Abstract
Here we discuss the spatio-temporal organization of replication in eubacteria and eukaryotes. Although there are significant differences in how replication is organized in cells that contain nuclei from those that do not, you will see that organization of replication in all organisms is principally dictated by the structured arrangement of the chromosome. We will begin with how replication is organized in eubacteria with particular emphasis on three well studied model organisms. We will then discuss spatial and temporal organization of replication in eukaryotes highlighting the similarities and differences between these two domains of life.
Compared to bacteria, eukaryotes face three challenges when replicating their genomes: the replication of many chromosomes, the compaction of DNA into higher-order chromatin, and the preservation of epigenetic information.
During the bacterial cell cycle, the chromosome exists as a condensed and highly ordered structure that is known as the nucleoid. Most bacteria that have been actively studied have a single circular chromosome that ranges in size from 2 Mb to 8 Mb. DNA replication initiates at a unique origin (oriC) and proceeds bidirectionally around the chromosome to a site opposite oriC called the replication terminus (ter). Unlike eukaryotes, segregation of the newly replicated DNA occurs in lockstep with replisome progression. After a period of cohesion, the replicated DNA is resolved into spatially separate lobes that become fully segregated on the completion of DNA synthesis.
Temporal control of replication initiation is dictated by growth rate and is principally mediated by nutrient dependent changes in the synthesis and activity of the replication initiator protein DnaA. Accumulation of ATP-bound DnaA above a critical threshold triggers initiation (Katayama et al. 2010; Leonard and Grimwade 2011). On origin firing, a diverse collection of control mechanisms ensure that cells do not prematurely reinitiate a new round of synthesis and serve to “reset” DnaA to appropriately monitor nutrient status (Boye et al. 2000; Wang and Levin 2009). In cases in which new rounds of replication initiate prior to the completion of an earlier round (so called, multiforked replication), origins fire synchronously (Skarstad et al. 1986). Thus, unlike eukaryotes in which there is temporal organization of initiation among the myriad of chromosomal origins (see below), in bacteria, spatio-temporal organization of replication is reduced to spatial organization. The exception to this rule is Vibrio cholerae, which contains two unique chromosomes. In this bacterium, it appears that replication of the two chromosomes is initiated at different but stereotypical times in the cell cycle (Rasmussen et al. 2007). The molecular mechanisms underlying the difference in origin firing is only beginning to be unraveled (Duigou et al. 2008), and will not be discussed in this article. Here we focus on the spatial organization of bacterial replication.
Early cytological experiments suggested that replication in bacteria occurs at a specific mid-cell position akin to the replication factories observed in eukaryotes (see below) (Lemon and Grossman 1998, 2000). However, accumulating evidence over the last decade suggests that after replication initiation, the replisomes are not stationary nor do they cluster (Jensen et al. 2001; Bates and Kleckner 2005; Berkmen and Grossman 2006; Reyes-Lamothe et al. 2008). Instead, it appears that the nucleoid serves as both the template and track for the replication machinery. Accordingly, our current view is that the nucleoid itself is the major organizer of DNA replication in bacteria. Our premise is that understanding the spatial organization of bacterial replication requires an understanding of the structure and organization of the bacterial chromosome. We will, therefore, begin by describing how the chromosome is organized within the bacterial nucleoid and how the nucleoid is organized within the cell with particular emphasis on the three model organisms for which we have the most information: Escherichia coli, Bacillus subtilis, and Caulobacter crescentus. We will then discuss spatial organization of replication in the context of this highly organized structure.
