Abstract
Bcl-2 family members are key modulators of apoptosis that have recently been shown to also regulate autophagy. It has been previously reported that Bcl-2 and Bcl-XL bind and inhibit BECN1, an essential mediator of autophagy. Bcl-B is an anti-apoptotic member of the Bcl-2 family that possesses the four BH (Bcl-2 homology) domains (BH1, BH2, BH3 and BH4) and a predicted C-terminal trans-membrane domain. Although the anti-apoptotic properties of Bcl-B are well characterized, its physiological function remains to be established. In the present study, we first established that Bcl-B interacts with the BH3 domain of BECN1. We also showed that Bcl-B overexpression reduces autophagy triggered by a variety of pro-autophagic stimuli. This impairment of autophagy was closely related to the capacity of Bcl-B to bind to BECN1. Importantly, we have demonstrated that Bcl-B knockdown triggers autophagic cell death and sensitizes cells to amino acid starvation. The cell death induced by Bcl-B knockdown was partially dependent on components of the autophagy machinery (LC3; BECN1; ATG5). These findings reveal a new role of Bcl-B in the regulation of autophagy.
Keywords: BECN1, Bcl-B, amino acid starvation, apoptosis, autophagy
Introduction
Cell death can occur by both apoptotic and nonapoptotic mechanisms.1 Apoptosis, called type 1 cell death, is a genetically regulated cellular suicide mechanism that plays critical roles in normal development, tissue homeostasis, and the elimination of infected or damaged cells.2 Defective apoptosis represents an important feature of human cancer.3 Traditional cancer therapy triggers malignant cell elimination by inducing apoptosis. However, the apoptotic resistance inherent in cancer cells has been an important barrier to efficient chemotherapy.4 Autophagic cell death, also called type 2 nonapoptotic cell death, has recently been recognized as a mechanism contributing to tumor cell eradication induced by chemotherapeutics under some circumstances.5,6 Autophagy is a conserved catabolic process by which long-lived proteins, cytoplasmic organelles, and intracellular pathogens undergo lysosome-dependent degradation.7 Autophagy is upregulated by conditions such as nutrient starvation and hypoxia, in which it contributes to cell survival by providing substrates for maintaining ATP levels. However, when deregulated, components of the autophagy machinery contribute to nonapoptotic cell death (reviewed in ref. 7).
The anti-apoptotic proteins Bcl-2 and Bcl-XL have been shown to inhibit autophagy through their binding to the BH3-only protein BECN1, a critical inducer of autophagy.8-11 Thus, Bcl-2 family proteins represent a point of convergence of the apoptotic and autophagic processes.12 Among the six human anti-apoptotic Bcl-2 family proteins [Bcl-2 (BCL2), Bcl-XL (BCL2L1), Mcl-1 (MCL1), Bcl-W (BCL2L2), Bfl-1 (BCL2A1) and Bcl-B (BCL2L10)], Bcl-B was the last anti-apoptotic member to be identified.13,14 Bcl-B contains conserved BH1, BH2, BH3-like, and BH4 domains, and a COOH-terminal transmembrane domain, as is typical of anti-apoptotic Bcl-2 family members.13,14 A strong correlation exists between the ability of Bcl-B to bind pro-apoptotic Bcl-2 family proteins and its anti-apoptotic capacity.15 Moreover, Krajewska et al. observed that, in normal tissues, human Bcl-B protein is mainly expressed in plasma cells and that its expression is upregulated in multiple myeloma and some solid tumors, including breast, prostate, colorectal, and lung adenocarcinomas.16 Nevertheless, little is known about the in vivo function of human Bcl-B, essentially because its mouse ortholog Boo/Diva shares with Bcl-B only 49% amino acid identity and is expressed only in ovary and testis.
In this study, we revealed a new function of the Bcl-B protein in the regulation of autophagy. We first established that the Bcl-B protein interacts directly with BECN1, mainly at the endoplasmic reticulum (ER). We also showed that Bcl-B overexpression reduces autophagic cell death, an effect closely related to its capacity to bind BECN1. Further, we demonstrated that Bcl-B silencing promotes autophagic cell death and sensitizes cells to cell death induced by amino acid starvation.
Results
Bcl-B binds BECN1
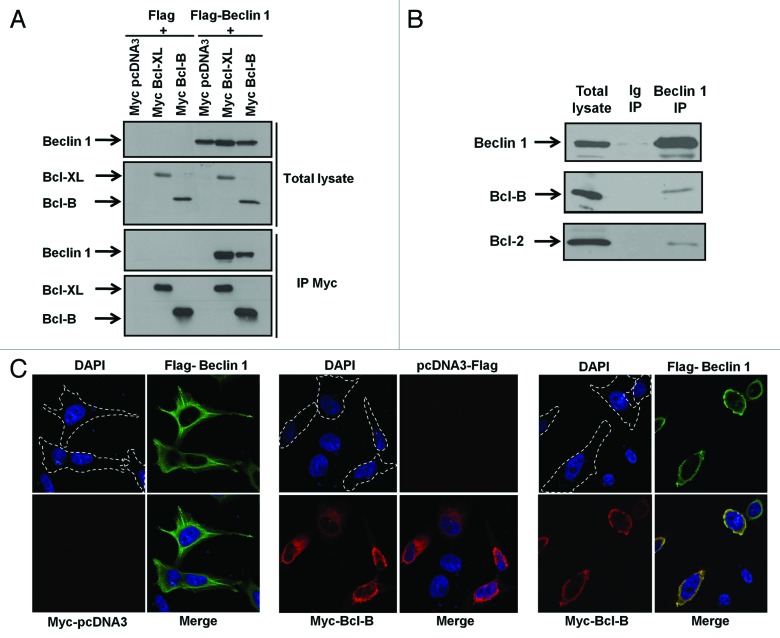
Previous studies have identified the Bcl-2 family members Bcl-2 and Bcl-XL as key regulators of autophagy.8,9 Bcl-2 and Bcl-XL have been reported to inhibit autophagy induced by amino acid (AA) starvation through their capacity to bind BECN1 protein. While Bcl-B is known to regulate mitochondrial apoptosis,13-15 its effect on autophagy is unknown. To determine whether Bcl-B binds BECN1, we first performed co-immunoprecipitation experiments in 293T recipient cells transfected with plasmids encoding either Bcl-XL or Bcl-B in combination with a plasmid encoding BECN1 (Fig. 1A). Exogenous BECN1 was not only pulled down by Bcl-XL but also by Bcl-B. To address whether endogenous Bcl-B also interacts with BECN1, we performed co-immunoprecipitation experiments in HeLa cells that express detectable endogenous levels of both Bcl-B and BECN1. We found that endogenous Bcl-B binds endogenous BECN1 (Fig. 1B).
Figure 1. Bcl-B binds to BECN1. (A) 293T cells were co-transfected with plasmids encoding either Flag or Flag-BECN1 in combination with plasmids encoding Myc-tagged versions of Bcl-XL (as a control for BECN1 binding) and Bcl-B. After 24 h, immunoprecipitations (IPs) were performed using an anti-Myc antibody and the immune complexes were analyzed by immunoblotting using anti-BECN1 or anti-Myc antibodies (bottom panel). The cell lysates (50 μg) were analyzed directly by SDS-PAGE to confirm the protein expression (top panel). (B) IPs were performed from HeLa cells using either mouse Ig (second lane) or mouse anti-BECN1 (third lane) antibodies and endogenous immune-complexes were analyzed by immunoblotting using anti-BECN1 or anti-Bcl-B antibodies. The cell lysates (50 μg) were analyzed directly (first lane). (C) HeLa cells were co-transfected with plasmids encoding Flag-BECN1 and Myc-pcDNA3 (left panel); pcDNA3-Flag and Myc-Bcl-B (center panel); or Flag-BECN1 and Myc-Bcl-B (right panel). After 24 h, the cells were fixed, permeabilized, and successively incubated with anti-Myc and BECN1 antibodies followed by secondary antibodies conjugated to red or green fluorochromes, respectively. The nuclei were stained with DAPI reagent (5 μg/ml). The antibody localization was visualized by confocal UV microscopy. The panels represent the UV (top left), red (bottom left), green (top right), and merge (bottom right) channels.
