIHR), the Wellcome Trust, NSERC and the BBSRC. K.P. contributed all major figures, wrote the manuscript and contributed ideas. J.A.-A. contributed Figure 2, ideas and edited the manuscript, L.P. contributed Figure 3A–E, A.W. performed some of the experiments in Figures 4 and 5, and edited the manuscript, S.A. contributed Figure 4A, A. McM. contributed ideas, blood and funding, P.K. obtained funding for salaries and consumables and contributed ideas, K.S. obtained funding for ImageStream, formulated the initial idea, supervised the project, and wrote the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/18935
References
- 1.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 4.Shvets E, Fass E, Elazar Z. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008;4:621–8. doi: 10.4161/auto.5939. [DOI] [PubMed] [Google Scholar]
- 5.Eng KE, Panas MD, Karlsson Hedestam GB, McInerney GM. A novel quantitative flow cytometry-based assay for autophagy. Autophagy. 2010;6:634–41. doi: 10.4161/auto.6.5.12112. [DOI] [PubMed] [Google Scholar]
- 6.Effros RB. Replicative senescence of CD8 T cells: potential effects on cancer immune surveillance and immunotherapy. Cancer Immunol Immunother. 2004;53:925–33. doi: 10.1007/s00262-004-0508-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–40. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madeo F, Tavernarakis N, Kroemer G. Can autophagy promote longevity? Nat Cell Biol. 2010;12:842–6. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- 9.Potestio M, Pawelec G, Di Lorenzo G, Candore G, D'Anna C, Gervasi F, et al. Age-related changes in the expression of CD95 (APO1/FAS) on blood lymphocytes. Exp Gerontol. 1999;34:659–73. doi: 10.1016/S0531-5565(99)00041-8. [DOI] [PubMed] [Google Scholar]
- 10.Akbar AN, Henson SM. Are senescence and exhaustion intertwined or unrelated processes that compromise immunity? Nat Rev Immunol. 2011;11:289–95. doi: 10.1038/nri2959. [DOI] [PubMed] [Google Scholar]
- 11.Mah LJ, El-Osta A, Karagiannis TC. gammaH2AX: a sensitive molecular marker of DNA damage and repair. Leukemia. 2010;24:679–86. doi: 10.1038/leu.2010.6. [DOI] [PubMed] [Google Scholar]
- 12.Beum PV, Lindorfer MA, Hall BE, George TC, Frost K, Morrissey PJ, et al. Quantitative analysis of protein co-localization on B cells opsonized with rituximab and complement using the ImageStream multispectral imaging flow cytometer. J Immunol Methods. 2006;317:90–9. doi: 10.1016/j.jim.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–44. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid D, Munz C. Innate and adaptive immunity through autophagy. Immunity. 2007;27:11–21. doi: 10.1016/j.immuni.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 16.Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walsh CM, Edinger AL. The complex interplay between autophagy, apoptosis, and necrotic signals promotes T-cell homeostasis. Immunol Rev. 2010;236:95–109. doi: 10.1111/j.1600-065X.2010.00919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007;282:25464–74. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- 19.Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci USA. 2010;107:832–7. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–79. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–6. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- 22.Freundt EC, Czapiga M, Lenardo MJ. Photoconversion of Lysotracker Red to a green fluorescent molecule. Cell Res. 2007;17:956–8. doi: 10.1038/cr.2007.80. [DOI] [PubMed] [Google Scholar]
- 23.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–76. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanida I. Autophagy basics. Microbiol Immunol. 2011;55:1–11. doi: 10.1111/j.1348-0421.2010.00271.x. [DOI] [PubMed] [Google Scholar]
- 25.Brenchley JM, Karandikar NJ, Betts MR, Ambrozak DR, Hill BJ, Crotty LE, et al. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood. 2003;101:2711–20. doi: 10.1182/blood-2002-07-2103. [DOI] [PubMed] [Google Scholar]
- 26.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 27.Sachdeva UM, Thompson CB. Diurnal rhythms of autophagy: implications for cell biology and human disease. Autophagy. 2008;4:581–9. doi: 10.4161/auto.6141. [DOI] [PubMed] [Google Scholar]
- 28.Pua HH, He YW. Maintaining T lymphocyte homeostasis: another duty of autophagy. Autophagy. 2007;3:266–7. doi: 10.4161/auto.3908. [DOI] [PubMed] [Google Scholar]
- 29.Gerland LM, Genestier L, Peyrol S, Michallet MC, Hayette S, Urbanowicz I, et al. Autolysosomes accumulate during in vitro CD8+ T-lymphocyte aging and may participate in induced death sensitization of senescent cells. Exp Gerontol. 2004;39:789–800. doi: 10.1016/j.exger.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 30.Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, et al. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest. 2006;116:2161–72. doi: 10.1172/JCI26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yousefi S, Simon HU. Autophagy in cells of the blood. Biochim Biophys Acta. 2009;1793:1461–4. doi: 10.1016/j.bbamcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 32.Lee DY, Sugden B. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene. 2008;27:2833–42. doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- 33.Biederbick A, Kern HF, Elsasser HP. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995;66:3–14. [PubMed] [Google Scholar]
- 34.Cuervo AM, Dice JF. When lysosomes get old. Exp Gerontol. 2000;35:119–31. doi: 10.1016/S0531-5565(00)00075-9. [DOI] [PubMed] [Google Scholar]
- 35.Kurz T, Terman A, Gustafsson B, Brunk UT. Lysosomes in iron metabolism, aging and apoptosis. Histochem Cell Biol. 2008;129:389–406. doi: 10.1007/s00418-008-0394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin J, Epel E, Cheon J, Kroenke C, Sinclair E, Bigos M, et al. Analyses and comparisons of telomerase activity and telomere length in human T and B cells: insights for epidemiology of telomere maintenance. J Immunol Methods. 2010;352:71–80. doi: 10.1016/j.jim.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011 doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terman A, Gustafsson B, Brunk UT. Autophagy, organelles and aging. J Pathol. 2007;211:134–43. doi: 10.1002/path.2094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.