ORGANIZATION OF THE BACTERIAL CHROMOSOME
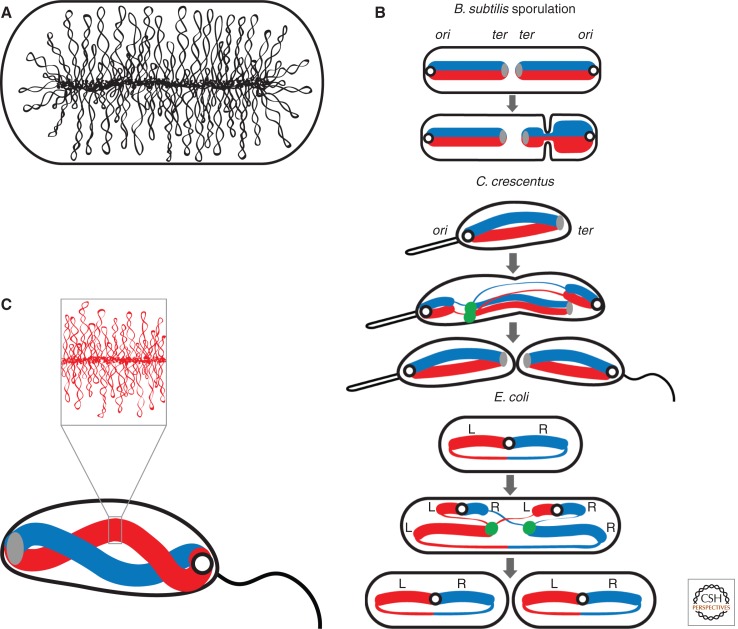
The bacterial chromosome is compacted ∼1000-fold within the bacterial cell. The principal mechanism of chromosome condensation is through DNA supercoiling. In eukaryotes, nearly all supercoils are constrained by the wrapping of DNA around histone octamers called nucleosomes. In bacteria, approximately half of the DNA supercoils are constrained by abundant nucleoid associated proteins (NAPs) that bend or twist the DNA. The rest of the supercoils are unconstrained arising from under-winding the DNA helix (Vologodskii and Cozzarelli 1994; Holmes and Cozzarelli 2000; Dillon and Dorman 2010). The resulting interwound loops or superhelical branches play a key role in compacting and organizing the genome. Evidence suggests that the smallest unit of organization of the bacterial chromosome is an independent supercoiled loop. In E. coli, these unconstrained supercoiled domains have an average size of ∼10 kb (Postow et al. 2004; Stein et al. 2005). Accordingly, its 4.6 Mb genome is thought to contain ∼450 of these topologically isolated domains. Our current view is that these plectonemic loops emanate from a central chromosome core like the bristles of a bottlebrush (Fig. 1A). In addition to their role in the condensation of the chromosome, these topological domains protect the chromosome from DNA relaxation, assist in decatenation of chromosomal links, and have been proposed to aid in the repair of double strand breaks by maintaining broken ends in close proximity.
Figure 1.
The bacterial nucleoid. (A) Diagram of the bottlebrush model of the nucleoid. The image shows a schematic representation of the interwoven super-helical loops emanating from a chromosome core. These flexible, topologically isolated domains are thought to be on average 10 kb in size. (B) Chromosome organization in B. subtilis during spore formation, and C. crescentus and slow growing E. coli during the vegetative cell cycle (gray arrows). Replication origins are illustrated as open circles, termini as solid gray ovals, the left (L) and right (R) replication arms as thick blue and red lines, the E. coli terminus region and newly replicated DNA as thinner lines, and the replisomes as green dots. (C) A model for the gently twisted arms of the C. crescentus chromosome based on recent chromosome conformation capture analysis. The inset shows the plectonemic loops within one of the arms.
There do not appear to be specific sequence elements that define the domain boundaries for these supercoiled loops making it a challenge to characterize them molecularly. It is thought that abundant nucleoid associated proteins with relaxed sequence specificity are responsible for constraining the DNA loops at the chromosome core (Hardy and Cozzarelli 2005). However, definitive evidence for this has been difficult to obtain. RNA polymerase and the SMC/Muk condensin complexes have also been suggested to serve as boundary proteins (Deng et al. 2005; Cui et al. 2008). The current view is that these topologically isolated loops and their boundaries are highly dynamic, changing in response to DNA transactions that occur within and between them. Thus, these small fluid domains provide structure to the chromosome without imposing rigidity.
The next level of organization within the nucleoid involves chromosomal regions that are physically insulated from each other. These regions, called macrodomains, are 800 kb–1 Mb in size and have, thus far, only been described in E. coli. We suspect that this higher-order organization exists in many bacteria. Organization of the chromosome in macrodomains was first described by Niki and colleagues based on cytological observations (Niki et al. 2000) using fluorescence in situ hybridization (FISH). Loci in large regions flanking the origin and terminus had specific localizations distinct from the rest of the chromosome suggesting that loci in these regions cluster and that these large domains are spatially separate. A genetic chromosome conformation assay involving recombination to monitor the frequency of random collisions between different sites on the chromosome independently identified four large regions of the chromosome that do not interact with each other (Valens et al. 2004). Strikingly, this genetic assay identified the same macrodomains surrounding the origin and terminus. It also defined two additional physically insulated domains. These domains flank the Ter macrodomain and are called the left and right macrodomains. Finally, two flexible or unstructured regions were identified that lie to the left and right of the Ori macrodomain. Loci in these unstructured regions can interact with loci in adjacent domains but not with other regions of the chromosome (Valens et al. 2004).