To determine whether a Bcl-B/BECN1 complex is formed in intact cells by another method, we used confocal microscopy. When HeLa cells were transfected with a plasmid encoding Flag-BECN1 (Fig. 1C, left panel), we observed that exogenous BECN1 exhibited a diffuse pattern of expression, which is compatible with its reported cytosolic localization.17,18 When HeLa cells were transfected with a plasmid encoding Myc-Bcl-B (Fig. 1C, middle panel), we observed that Bcl-B exhibited a punctate perinuclear pattern, which is consistent with its reported mitochondrial and ER localizations13,19 (and data not published). When the proteins were co-expressed, BECN1 adopted a perinuclear pattern that was superposable to that of Bcl-B (Fig. 1C, right panel). Because Bcl-B is predominantly found on the outer mitochondrial and ER membranes, we performed confocal microscopy experiments using ER and mitochondrial markers to determine the subcellular localization of the Bcl-B/BECN1 complex (Fig. S1). In ≥ 80% of cells, the Bcl-B/BECN1 complex was present in the ER compartment (calreticulin staining), whereas Bcl-B/BECN1 colocalization with mitochondria (Hsp60 staining) was far less common. All together, these results demonstrate that Bcl-B and BECN1 colocalize mostly at the ER compartment.
The Bcl-B BH1 domain contributes to binding to BECN1
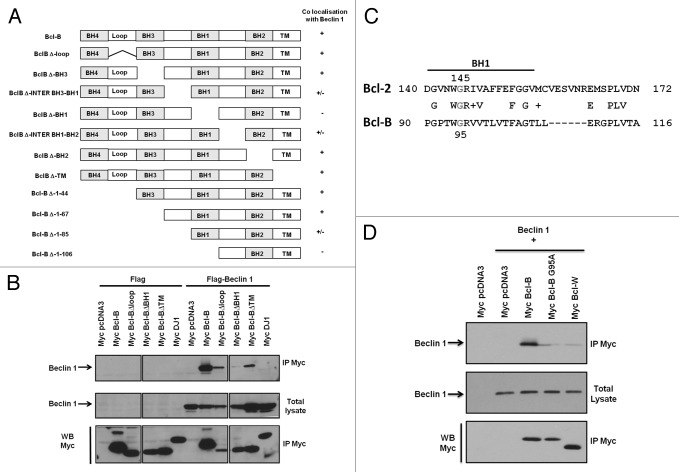
It has been previously reported that the BH1 domains of Bcl-2 and Bcl-XL are necessary for their direct binding to BECN1.8,9 To identify the domains of Bcl-B crucial for its interaction with BECN1, we generated Bcl-B deletion mutants lacking BH, inter-BH, loop or transmembrane domains (Fig. 2A, left panel). Then, we transfected into HeLa cells a GFP-BECN1 plasmid, together with plasmids encoding either WT Bcl-B or Bcl-B deletion mutants, and performed confocal microscopy experiments to determine their colocalization profiles (Fig. 2A, right column; Fig. S2). The colocalization was lost when BECN1 was co-expressed with Bcl-B mutants lacking the BH1 domain. Importantly, Bcl-B mutants lacking the regions surrounding the BH1 domain (Bcl-B Δinter BH3-BH1 and Bcl-B Δinter BH1-BH2) partially lost their capacity to colocalize with BECN1.
Figure 2. The Bcl-B BH1 domain contributes to binding to BECN1. (A) The scheme represents the protein sequences of different Bcl-B deletion mutants. Plasmids encoding Bcl-B mutants were co-transfected in HeLa cells in combination with a plasmid encoding GFP-BECN1 protein. After 24 h, the cells were fixed, permeabilized, and successively incubated with an anti-Myc antibody followed by secondary antibodies conjugated to a red fluorophore. The antibody localization was visualized by confocal UV microscopy to determine the colocalization of the Bcl-B mutants with BECN1. (+): co-localization with BECN1; (-): absence of colocalization with BECN1; (+/−): partial colocalization with BECN1. (B) 293T cells were co-transfected with plasmids encoding either Flag or Flag-BECN1 in combination with plasmids encoding Bcl-B deletion mutants. After 24 h, IPs were performed using an anti-Myc antibody and the immune complexes were analyzed by immunoblotting using anti-BECN1 (top panel) or anti-Myc antibodies (bottom panel). The cell lysates (50 μg) were analyzed directly in the gels to confirm the protein expression (middle panel). (C) The scheme represents the protein sequence alignment of the Bcl-2 and Bcl-B BH1 domains. Note that glycine 145 of the Bcl-2 BH1 domain (reported to be important for the binding to BECN1) is also present in the Bcl-B BH1 domain in position 95. (D) 293T cells were co-transfected with plasmids encoding Flag-BECN1 in combination with plasmids encoding Myc-tagged versions of Bcl-B, Bcl-B G95A or Bcl-W. After 24 h, IPs were performed using an anti-Myc antibody and the immune complexes were analyzed by immunoblotting using anti-BECN1 (top panel) or anti-Myc antibodies (bottom panel). The cell lysates (50 μg) were analyzed directly in the gels to confirm the protein expression (middle panel).
To confirm these results, we performed co-immunoprecipitation experiments with cellular extracts prepared from HeLa cells transfected with plasmids encoding BECN1 and the four representative Bcl-B mutants (Fig. 2B). Of the four Bcl-B mutants, Bcl-B lacking the BH1 domain lost its ability to pull down BECN1, confirming the importance of the Bcl-B BH1 domain for the interaction with BECN1. To characterize further the interaction between Bcl-B and BECN1, we identified the minimal amino acid region of the Bcl-B BH1 domain necessary for interaction with BECN1. Figure 2C shows the amino acid sequence alignment of the Bcl-2 and Bcl-B BH1 domains. Interestingly, the glycine in position 145 of Bcl-2, which is necessary for the interaction between Bcl-2 and BECN1, is conserved in the Bcl-B BH1 domain in position 95. To investigate whether glycine 95 of Bcl-B is required for the interaction with BECN1, we generated the Bcl-B G95A mutant. We transfected 293T cells with Bcl-B, Bcl-B G95A, or Bcl-W (which serves as a negative control) in combination with BECN1. We found that the binding of Bcl-B G95A to BECN1 was markedly decreased, demonstrating that glycine 95 contributes to the interaction of Bcl-B with BECN1. In contrast, Bcl-B (G95A) still retained its ability to bind the pro-apoptotic protein BimEL, although the interaction appeared to be weakened (Fig. S3A). These results demonstrate that the Bcl-B BH1 domain and, more particularly, glycine 95, contribute to the Bcl-B/BECN1 interaction.
The BECN1 BH3 domain is necessary for binding to Bcl-B
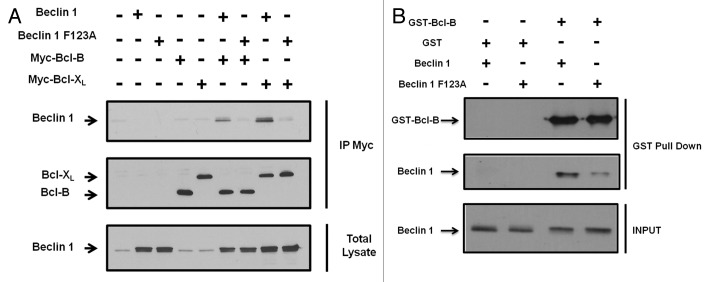
It has been previously reported that BECN1 protein carries a BH3 domain, which is implicated in binding to Bcl-2 family members (Bcl-2 and Bcl-XL). Pattingre et al. showed that a single mutation in the BECN1 BH3 domain (BECN1 F123A) was sufficient to abrogate its binding to Bcl-2.9 To investigate whether the BECN1 BH3 domain is also necessary for its binding to Bcl-B, we performed co-immunoprecipitation experiments using 293T cells transfected with either Bcl-B or Bcl-XL in combination with either wild-type (WT) BECN1 or BECN1 F123A (Fig. 3A). The interaction of BECN1 F123A with both Bcl-XL and Bcl-B was significantly diminished compared with its WT counterpart. To confirm these results, we performed a GST pulldown assay in which recombinant GST-tagged Bcl-B protein was incubated with BECN1 or BECN1 F123A proteins obtained following transcription/translation in a cell-free system (Fig. 3B). As expected, we found that GST-Bcl-B protein pulled down BECN1 with greater efficiency than the BECN1 F123A mutant. These experiments confirm that BECN1 and Bcl-B directly interact, and they also demonstrate that the BH3 domain of BECN1 is necessary for binding to Bcl-B.