Further support for the presence of structured macrodomains and unstructured flexible chromosomal regions came from time-lapse imaging of fluorescently labeled chromosomal loci (Espeli et al. 2008). These studies revealed that macrodomains and the unstructured regions have strikingly different dynamic behavior. Loci in the flexible regions were significantly more mobile than those found in the macrodomains. The larger territory occupied by loci in the unstructured regions is consistent with the ability of these regions to support long distance interactions with loci present in flanking macrodomains.
Until recently, macrodomains, like the smaller topologically isolated loops, were inferred to exist based on indirect assays. The molecular mechanism underlying this higher-order organization was lacking. In a triumph of bioinformatics analysis and the careful delineation of the Ter macrodomain, Merceir et al. (2008), identified a sequence motif (matS) that was present 23 times in this region of the chromosome but was virtually absent in the rest of the genome. This motif led to discovery of the site-specific DNA binding protein MatP that interacts with it. MatP binds to the matS sites in the Ter macrodomain in vivo. Moreover, cytological studies indicate that MatP localizes as a focus that overlaps loci in the Ter macrodomain, suggesting that it gathers or organizes matS sites together. In support of the idea that MatP is indeed the Ter macrodomain organizer, loci in this domain become more mobile in cells lacking MatP. Moreover, these loci recombine with neighboring domains more frequently when MatP is absent.
Although no experiment has connected the smaller flexible supercoiled domains with the larger constrained macrodomains, we and others imagine that the plectonemic loops (the bottlebrush bristles that emanate from the nucleoid core) are organized-gathered together via site-specific DNA binding proteins like MatP bridging these topologically isolated loops. Based on their size, individual macrodomains are expected to contain ∼100 plectonemic loops. matS sites are well-spaced throughout the Ter macrodomain: there is approximately one every 40 kb. Thus, MatP is well-positioned to gather many of these dynamic bottlebrush bristles into a spatially distinct chromosome lobe. We suspect that analogous DNA binding proteins organize the other macrodomains in E. coli and that similar, yet to be discovered, architectural proteins function in higher-order organization in other bacteria.
The emergence of fluorescence microscopy in bacteria has had a transformative effect on the study of bacterial chromosome organization and the spatial organization of replication. Prior to the cytological studies, it was appreciated that the bacterial chromosome was highly condensed and likely structured but how this organization related to the body plan of the cell was unclear. The ability to visualize individual chromosomal loci in live cells using fluorescent fusions to DNA binding proteins and their target sequences revealed a degree of spatial organization of the chromosome that had not been previously appreciated (Robinett et al. 1996; Straight et al. 1996; Michaelis et al. 1997; Webb et al. 1997; Li and Austin 2002; Lau et al. 2003; Viollier et al. 2004).
The earliest cytological studies aimed at defining the localization of chromosomal loci within the bacterial cell were performed in B. subtilis. These studies revealed that replication initiates at or near mid-cell and that the newly replicated origins move apart to the outer edges of the nucleoid (Lin et al. 1997; Webb et al. 1997; Lemon and Grossman 1998). Analysis of four chromosomal loci suggests that the chromosome adopts an organization in which the replicated origins reside at the cell quarters, the termini at mid-cell, and left and right chromosome arms lie between them (Teleman et al. 1998). The proposed “ori-ter ter-ori” organization of the sister chromatids requires further investigation; however, a similar layout has been established during spore-formation in B. subtilis using an elegant genetic strategy (Wu and Errington 1998). During sporulation, the replicated chromosomes adopt an elongated structure that extends from one cell pole to the other (Fig. 1B) (Lin et al. 1997; Webb et al. 1997; Ben-Yehuda et al. 2003). Within this structure, the replicated origins are anchored at opposite cell poles and the termini reside at mid-cell.
More recent analysis of chromosome organization in C. crescentus and E. coli benefited from new tools, improved technology, and system-wide approaches. In C. crescentus, analysis of 112 individual loci revealed that the physical position of a locus inside the cell recapitulates the genetic map (Viollier et al. 2004). Prior to replication initiation, the origin and terminus reside near opposite cell poles while other loci lie along the long axis of the cell in an order that correlates with their position in the genome (Fig. 1B). On replication initiation, one of the origins is segregated to the opposite cell pole. When replication is complete, the replicated chromosomes reside in an ori-ter ter-ori organization.
In E. coli, similar systematic analyses were performed using slow-growing cells in which newborn cells have a single copy of the chromosome and a G1-like period prior to the initiation of replication. Strikingly, these studies revealed that the E. coli chromosome has a markedly different organization from C. crescentus. In cells with a single chromosome, the origin resides at mid-cell with the left and right chromosome arms in opposite cell halves (Fig. 1B) (Nielsen et al. 2006; Wang et al. 2006). The terminus region appears to connect the two arms to complete the circle. After chromosome replication and prior to cytokinesis, the replicated chromosomes reside in a left-ori-right left-ori-right organization. The distinct spatial localization of the left and right chromosome arms observed in these studies is consistent with low frequency of recombination between loci in the right and left macrodomains (Valens et al. 2004). However, the cytological analysis that indicates that the ter region connects the left and right arms appears to be at odds with a structured Ter macrodomain. How the Ter macrodomain fits in the context of the cellular organization of the chromosome remains to be discovered.
The picture that emerges of this highly condensed and structured nucleoid is of a bottlebrush with plectonemic loops emanating along its length in an order that corresponds with the linear order of loci on the chromosome. In E. coli during slow growth, in which the left and right arms are located in separate cell halves, we envision two bottlebrush arms flanking the origin. In the case of C. crescentus and B. subtilis, the localization studies are not of high enough resolution to say whether there is single core for the left and right chromosome arms or whether the left and right arms represent spatially distinct bottlebrushes.
Recent experiments aimed at defining the three-dimensional structure of the C. crescentus chromosome has shed light on this issue. Umbarger and colleagues used a high throughput chromosome conformation capture assay (called, 5C) to quantify thousands of interloci interaction frequencies (Umbarger et al. 2011). These frequencies were then used to infer the average three-dimensional distances between each locus. Computational modeling of the entire dataset generated the first three-dimensional model of the bacterial chromosome. In agreement with earlier cytological studies, the resulting model suggests that the C. crescentus chromosome is ellipsoidal, with left and right arms roughly symmetric about the ori-ter axis. The two highly condensed arms are spatially distinct but are wound around each other approximately one and half times (Fig. 1C). Thus, it appears that this bacterium has two bottlebrushes, one for each chromosome arm, that are gently wound around each other along their lengths. This rendering of the C. crescentus chromosome is reminiscent of the nucleoid structures observed by fluorescence microscopy in B. subtilis in which the nucleoid appears to gently twist around itself (Berlatzky et al. 2008).
SPATIAL ORGANIZATION OF REPLICATION
Now that we have presented a picture of how the bacterial chromosome is organized within the nucleoid (and how the nucleoid is arranged within the bacterial cell), we return to the central issue of how replication is organized. As mentioned at the beginning of this article, when bacterial cell biology emerged as a field and the technology and tools were in their infancy, replisomes were observed to localize in a mid-cell focus and it was proposed that DNA synthesis occurred in eukaryotic-like replication factories. Experiments over the last decade have challenged this view and it is now generally accepted that after replication initiation, the polymerases that replicate the left and right chromosomal arms function independently of each other and use the chromosome as both a template and a track.
In accordance with the organization of the chromosome within the bacterial cell, replication in E. coli and B. subtilis initiates at or near mid-cell (Lemon and Grossman 1998; Bates and Kleckner 2005; Reyes-Lamothe et al. 2008), whereas in C. crescentus replication initiates at the cell pole (Jensen et al. 2001). The cellular “marks” that direct the replisomes to these positions appear to be the origin itself rather than extrinsic localization determinants. Experiments in B. subtilis in which the origin region was tethered to the cell pole resulted in replication initiation near the cell poles (Berkmen and Grossman 2006). Similarly, in E. coli MukB mutants, the origin localizes close to the cell pole and replication initiates at this position (Danilova et al. 2007). Although assembly of a prereplicative complex is dictated by the localization of oriC, there is strong evidence, at least in B. subtilis, that origin firing occurs at the cytoplasmic membrane. Membrane fractions are enriched in origin DNA compared to other regions of the chromosome and an essential component of the helicase loader is a peripherally associated membrane protein (Beeson and Sueoka 1979; Hoshino et al. 1987; Rokop et al. 2004). We envision that the subcellular localization of oriC directs the initiator protein DnaA to this position. However, once DnaA assembles at this site, transient association of the complex with the membrane triggers origin unwinding and replication initiation. Initiation of replication at the membrane ensures that newly replicated origins are generated at the nucleoid periphery. We hypothesize that this facilitates origin segregation and ensures compartmentalization of replicated and unreplicated DNA.