Figure 3. The BECN1 BH3 domain is necessary for binding to Bcl-B. (A), 293T cells were co-transfected with plasmids encoding either Flag-BECN1 or Flag-BECN1 F123A in combination with plasmids encoding Myc-tagged versions of either Bcl-B or Bcl-XL (a Bcl-2 member that binds to BECN1). After 24 h, IPs were performed using an anti-Myc antibody and the immune complexes were analyzed by immunoblotting using either anti-BECN1 or anti-Myc antibodies (bottom panels). The cell lysates (50 μg) were analyzed directly in the gels to confirm the protein expression (top panels). (B), GST-fusion Bcl-B protein (2 μg) was incubated overnight at 4°C with either S35 transcribed/translated BECN1 or BECN1 G95A protein. The expression of S35 BECN1 proteins was confirmed by SDS-PAGE immunoblot using a BECN1 antibody (bottom panel). The GST-Bcl-B protein was recovered on glutathione-sepharose and the associated proteins were analyzed by SDS-PAGE immunoblot using a Bcl-B (top panel) or BECN1 (middle panel) antibody.
Bcl-B overexpression inhibits LC3-II accumulation, autophagic flux and cell death induced by amino acid starvation
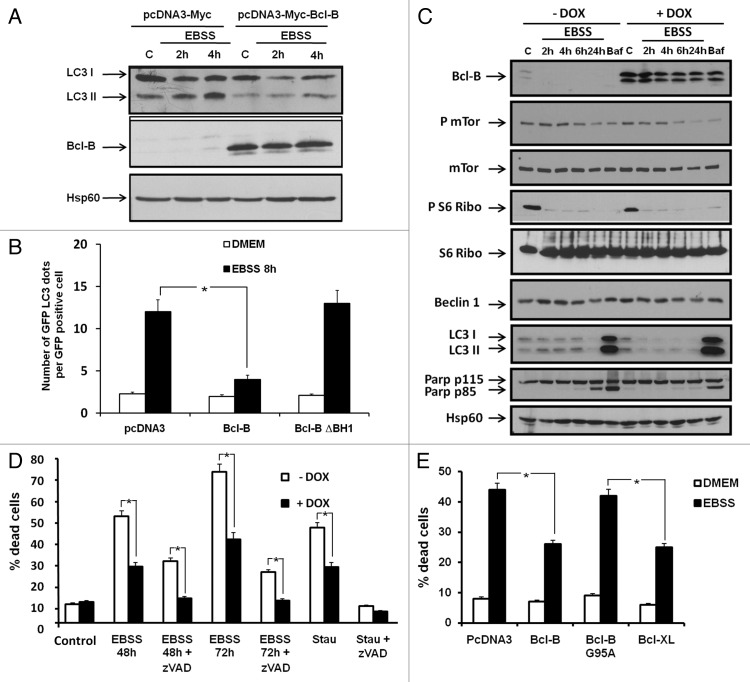
Bcl-2 inhibits autophagy mediated by AA starvation and other stimuli via its binding to BECN1.9,20 The LC3 protein is a commonly used marker of autophagy. After proteolytic processing by Atg4, LC3 conjugation with phosphatidylethanolamine allows its insertion into autophagic vesicle membranes, thereby promoting autophagy. The ratio of unconjugated to conjugated LC3 serves as a surrogate indicator of autophagic activity. AA starvation induced LC3-II accumulation in HeLa cells transfected with a plasmid encoding Myc-Tag but not Myc-Bcl-B (Fig. 4A). Similar results were obtained using the 293T and HT29 cell lines (not shown).
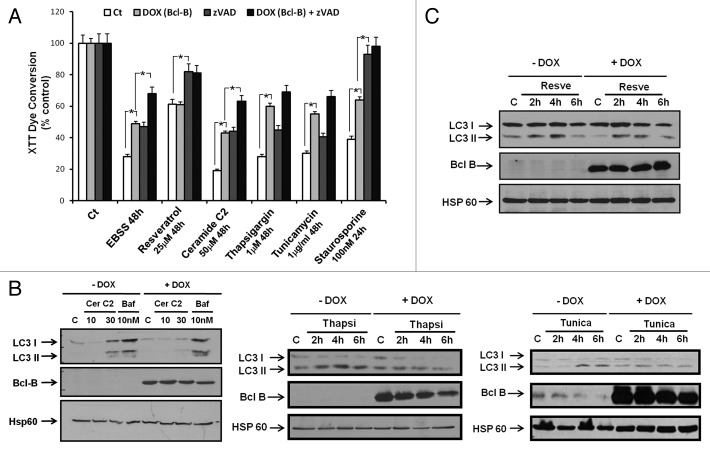
Figure 4. Bcl-B overexpression impairs LC3 processing and cell death induced by amino acid starvation. (A) HeLa cells were transfected with plasmids encoding either Myc-tag or Myc-Bcl-B. After 24 h, the cells were placed in DMEM (complete media) or EBSS (amino acid-free media) for 2 or 4 h. Next, the cells were washed and the cell lysates (30 μg) were subjected to SDS-PAGE immunoblot using LC3B, Bcl-B or Hsp60 antibodies. (B) HeLa cells were transfected with plasmids encoding Myc-Tag, Myc-Bcl-B or Bcl-B ΔBH1 in combination with a plasmid encoding GFP-LC3. After 24 h, the cells were placed in DMEM, EBSS for 8 h or stimulated in DMEM for 24 h in the presence of bafilomycin A1 (10 nM). Confocal microscopy quantification of autophagy was performed. (C) HeLa-TET-Bcl-B cells were incubated with or without doxycycline (1 μg/ml) for 48 h to induce Bcl-B expression. The cells were placed in DMEM or EBSS medium for 2, 4, 6 or 24 h. Bafilomycin stimulation for 24 h serves as a positive control for LC3 cleavage. Next, the cells were washed and the cell lysates (30 μg) were subjected to SDS-PAGE immunoblot using the indicated antibodies. (D) HeLa-TET-Bcl-B cells were incubated with or without doxycycline for 48 h. Next, the cells were placed in DMEM or EBSS medium for 48 or 72 h in the presence or absence of Z-VAD-fmk (50 μM). The cells were washed and incubated with propidium iodide (1 μg/ml), and the percentage of dead cells was assessed by flow cytometry. Staurosporine stimulation for 24 h serves as a positive control for the induction of apoptosis. (E) HeLa cells were transfected with plasmids encoding Bcl-B, Bcl-B G95A or Bcl-XL. After 24 h, the cells were placed in DMEM or EBSS medium for 48 h. The percentage of dead cells was determined as described by flow cytometry. Where indicated, the cumulative data ± SD from three independent experiments are shown. *p < 0.05 in two-sided Student’s t-test.
To further evaluate the impact of Bcl-B on autophagy, we monitored LC3 protein localization by confocal microscopy, using cells transfected with GFP-LC3, and the number of GFP-LC3 puncta was evaluated (Fig. 4B). The overexpression of Bcl-B strikingly reduced GFP-LC3 localization to autophagic vesicles, as indicated by a reduced number of GFP-LC3 puncta. In contrast, Bcl-B lacking the BH1 domain had no effect on GFP-LC3 localization. To better characterize the autophagy-inhibitory effect of Bcl-B, we generated HeLa-TET-Bcl-B cells in which Bcl-B expression can be induced using doxycycline. Bcl-B expression was induced for 48 h before AA starvation. In the absence of Bcl-B, AA deprivation triggered the dephosphorylation of mTor and S6 ribosomal protein and LC3-II conversion, a hallmark of autophagy (Fig. 4C). Although AA starvation similarly inhibited mTor and S6 phosphorylation in Bcl-B-overexpressing HeLa cells, we observed a decrease in levels of LC3-I and LC3-II, suggesting that Bcl-B could modulate the autophagy induced by AA deprivation. In addition, Bcl-B overexpression reduced the cleavage of PARP, a marker of apoptosis, in HeLa cells subjected to AA deprivation (Fig. 4C).
We next investigated whether the autophagic inhibitory effect of Bcl-B could be due to a blockade of autophagic flux. In DMEM complete media conditions, the specific inhibitor of vacuolar H+ ATPase bafilomycin A1 (Baf) induced a comparable accumulation of LC3-II in cells overexpressing Bcl-B and control cells (Fig. S4A, lanes 2 and 6), suggesting that Bcl-B does not block basal autophagic flux. In accordance with our previous results, we observed that Bcl-B overexpression inhibited the LC3-II accumulation induced by EBSS (Fig. S4A, lanes 3 and 7). In EBSS media, Baf induced a significant increase in LC3-I and LC3-II accumulation (compare with DMEM + Baf) only in cells that did not overexpress Bcl-B (Fig. S4A, lanes 4 and 8). These results suggest that Bcl-B blocks the autophagic flux induced by EBSS. A quantification of LC3-II protein levels from three independent experiments is given in Figure S4B. Similar results were obtained in 293T cells transfected with either control or Bcl-B plasmids (Fig. S4C and S4D). The western blot data were also confirmed by confocal microscopy, using HeLa cells transfected with GFP-LC3. The number of GFP-LC3 puncta was evaluated under either DMEM or EBSS media in the presence or the absence of Baf (Fig. S4E). These results show that Bcl-B overexpression blocks the autophagic flux induced by AA starvation in two different cell lines.
To investigate the consequences of Bcl-B overexpression on cell death, HeLa cells were first treated for 48 h with doxycycline and then maintained for 48 or 72 h in the absence of AA (Fig. 4D). Cell death induced by AA deprivation was significantly reduced by Bcl-B overexpression and by the pan-caspase inhibitors Z-VAD-fmk (Fig. 4D) or qVD-OPH (not shown). Even more cell death inhibition was obtained when Bcl-B was overexpressed and caspases were simultaneously inhibited. As a control of apoptosis induction, we also examined cell death induced by staurosporine. Staurosporine-induced cell death was fully inhibited by Z-VAD-fmk and partly inhibited by Bcl-B overexpression. The inhibition of EBSS-induced cell death by Bcl-B expression was also confirmed in clonogenic assays (Fig. S5). In another experimental design, HeLa cells were transiently transfected with plasmids encoding Bcl-B, Bcl-B (G95A), or Bcl-XL. Cell death induced by AA deprivation was inhibited by Bcl-B or Bcl-XL overexpression (Fig. 4E). However, the overexpression of Bcl-B (G95A) failed to reduce the cell death induced by AA starvation, confirming the requirement of BECN1 in this process. In contrast, Bcl-B (G95A) still retained its ability to inhibit staurosporine- and etoposide-induced apoptosis (Fig. S3B), confirming that BECN1 interaction is not required for Bcl-B anti-apoptotic function.
Bcl-B overexpression inhibits cell death induced by other pro-autophagic and pro-apoptotic stimuli
To better characterize the influence of Bcl-B overexpression on cell fate, we tested molecules that have already been reported to induce apoptosis and/or autophagy on HeLa-TET-Bcl-B cells (Fig. 5A). To this end, the effects of EBSS, resveratrol, C2 ceramide, thapsigargin and tunicamycin were assessed using the XTT dye-reduction viability assay. Resveratrol is a natural polyphenolic compound that was reported to trigger autophagic cell death in CML cells via both JNK-mediated p62 overexpression and AMPK activation.21 The cell-permeable sphingolipid C2 ceramide was reported to induce apoptosis and autophagic cell death. Like AA starvation, C2 ceramide triggers autophagy by interfering with the mTOR signaling pathway and by dissociating the BECN1/Bcl-2 complex.20 The ER-stress inducers thapsigargin and tunicamycin were reported to stimulate autophagy through the PERK-eIF2α pathway, the IRE1-JNK1-BECN1 pathway and Ca2+ release.22 We also pretreated cells with Z-VAD-fmk, in some cases, to determine the role of caspases (Fig. 5A). AA starvation for 48 h greatly reduced metabolically active cells (XTT), which was partially counteracted by Bcl-B (induced by doxycycline) or by caspase inhibition. Combining Bcl-B overexpression and caspase inhibition further improved the protection against some stimuli (EBSS, C2 ceramide, thapsigargin, tunicamycin), but the results reached statistical significance only for EBSS and C2 ceramide. Nevertheless, these data suggest that Bcl-B affords a component of caspase-independent (i.e., non-apoptotic) protection from cell death induced by pro-autophagic stimuli. These results indicate that the capacity of Bcl-B to protect cells from caspase-independent cell death can be extended to other pro-autophagic stimuli aside from AA deprivation alone. Interestingly, cells treated with resveratrol, a molecule capable of inducing both autophagy and apoptosis,23 were protected by Z-VAD-fmk but not by Bcl-B overexpression. Next, we determined whether the protective effect of Bcl-B overexpression was correlated with an alteration in LC3 processing, monitoring the LC3-I and LC3-II proteins by immunoblotting. LC3-II levels were lower in Bcl-B overexpressing cells stimulated by C2 ceramide, thapsigargin or tunicamycin (Fig. 5B) but not by resveratrol (Fig. 5C). These results suggest that Bcl-B overexpression reduces autophagy, correlating with its prosurvival effect.
Figure 5. Bcl-B overexpression inhibits LC3 processing and reduces cell death induced by diverse pro-autophagic stimuli. (A), HeLa-TET-Bcl-B cells were incubated with or without doxycycline in the presence or absence of Z-VAD-fmk for 48 h. Then, the cells were placed in EBSS media or incubated with known inducers of autophagy for 48 h. Staurosporine treatment for 24 h was used as a positive control for the induction of apoptosis. The cell viability was assessed by an XTT cell metabolism assay. (B and C), HeLa-TET-Bcl-B cells were incubated with or without doxycycline for 48 h. Then, the cells were stimulated with rapamycin, C2 ceramide, thapsigargin or tunicamycin during the time indicated. The cells were washed and the cell lysates (30 μg) were subjected to SDS-PAGE immunoblot using an LC3B, Bcl-B, or Hsp60 antibody. Where indicated, the cumulative data ± SD from three independent experiments are shown. *p < 0.05 in two-sided Student’s t-test.
Bcl-B silencing induces both apoptotic and autophagic cell death
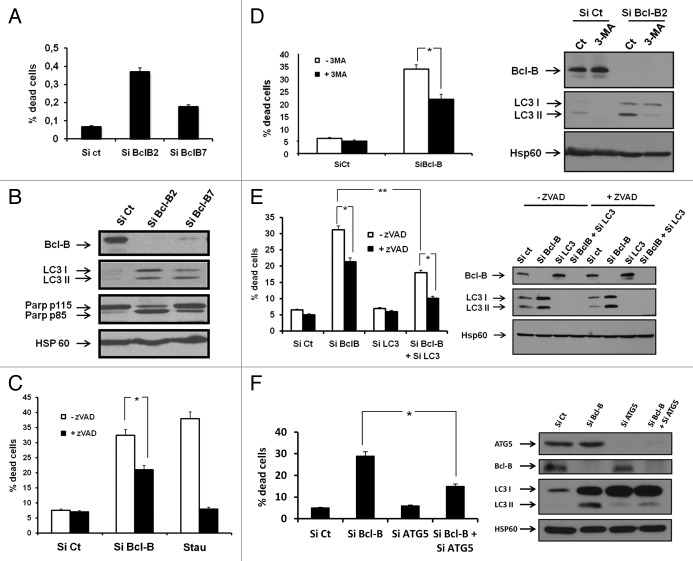
To characterize further the role of Bcl-B in cell survival in autophagy-promoting conditions, we knocked down Bcl-B using an siRNA approach. Both Bcl-B siRNAs tested increased PI staining (Fig. 6A), PARP cleavage, and LC3-II conversion and accumulation (Fig. 6B). These results were confirmed in the U266 myeloma cell line expressing endogenous Bcl-B (Fig. S6). Importantly, Z-VAD-fmk reduced but did not abrogate cell death induced by Bcl-B siRNA, showing that Bcl-B knockdown induced both caspase-dependent and caspase-independent cell death (Fig. 6C). To determine whether autophagy was necessary to trigger the cell death process mediated by Bcl-B knockdown, we established that the autophagy inhibitor 3-methyl-adenine (3-MA) significantly lowered the cell death induced by Bcl-B silencing (Fig. 6D). Further, silencing LC3 expression by siRNA significantly reduced the cell death mediated by Bcl-B silencing (Fig. 6E). Identical results were obtained when ATG5 expression was knocked down (Fig. 6F). Finally, using a BECN1 siRNA, we established that this protein contributes to the mediation of the cell death induced by Bcl-B siRNA (Fig. S7). Indeed, Bcl-B silencing alone induced 30–40% cell death, as judged by PI staining. The silencing of BECN1 alone (Fig. S7A and S7B) did not affect cell death in identical conditions, whereas extinction of both Bcl-B and BECN1 provided protection against cell death. This protection was confirmed with phase contrast microscopy images and cathepsin B and L activities (Fig. S7C and S7D). These results suggest that Bcl-B silencing triggers both apoptotic and autophagic cell death.