Once DNA synthesis has begun, the replisomes independently replicate the two arms moving out from mid-cell toward the cell poles and back again in E. coli and from one cell pole toward the other in C. crescentus (Jensen et al. 2001; Reyes-Lamothe et al. 2008). In B. subtilis, the movement of the replisome during a replication cycle has not been described, but we suspect that replication proceeds along the arms in a pattern that is dictated by the organization of the chromosome. In all cases, topologically isolated domains and the larger macrodomains must be disrupted and then reformed during passage of the replisomes, but the path of the polymerases is thought to follow the path of the DNA.
In line with this theme, termination occurs at the location of the terminus DNA. In E. coli, B. subtilis, and C. crescentus, this is at mid-cell. In B. subtilis and E. coli, the terminus DNA appears to remain at mid-cell throughout the replication cycle and only on its duplication do the two replicated chromosomes become spatially separated from each other (Teleman et al. 1998; Lau et al. 2003). In C. crescentus, the terminus starts out close to the cell pole opposite the origin, but at the time when replication is near complete, the replicated origins are present at opposite cell poles with the terminus DNA between them (Jensen et al. 2001).
In summary, the spatial organization of replication in bacteria mirrors the organization of the chromosome within the nucleoid. With the replication machinery following the track of the nucleoid, it stands to reason that the replication process itself could serve as a key player in setting up the organization of the resulting sister chromatids. Indeed, recent work in E. coli suggests that replication along the nucleoid and the coupled segregation and compaction of the newly replicated DNA helps to reproduce the same nucleoid pattern in the daughter cells (Nielsen et al. 2006; Wang et al. 2006).
SPATIO-TEMPORAL ORGANIZATION OF EUKARYOTIC REPLICATION
Unlike bacteria, eukaryotes temporally separate the replication of its genome (S phase) and its segregation (M phase). Accordingly, the discussion of spatial and temporal control of DNA replication in eukaryotes will be restricted to the S phase of the cell cycle. Eukaryotes face three major challenges in replicating their genomes that are not encountered by bacteria. First, the much larger genomes of eukaryotes are fragmented into genetically independent chromosomes, which must be replicated together during S phase. Second, eukaryotic genomes are folded into complex higher-order chromatin structures in which numerous DNA–protein interactions regulate function. This higher-ordered structure is replicated with an average replication fork rate of ∼1.5 kb/min, which is much slower than the bacterial rate of ∼40 kb/min. Thus, eukaryotic cells initiate replication at hundreds (e.g., yeasts) to many thousands (e.g., mammalian cells) of sites to complete synthesis within an appropriate time of their cell cycle. Third, replication must occur with a degree of precision that ensures not only genetic but also epigenetic information is preserved and transmitted to daughter cells. The requirement to activate replication at so many locations on the genome and also preserve the epigenetic code implies that DNA replication must be highly coordinated both temporally and spatially.
STRUCTURING THE REPLICATION PROGRAM
Seminal experiments performed in the 1960s showed two key features of the mammalian replication process: the existence of replicon clusters and that these clusters fire at stereotypical times in the cell cycle. DNA fiber autoradiography showed that replicons in eukaryotes were often activated in groups in which clusters of adjacent replicons (typically 4–10) initiated replication at very similar times (Huberman and Riggs 1966; Edenberg and Huberman 1975). This implied that although individual replicons are discrete synthetic units, the higher-order targets for replication initiation are replicon clusters. Replicon clusters contain ∼1 Mb of contiguous DNA and are typically 5- to 10-fold smaller than cytogenetically defined subchromosomal bands, which were also shown around this time to replicate at predictable times during the S phase.
THE NUCLEAR MATRIX AND NUCLEOIDS
How might spatial information contribute to the replication process? During the 1970s, two groups developed biochemical approaches to address this question. In seminal work, Berezney and Coffey prepared a subnuclear structural framework, which they called the nuclear matrix (Berezney and Coffey 1974); nuclei were extracted with 2M NaCl after first fragmenting DNA with DNase. This nuclear remnant was highly enriched in pulse-labeled nascent DNA (Berezney and Coffey 1975) and pulse-chase experiments inferred that synthesis was taking place within complexes that were associated with the matrix during synthesis (Pardoll et al. 1980). The idea that sites of active DNA synthesis were associated with the nuclear substructure was supported by work from Cook and colleagues (McCready et al. 1980), who prepared “nucleoids” from HeLa cells by direct lysis in a hypertonic (2M NaCl) lysis mix. Nucleoids contain all nuclear DNA, which can be revealed as a “halo” of supercoiled (hence intact) DNA when nucleoids are spread on a water meniscus and visualized by electron microscopy. Using nucleoids, pulse-labeled nascent DNA was shown to be bound to the nuclear substructure—called the nuclear cage—and then move away from the structure as synthesis proceeds (McCready et al. 1980).
Nucleoids provide an analytical tool, which because of the extraction used may or may not faithfully represent the structures present in vivo. To address this point, physiological extraction protocols were developed using cells grown in agarose microbeads (Jackson and Cook 1986). Critically, with this approach, the conditions used preserve the natural chromatin architecture and all synthetic activity that was present in the cell at the time of lysis. As with studies using hypertonic extraction, the nuclear framework—here called the “nucleoskeleton” to reflect the physiological entity—was shown to provide an organizing center on which the active sites of nascent replication could be revealed as replication factories (Hozak et al. 1993). Within these active sites, many replicons were labeled together and this correlated with the coreplication of adjacent replicons within clusters (see above). Importantly, this concept imposes structural constraints on the replication process in which the chromatin must move during synthesis while the synthetic machinery remains fixed. Though this model is clearly different from that seen in bacteria, the basic idea has been supported by live cell imaging of tagged genomic loci in budding yeast (Kitamura et al. 2006) in which factories are also observed (Meister et al. 2006). Collectively, these early studies provided the first evidence for spatial organization of replication within the nucleus and the idea that active sites of DNA synthesis associate with the nuclear substructure.
VISUALIZING REPLICATION IN THE NUCLEUS
The structural organization of replication described above likely reflects a topological imperative to maintain genetic integrity when many thousands of initiation events are required for efficient DNA synthesis. However, the synthetic process in higher eukaryotes must also replicate the epigenetic features of chromatin that maintain cell differentiation. This appears to be achieved by establishing a strict temporal order of DNA replication.
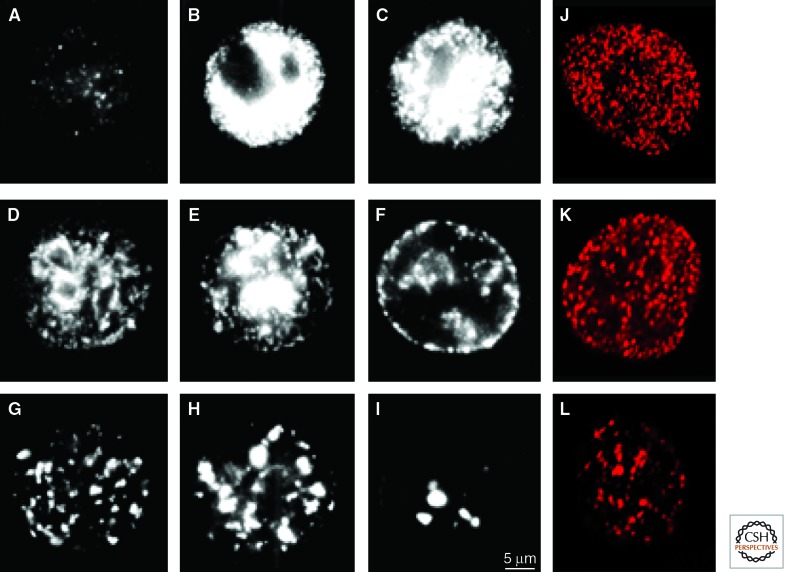
The ability to analyze replication of specific regions of the mammalian genome at defined times during the S phase was facilitated by our ability to label sites of DNA replication and visualize these sites in fixed or living cells both in nuclei and mitotic chromosomes. In early studies, the analysis of chromosomes prepared from cells labeled with the thymidine analog bromo-deoxyuridine (BrdU)—in the previous S phase—and visualized by indirect immuno-labeling showed that euchromatin was always replicated before the heterochromatin (Holmquist and Ashley 2006). Using this approach, indirect immunofluorescence was used to visualize replication sites within nuclei of rat fibroblasts (Nakamura et al. 1986). Early in S phase, these cells were found to contain ∼125 nascent replication sites, each with an estimated ∼1 Mb of DNA. These sites were proposed to represent domains of replicon clusters that engaged synthesis over periods of ∼1 h before synthesis terminated and switched to adjacent sites. In subsequent studies, this labeling approach was used to define a strict temporal program of S phase synthesis (Fig. 2) (Zink 2006).