Figure 6. Bcl-B silencing induces both apoptotic and autophagic cell death. (A) HeLa cells were transfected with either control siRNA or with two different Bcl-B siRNAs for 72 h. One portion of the cells was incubated with propidium iodide, and the percentage of dead cells was assessed by flow cytometry. (B) The second portion of the cells was lysed, and the protein extracts were subjected to SDS-PAGE immunoblot using Bcl-B, LC3, PARP and Hsp60 antibodies (bottom). (C) HeLa cells were transfected with control or Bcl-B siRNA for 72 h, or stimulated for 24 h with staurosporine (100 nM) in either the presence or the absence of Z-VAD-fmk. The cells were next incubated with propidium iodide, and the percentage of dead cells was assessed by flow cytometry. (D) HeLa cells were transfected with either control or Bcl-B siRNA in presence or absence of 3-MA (2 mM) for 72 h. The percentage of dead cells was assessed by flow cytometry (left panel), and the protein expression was checked by SDS-PAGE immunoblot using Bcl-B, LC3 or Hsp60 antibody (right panel). (E) HeLa cells were transfected with control, LC3 or Bcl-B siRNA for 72 h in the presence or absence of Z-VAD-fmk. The percentage of dead cells was assessed by flow cytometry (left panel), and the protein expression was verified by SDS-PAGE immunoblot using Bcl-B, LC3 or Hsp60 antibody (right panel). (F) HeLa cells were transfected with control, ATG5 or Bcl-B siRNA. After 72 h, the cells were harvested and washed. The cell lysates were prepared and subjected to SDS-PAGE immunoblot (right panel). The protein knockdown was verified using a Bcl-B or ATG5 antibody. Autophagy was monitored using LC3 B antibody. The cells were incubated with propidium iodide to determine the percentage of dead cells by flow cytometry (right panel). Where indicated, the cumulative data ± SD from three independent experiments are shown. *p < 0.05, **p < 0.005 in two-sided Student’s t-test.
Bcl-B silencing sensitizes cells to amino acid starvation
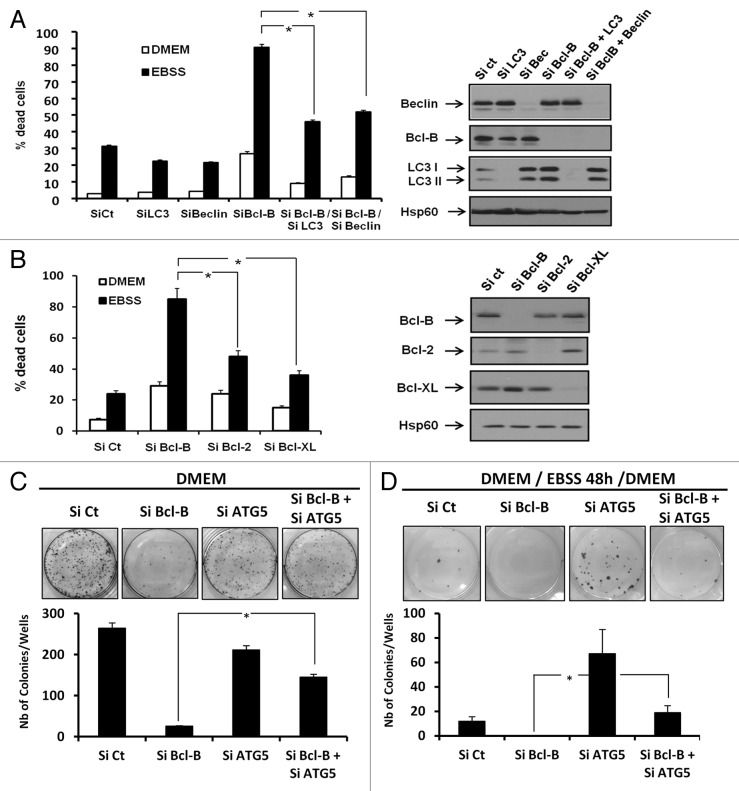
To strengthen our data showing that Bcl-B overexpression inhibits cell death induced by AA deprivation, we analyzed the consequences of Bcl-B knockdown on cell death induced by AA starvation. To this end, cells were first transfected with a Bcl-B siRNA in combination with either an LC3 or BECN1 siRNA for 72 h and then cultured for 24 h in either DMEM or EBSS media (Fig. 7A). AA deprivation-induced cell death was markedly elevated in Bcl-B-depleted cells compared with cells transfected with control siRNA or either LC3 or BECN1 siRNA. Thus, Bcl-B loss dramatically sensitizes cells to cell death induced by AA deprivation. Importantly, both LC3 and BECN1 siRNA reduced the cell death induced by the combination of AA deprivation and Bcl-B siRNA, suggesting that the cell death observed is dependent on components of the autophagy machinery. The protein knockdown efficiency was confirmed by immunoblot analysis (Fig. 7A, right panel). Finally, we compared the capacity of Bcl-B, Bcl-2 and Bcl-XL siRNA to modulate cell survival during AA starvation (Fig. 7B). PI staining indicated that the knockdown of these 3 anti-apoptotic proteins induced similar cell death responses when cells were incubated in complete media. However, upon AA starvation, Bcl-B knockdown triggered ≈90% cell death, whereas Bcl-2 and Bcl-XL knockdown had less impact on the ability of HeLa cells to die (Fig. 7B). Immunoblotting showed that the siRNA treatments effectively and selectively knocked down Bcl-B, Bcl-2 and Bcl-XL protein expression (Fig. 7B, left panel). All together, these results show that Bcl-B protein knockdown sensitizes cells to the cell death induced by AA starvation by triggering both apoptosis and autophagy.
Figure 7. Bcl-B knockdown sensitizes tumor cells to amino acid starvation. (A) HeLa cells were transfected with control, LC3, BECN1 or Bcl-B siRNA for 72 h. The cells were incubated either in DMEM or in EBSS medium for 24 h. The percentage of dead cells was assessed by flow cytometry (Left panel) and protein expression was assessed by SDS-PAGE immunoblot using the indicated antibodies (right panel). (B) HeLa cells were transfected with control, Bcl-B, Bcl-2 or Bcl-XL siRNA for 72 h. The cells were incubated in either DMEM or EBSS medium for 24 h. The percentage of dead cells was assessed by flow cytometry (left panel), and the protein expression was analyzed by SDS-PAGE immunoblot using the indicated antibodies (right panel). (C and D), HeLa cells (5 x 102 per well) were transfected with control, ATG5 or Bcl-B siRNA. After 72 h, the cells were washed and incubated in DMEM complete medium (C) or EBSS medium (D). After 2 d, the media were removed and replaced with DMEM for an additional 5-d period. The cells were then fixed, and the colonies were detected by adding aqueous crystal violet solution (top panels). The number of colonies was scored by ImageJ quantification software (bottom panels). Where indicated, the cumulative data ± SD from three independent experiments are shown. *p < 0.05 in two-sided Student’s t-test.
Finally, to evaluate the implications of autophagy in a cell-growth capacity, we performed clonogenic assays in Hela cells incubated with control, Bcl-B, ATG5 or the combination of Bcl-B and ATG5 siRNAs. After 72 h of siRNA transfection, the DMEM media was removed and replaced with either DMEM (Fig. 7C) or EBSS media (Fig. 7D). Two days later, the media were removed and replaced with DMEM for an additional 5-d incubation period. In the cells maintained continuously in DMEM complete medium (Fig. 7C), the ATG5 siRNA did not significantly affect the number of colonies compared with the control siRNA. As expected, Bcl-B silencing drastically lowered the capacity of the cells to form colonies. Importantly, the extinction of both Bcl-B and ATG5 rescued the clonogenic capacity of Hela cells (Fig. 7C bottom panel). These results confirm that the cell death induced by Bcl-B depletion is partially dependent on components of the autophagy process. In AA-deprivation conditions (Fig. 7D), ATG5 knockdown significantly enhanced colony number compared with control siRNA, meaning that EBSS-induced autophagic cell death is dependent on ATG5. Conversely, Bcl-B siRNA abolished colony formation, confirming that Bcl-B depletion sensitizes cells to the cell death induced by AA deprivation. A significant rescue of colony formation was also observed in AA-depleted HeLa cells transfected with both ATG5 and Bcl-B siRNAs. Taken together, these results indicate that autophagy contributes to the mediation of Bcl-B silencing-induced cell death.
Discussion
In this study, we elucidate a new function of the Bcl-B anti-apoptotic protein as a regulator of autophagy. We showed that Bcl-B overexpression prevents the autophagy induced by a variety of pro-autophagic stimuli. The use of Bcl-B mutants that have lost their ability to bind to BECN1 revealed that this inhibitory effect is closely related to the capacity of Bcl-B to interact with BECN1. Moreover, we demonstrated that the loss of function of Bcl-B induces autophagic cell death and sensitizes cells to AA starvation.
Among the 6 anti-apoptotic Bcl-2 members (Bcl-2, Bcl-XL, Mcl-1, Bcl-W, Bfl-1 and Bcl-B), only Bcl-2 and Bcl-XL have been consistently reported to interact with BECN1 and to inhibit autophagy.8-11 It is known that not all anti-apoptotic Bcl-2 family members are capable of antagonizing autophagy by binding to BECN1.24,25 We show, in this study, that Bcl-B interacts with BECN1 and suppresses autophagy. We also established that the BECN1/Bcl-B interaction requirements were similar to those of Bcl-2/Bcl-XL, requiring the BH1 and BH3 domains of Bcl-B and BECN1, respectively. These results suggest that the molecular basis of autophagy regulation by Bcl-B protein is similar to that of Bcl-2 and Bcl-XL. Bcl-W and, to a lesser extent, Mcl-1 have also been reported to bind to BECN1,24 but this observation was not linked to autophagy inhibition24,25 (and unpublished data). These observations imply that the binding of anti-apoptotic Bcl-2 members to BECN1 is necessary but not sufficient to inhibit autophagy and that further intrinsic properties shared only by Bcl-2, Bcl-XL and Bcl-B are also required. Regarding previous studies9,10 and our own results, we hypothesize that the ability of Bcl-2, Bcl-XL and Bcl-B to maintain BECN1 at the ER could explain their anti-autophagic properties. This notion is supported by a prior report showing that the anti-autophagic function of Bcl-2 requires its targeting to the ER but not the mitochondrial compartment.9 In this regard, we observed a clear colocalization of Bcl-B and BECN1 on the ER membranes. However, further work is required to determine whether Bcl-B localization to the ER is an important determinant of its anti-autophagic activity. It is possible, for example, that either post-translational modifications or interactions with other proteins are required.
We demonstrated that Bcl-B overexpression inhibits LC3 processing and cell death induced by AA starvation, a process that has been consistently described to be dependent on BECN1.26 These observations are in agreement with studies in the literature reporting similar effects caused by Bcl-2 and Bcl-XL overexpression.9,10 Moreover, the use of Bcl-B mutants (Bcl-B ΔBH1 and Bcl-B G95A) lead us to conclude that, similar to Bcl-2 and Bcl-XL, the anti-autophagic function of Bcl-B is strictly related to its capacity to bind BECN1. These functional similarities between Bcl-2, Bcl-XL and Bcl-B confirm that the mechanism of the anti-autophagic function of Bcl-B is comparable to those of Bcl-2 and Bcl-XL.
We also showed that AA starvation triggered both apoptosis and autophagic cell death. We also obtained evidence, using siRNAs targeting autophagy components, that Bcl-B promotes cell survival by blocking both apoptotic (caspase-dependent) and nonapoptotic (caspase-independent) cell death mechanisms. At least in the cell lines used, AA starvation-mediated cell death proceeds via both apoptotic and autophagic cell death, and Bcl-B suppresses both forms of cell death.
To further investigate the anti-autophagic function of Bcl-B, we also tested its capacity to antagonize the cell death induced by various agents previously described to induce both apoptosis and autophagy. We found that Bcl-B overexpression protected cells from the cytotoxicity induced by C2 ceramide, tunicamycin, and thapsigargin by both caspase-dependent (Z-VAD inhibitable) and caspase-independent mechanisms. These results are in agreement with reports showing that Bcl-2 is a potent inhibitor of C2 ceramide- and thapsigargin-induced autophagy.20,27 Interestingly, Bcl-B overexpression failed to protect cells from resveratrol-induced cell death. We speculate that this could be because AA starvation, C2 ceramide, thapsigargin, and tunicamycin all induce autophagy via the BECN1 pathway,20,27 whereas resveratrol is thought to initiate autophagy independently of BECN1.23 This observation further strengthens the notion that the anti-autophagic activity of Bcl-B depends on binding the BECN1 protein.
Controversy exists as to whether autophagy promotes cell survival vs. cell death in various scenarios relevant to cancer biology.28 For example, in contrast to several studies that reported that silencing or knocking out certain autophagy genes, such as BECN1, reduces nonapoptotic cell death,29 Priault et al. showed that, in colorectal cancer cells, Bcl-2 and Bcl-XL are required for starved cells to display a fully functional autophagic pathway.25 They concluded that Bcl-2 and Bcl-XL aid cell survival by assisting with the formation of autophagosomes. In our study, Bcl-B silencing apparently induced autophagic cell death, inasmuch as siRNA-mediated silencing of LC3 or BECN1 improved cell survival when Bcl-B is depleted. Altogether, our results demonstrate that, at least in our cell line model in which autophagy is linked to cell death, the Bcl-B protein acts as a survival factor through both its anti-autophagic and anti-apoptotic functions.
Depending on the setting, BECN1 has been reported to be engaged in different protein interactions. The class III PI3-kinase Vps34 is a major partner of BECN1.30,31 The Vps34/BECN1 complex is involved in the initiation of autophagosome formation during autophagy.30 It has been proposed that Bcl-2 disrupts BECN1 interaction with Vps34,9 sequestering BECN1 away from the autophagy core complex. When nutrients are abundant, Bcl-2/BECN1 binding is stabilized at the expense of BECN1/Vps34 interaction. Conversely, under autophagic conditions, BECN1/Vps34 interaction is stabilized at the expense of Bcl-2/BECN1 binding. This model is, however, controversial because other studies also reported that Bcl-2 is unable to displace the BECN1/Vps34 complex25,32,33 and because the BECN1/Bcl-2 complex is not disrupted under autophagic conditions.25 Further work is needed to elucidate the anti-autophagic mechanism of Bcl-B as pertains to BECN1.34
The role of Bcl-B in cancer is poorly understood. Some tumors overexpress Bcl-B, which in some cases correlates with more aggressive disease.16 In HeLa cells, Bcl-B silencing triggers both apoptotic and autophagic cell death. This observation is in agreement with recent reports in the literature describing the Bcl-B protein as a survival factor.13,15 The survival-promoting effects of Bcl-B may, therefore, be multifactorial, including binding pro-apoptotic Bcl-2 family proteins such as Bax35 and the pro-autophagic protein BECN1. Moreover, other studies have established that the closest mouse homolog, BOO/DIVA, may also exhibit proapoptotic activity by interfering with APAF-1 activation downstream of the mitochondria.36,37
Redundancy among anti-apoptotic Bcl-2 family members has been frequently documented in human cancers, where not one but several Bcl-2 family proteins are overexpressed simultaneously. Despite the potential for redundancy of Bcl-B, Bcl-2, and Bcl-XL as pertains to autophagy suppression, we observed an apparently dominant role for Bcl-B in HeLa cells, in which the silencing of Bcl-B was substantially more cytotoxic than the silencing of either Bcl-2 or Bcl-XL in the context of AA deprivation. This observation suggests that Bcl-B exhibits an anti-autophagic function that Bcl-2 and Bcl-XL cannot fully compensate in this particular tumor cell line. Though these observations deserve to be confirmed in additional tumor models where Bcl-2, Bcl-XL, and Bcl-B are expressed, our findings suggest that Bcl-B protein levels contribute to cancer cell survival by impacting both apoptotic and autophagic cell death mechanisms.
Materials and Methods
Cell lines
Human HeLa, 293T, and HT29 cell lines were provided by ATCC and were grown at 37°C under 5% CO2 in DMEM medium (Gibco BRL, Paisley, UK) supplemented with 10% fetal calf serum (Gibco BRL, 10270), 50 units/ml penicillin, 50 µg/ml streptomycin (Lonza, DE17-602E) and 1 mM sodium pyruvate (Lonza, BE13-115E). Human HeLa-TET-Bcl-B cells were a kind gift from Dr. Reed (Sanford-Burnham Medical Research Institute). To induce Bcl-B expression, HeLa-TET-Bcl-B cells are maintained in DMEM media (Gibco BRL, 31966) supplemented with 10% fetal calf serum (Gibco BRL, 10270) and 1 μg of doxycycline for 48 h. The human myeloma RPMI 8226 cell line was purchased from ATCC and was grown at 37°C under 5% CO2 in RPMI 1640 media (Gibco BRL, 61870) supplemented with 10% fetal calf serum (Gibco BRL, 10270), 50 units/ml penicillin, 50 µg/ml streptomycin and 1 mM sodium pyruvate.
Reagents and antibodies
Sodium fluoride (S7920), sodium orthovanadate (220590), phenylmethylsulfonyl fluoride (PMSF) (P7626), aprotinin (A162-B), leupeptin (SP-04-2217-B), Triton X-100 (N150), Earle’s Balanced Salt Solution (EBSS) (E3024), resveratrol (R5010), C2 ceramide (A7191), thapsigargin (T9033), tunicamycin (T7765), and 3-methyladenine (M9281) were purchased from Sigma. zVAD-fmk was from PeptaNova (3188V). Bafilomycin A1 was from Tocris Bioscience (1334). Anti-Hsp60 (sc-1722) and anti-c-Myc (sc-40) antibodies were purchased from Santa Cruz Biotechnology. Anti-BECN1 (3738), anti-LC3 (2775), anti-mTor (2972), anti-phospho-mTor (2971), anti-S6 Ribosomal (2217), anti-phospho-S6 Ribosomal (4856), anti-Bcl-B (3869), anti-Bcl-2 (2872), anti-Bcl-XL (2764), anti-PARP (9542) and Hrp conjugated anti-rabbit (7074) antibodies were purchased from Cell Signaling Technology. Hrp-conjugated anti-mouse (P0260) and anti-goat (P0449) antibodies were from Dakopatts. An anti-calreticulin antibody was purchased from Enzo Life Science (SAP-600).
Immunoblotting
After stimulation, cells were lysed at 4°C in lysis buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 20 mM EDTA, 100 µM NaF, 10 μM Na3VO4, 1 mM PMSF, 10 µg/ml leupeptin, 10 µg/ml aprotinin, and 1% Triton X-100). The lysates were centrifuged at 16,000 × g for 15 min at 4°C, and the supernatants were supplemented with concentrated SDS sample buffer. A total of 30 µg of protein was separated on a 12% polyacrylamide gel and transferred onto a PVDF membrane (Immobilon-P, Millipore, IPVH00010) in a 20 mM Tris, 150 mM glycine and 20% ethanol buffer at 250 mA for 1 h 30 min at 4°C. After blocking nonspecific binding sites in saturation buffer (50 mM Tris pH 7.5, 50 mM NaCl, 0.15% Tween, and 5% BSA), the membranes were incubated with specific antibodies. The membranes were washed three times using TNA-1% NP-40 (50 mM Tris pH 7.5, and 150 mM NaCl) incubated further with HRP-conjugated antibody for 1 h at room temperature. The immunoblots were revealed using the enhanced chemiluminescence detection kit (Pierce, 32106).
Co-immunoprecipitations
After transfection, cells were suspended in lysis buffer [50 mM TRIS-HCl, pH 7.4, 150 mM NaCl, 20 mM EDTA, 50 μM NaF, 0.5% NP-40, 10 μM Na3VO4, 20 µg ml−1 leupeptin, 20 µg ml−1 aprotinin, 1 mM dithiothreitol and 1 mM phenylmethyl sulfonyl fluoride (PMSF)]. The lysates (500 µl) were then incubated with 1.5 μg of mouse anti-BECN1 antibody (Santa Cruz Biotechnology, sc-48341) and 30 µl protein G Sepharose (Zymed, 10-1242) at 4°C overnight. The beads were washed five times with 1 ml lysis buffer before boiling in Laemmli sample buffer. The immunoprecipitates were analyzed by western blotting using rabbit anti-BECN1 antibody (Cell Signaling Technology, 3738) and monoclonal anti-myc antibodies (Santa Cruz Biotechnology, sc-40).
Knockdown by siRNA
Stealth small interfering RNAs (siRNA) targeting BECN1 (HSS 112741), ATG 5 (HSS 114103), Bcl-XL (HSS 141362), Bcl-2 (HSS 100956), and Mcl-1 (HSS 181042) were purchased from Invitrogen. LC3 siRNA was purchased from Thermo Scientific (J-012846-05 and J-012846-07), and Bcl-B siRNAs were purchased from Applied Biosystems (16810 and 16210). HeLa, 293T, and HT29 cells were transfected with different siRNAs at the final concentration of 50 nM using the Lipofectamine RNAimax protocol (Invitrogen, 13778-150). Next, 48 h after transfection, cells were treated or not with effectors for 1, 2 or 3 supplemental days. The transfection of RPMI 8226 cells was performed using a Nucleofector system (Lonza, VCA-1003) as described previously.19 Briefly, 2.5 million cells were electroporated with different siRNAs at a final concentration of 100 nM using a Nucleofector kit V and program G-015. Subsequently, the cells were plated in 5 ml of RPMI 10% FCS media and incubated for 72 h at 37°C until experimental analysis.
Confocal microscopy
The cells prepared for fluorescence staining were grown on glass coverslips. Flag-BECN1 and Myc-Bcl-B constructs were transiently transfected into HeLa cells for 48 h using the Jet-PEI reagent (Polyplus-Transfection Inc., 101-10) according to the manufacturer’s instructions. After treatment, the cells were washed with ice-cold PBS and were successively fixed and permeabilized with 1% PFA and PBS plus 0.1% Triton X-100, respectively. Then, the cells were incubated for 1 h with monoclonal mouse anti-Myc antibody and with polyclonal rabbit anti-BECN1 antibody. Next, the cells were incubated with anti-mouse Alexa 594- and with anti-rabbit Alexa 488-coupled antibodies. Finally, the cells were incubated with 1 μg/ml DAPI, mounted on glass slides in Fluoromount-G (Southern Biotechnology Associates, 0100-01) and photographed with a confocal laser microscope (Carl Zeiss, LSM-510-Meta).
Cell death assay
Cell viability was measured by a propidium iodide (PI) dye-exclusion assay. Briefly, after treatment, both floating and adherent HeLa cells were collected and incubated with PI (10 μg/ml) for 5 min. The percentage of PI-positive cells was analyzed by flow cytometry using a MACSQUANT Analyzer (Myltenyi Biotech, 130-092).
Measurement of cell metabolism (XTT)
HeLa-TET-Bcl-B Cells (20 × 103 cells/100 µl) were incubated in a 96-well plate with the indicated concentration of cell death inducers for 24 or 48 h, and 50 µl of XTT reagent (Roche Applied Science, 11-465-015) (sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro) benzene sulfonic acid hydrate) was added to each well. The assay is based on the cleavage of the yellow tetrazolium salt XTT to form an orange formazan dye in metabolically active cells. The absorbance of the formazan product, reflecting cell viability, was measured at 490 nm. Each assay was performed in quadruplicate.
Autophagy assay
Autophagy in HeLa cells was measured by light microscopic quantitation of cells transfected with a GFP-LC3 construct (a kind gift from Dr. Maria I. Colombo at the Universidad Nacional de Cuyo). Briefly, cells were transiently transfected for 48 h with GFP-LC3 in combination with Myc-Bcl-B constructs using Jet-PEI reagent. After treatment, the cells were washed with ice-cold PBS and were successively fixed and permeabilized with 1% PFA and PBS plus 0.1% Triton X-100, respectively. Then, cells were successively incubated for 1 h with a monoclonal mouse anti-Myc antibody and with an anti-rabbit Alexa 594-coupled antibody. The number of GFP-LC3 puncta per cell in the GFP-LC3/Myc-positive cells was determined. A minimum of 50–100 cells per sample was counted for triplicate samples per condition per experiment. Prior to the analysis, the cells were starved for 8 h in Earle’s balanced salt solution (EBSS; starvation medium) or maintained in DMEM with 10% fetal calf serum.
Recombinant protein production and purification
GST-fusion proteins containing Bcl-B lacking their C-terminal transmembrane domains (~last 20 amino-acids) (“ΔTM”) were expressed from a pGEX 4T-1 plasmid in XL-1 Blue cells (Stratagene, Inc., 200249). The Bcl-B-GST fusion proteins were produced in bacteria and purified by affinity chromatography using glutathione-Sepharose, as described previously.38
WT and F123A BECN1 were transcribed and translated by means of the TNT-coupled reticulocyte lysate system in the presence of radiolabeled S35-methionine (ICN) according to the manufacturer’s instructions (Promega, L4610).
Pulldown assay
GST or GST-Bcl-B (2 μg each) were pre-incubated separately with 10 μl of glutathione-Sepharose 4B resin for 2 h at 4°C in lysis buffer (10 mM HEPES, pH 7.4, 142.4 mM KCl, 5 mM MgCl2, 0.5 mM EGTA, 1 mM dithiothreitol) containing 2% CHAPS. The resins were washed once in lysis buffer, followed by the addition of 5 μl of transcribed and translated WT and F123A BECN1 reactions. The reactions were performed in 500 μl of lysis buffer with overnight incubation at 4°C. The resins were washed with lysis buffer three times, boiled, and analyzed by SDS-PAGE/immunoblotting. Proteins associated with glutathione-Sepharose were analyzed by immunoblotting using anti-BECN1 or anti-Bcl-B antibody.
Plasmid constructions and directed mutagenesis
The plasmids pcDNA3-Myc, -Myc-Bcl-B, -Myc-Bcl-XL and -Myc-Bcl-W incorporating a Myc epitope tag have been previously described.19 The deletion mutants of Bcl-B and the F123A mutants of BECN1 were created by PCR using QuikChange Site-Directed Mutagenesis (Stratagene, 200518).
Colony formation assay
A total of 5 × 102 HeLa-TET-Bcl-B cells per well or 103 HeLa cells per well were pretreated with 1 µg/ml of doxycycline or transfected with different combinations of siRNA, respectively. After 72 h, both cell lines were washed and placed in DMEM or EBSS media. After 2 d, the cells were washed and replaced in DMEM complete medium for 5 more days. Finally, the cells were fixed and the colonies were detected by adding 1 ml of aqueous crystal violet solution. The number of colonies was scored by ImageJ quantification software (US. National Institutes of Health).
CB and CL activity measurement
HeLa cells were transfected with control, BECN1, Bcl-B or both siRNA. After 48 h, the cells were incubated with 10 µM of cathepsin B and L substrate Z-FR-R110 (Promokine, PK-CA707-10209) for 30 min at 37°C in 5% CO2. Then, the stained cells were washed with PBS and analyzed with a MACSQuant flow cytometer (Miltenyi).
Supplementary Material
Acknowledgments
This work was supported by the Association pour la Recherche sur le Cancer and the Fondation de France and by grants from the National Institutes of Health (NIH) (GM060554 ; CA55164; CA81543). The authors greatly acknowledge the C3M Imaging Core Facility (Microscopy and Imaging platform Côte d’Azur, Mica). GR is the recipient of a grant from the Fondation de France.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/autophagy/article/19084
References
- 1.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, et al. Nomenclature Committee on Cell Death 2009 Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed JC. Mechanisms of apoptosis. Am J Pathol. 2000;157:1415–30. doi: 10.1016/S0002-9440(10)64779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell 2006; 10:375-88. Write to the Help Desk NCBI | NLM | NIH Department of Health & Human Services Privacy Statement | Freedom of Information Act | Disclaimer Dec 18 2006 06:34:27. [DOI] [PubMed]
- 5.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 6.Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–7. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 8.Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282:13123–32. doi: 10.1074/jbc.M700492200. [DOI] [PubMed] [Google Scholar]
- 9.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–39. doi: 10.1038/sj.emboj.7601689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–96. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levine B, Sinha S, Kroemer G. Bcl-2 family members: dual regulators of apoptosis and autophagy. Autophagy. 2008;4:600–6. doi: 10.4161/auto.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ke N, Godzik A, Reed JC. Bcl-B, a novel Bcl-2 family member that differentially binds and regulates Bax and Bak. J Biol Chem. 2001;276:12481–4. doi: 10.1074/jbc.C000871200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Holzgreve W, De Geyter C. Bcl2-L-10, a novel anti-apoptotic member of the Bcl-2 family, blocks apoptosis in the mitochondria death pathway but not in the death receptor pathway. Hum Mol Genet. 2001;10:2329–39. doi: 10.1093/hmg/10.21.2329. [DOI] [PubMed] [Google Scholar]
- 15.Zhai D, Ke N, Zhang H, Ladror U, Joseph M, Eichinger A, et al. Characterization of the anti-apoptotic mechanism of Bcl-B. Biochem J. 2003;376:229–36. doi: 10.1042/BJ20030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krajewska M, Kitada S, Winter JN, Variakojis D, Lichtenstein A, Zhai D, et al. Bcl-B expression in human epithelial and nonepithelial malignancies. Clin Cancer Res. 2008;14:3011–21. doi: 10.1158/1078-0432.CCR-07-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arsov I, Li X, Matthews G, Coradin J, Hartmann B, Simon AK, et al. BAC-mediated transgenic expression of fluorescent autophagic protein Beclin 1 reveals a role for Beclin 1 in lymphocyte development. Cell Death Differ. 2008;15:1385–95. doi: 10.1038/cdd.2008.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luciano F, Krajewska M, Ortiz-Rubio P, Krajewski S, Zhai D, Faustin B, et al. Nur77 converts phenotype of Bcl-B, an antiapoptotic protein expressed in plasma cells and myeloma. Blood. 2007;109:3849–55. doi: 10.1182/blood-2006-11-056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2009;284:2719–28. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puissant A, Auberger P. AMPK- and p62/SQSTM1-dependent autophagy mediate Resveratrol-induced cell death in chronic myelogenous leukemia. Autophagy. 2010;6:655–7. doi: 10.4161/auto.6.5.12126. [DOI] [PubMed] [Google Scholar]
- 22.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puissant A, Robert G, Fenouille N, Luciano F, Cassuto JP, Raynaud S, et al. Resveratrol promotes autophagic cell death in chronic myelogenous leukemia cells via JNK-mediated p62/SQSTM1 expression and AMPK activation. Cancer Res. 2010;70:1042–52. doi: 10.1158/0008-5472.CAN-09-3537. [DOI] [PubMed] [Google Scholar]
- 24.Erlich S, Mizrachy L, Segev O, Lindenboim L, Zmira O, Adi-Harel S, et al. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy. 2007;3:561–8. doi: 10.4161/auto.4713. [DOI] [PubMed] [Google Scholar]
- 25.Priault M, Hue E, Marhuenda F, Pilet P, Oliver L, Vallette FM. Differential dependence on Beclin 1 for the regulation of pro-survival autophagy by Bcl-2 and Bcl-xL in HCT116 colorectal cancer cells. PLoS One. 2010;5:e8755. doi: 10.1371/journal.pone.0008755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mehrpour M, Esclatine A, Beau I, Codogno P. Overview of macroautophagy regulation in mammalian cells. Cell Res. 2010;20:748–62. doi: 10.1038/cr.2010.82. [DOI] [PubMed] [Google Scholar]
- 27.Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 28.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-Induced Expression of Noxa and Beclin-1 Promotes Autophagic Cell Death and Limits Clonogenic Survival. Mol Cell. [DOI] [PubMed]
- 30.Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001;2:330–5. doi: 10.1093/embo-reports/kve061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119:259–70. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 32.Ciechomska IA, Goemans GC, Skepper JN, Tolkovsky AM. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene. 2009;28:2128–41. doi: 10.1038/onc.2009.60. [DOI] [PubMed] [Google Scholar]
- 33.Maiuri MC, Criollo A, Tasdemir E, Vicencio JM, Tajeddine N, Hickman JA, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L) Autophagy. 2007;3:374–6. doi: 10.4161/auto.4237. [DOI] [PubMed] [Google Scholar]
- 34.Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene. 2010;29:1717–9. doi: 10.1038/onc.2009.519. [DOI] [PubMed] [Google Scholar]
- 35.Zhai D, Jin C, Huang Z, Satterthwait AC, Reed JC. Differential regulation of Bax and Bak by anti-apoptotic Bcl-2 family proteins Bcl-B and Mcl-1. J Biol Chem. 2008;283:9580–6. doi: 10.1074/jbc.M708426200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inohara N, Gourley TS, Carrio R, Muñiz M, Merino J, Garcia I, et al. Diva, a Bcl-2 homologue that binds directly to Apaf-1 and induces BH3-independent cell death. J Biol Chem. 1998;273:32479–86. doi: 10.1074/jbc.273.49.32479. [DOI] [PubMed] [Google Scholar]
- 37.Kim JH, Yoon S, Won M, Sim SH, Ko JJ, Han S, et al. HIP1R interacts with a member of Bcl-2 family, BCL2L10, and induces BAK-dependent cell death. Cell Physiol Biochem. 2009;23:43–52. doi: 10.1159/000204088. [DOI] [PubMed] [Google Scholar]
- 38.Zhai D, Jin C, Satterthwait AC, Reed JC. Comparison of chemical inhibitors of antiapoptotic Bcl-2-family proteins. Cell Death Differ. 2006;13:1419–21. doi: 10.1038/sj.cdd.4401937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.