Figure 2.
Patterns of DNA synthesis change throughout the S Phase. DNA replication precursor analogs can be used to monitor DNA synthesis. HeLa cells (A–I) were synchronized in mitosis using nocodazol and at the onset of the S phase using aphidicolin, a DNA synthesis inhibitor. After removing the inhibitor, cells were grown in medium and nascent sites of DNA replication visualized at 1-h intervals (A–I) during the ∼9-h S phase. At each time point, incorporation of biotin-dUTP into nascent sites was monitored in permeabilized cells by indirect immuno-labeling. Typical patterns seen at different times change as S phase proceeds, with classical early- (A–C), mid- (D–F), and late-S phase (G–I) sites correlating broadly with the synthesis of euchromatin, facultative heterochromatin, and constitutive heterochromatin within different nuclear compartments. Diploid human fibroblasts (J–L) show the same general patterns when nascent sites are visualized in unsynchronized cells after labeling with Cy3-dUTP in vivo.
Many studies have now confirmed the importance of higher-order chromatin domains as synthetic units during DNA synthesis and recent work has begun to reveal how different features of chromosome structure contribute to the S phase timing program. Protocols for labeling nascent sites of DNA synthesis with different halogenated replication precursors (e.g., CldU and IdU) provided early clues that the nascent sites within chromatin foci might be activated according to a temporal program (Manders et al. 1992, 1996). These observations were used to develop “next-in-line” models of S-phase progression and studies on mid/late replicating chromatin domains were used to develop this idea into a “domino hypothesis” (Sporbert et al. 2002). A direct test of this hypothesis, using replication domains in individual chromosome territories and DNA fibers, confirmed that the sequential activation of adjacent chromatin domains along chromosomes is a major mechanism of S-phase progression (Maya-Mendoza et al. 2010).
Over many years, the analysis of replication in fixed or live mammalian cells showed that clusters of adjacent replicons were replicated together throughout the S phase, and that these were contained in higher-order chromatin domains and most probably replicated by machinery within dedicated replication factories. If true, factory level control might explain how synchronous initiation of active replicons and suppression of potential origins within the same chromatin domains could be achieved. However, recent experiments using super-resolution light microscopy indicate that replication factories rather than being distinct structures might better be considered as nuclear domains with specific functional roles. Hence, the ∼1200 factories typically seen in diploid mammalian cells using confocal microscopy (Ma et al. 1998; Cseresnyes et al. 2009) could be resolved into 2–3 times more active sites using super-resolution systems (Cseresnyes et al. 2009; Baddeley et al. 2010).
CHROMOSOME STRUCTURE AND REPLICATION TIMING
In bacteria, the time of replication of any specific chromosomal locus will be defined by its proximity to oriC. In higher eukaryotes where replication initiates at ∼50,000 sites, initiation cannot be the only control point. In fact, in mammalian cells, while all potential origins are established well before the onset of the S phase (Blow and Dutta 2005), only a subset of these is selected for activation when the S phase begins. The analysis of replication foci in nuclei and replicon clusters on DNA fibers shows that the same loci are targeted for synthesis at the onset of the S phase with high efficiency (Jackson and Pombo 1998; Ma et al. 1998). However, it is important to note that at any time during the S phase, only about 15% of the genome is engaged in synthesis, so that initiation must occur throughout the S phase. As noted above, chromosome structure plays a key role in regulating origin activation in the downstream replication-timing program.
What are the basic features of this structure–function relationship? Individual chromosomes form chromosome territories (CT) that occupy discrete nuclear domains (Cremer and Cremer 2010). Within these structures, DNA binds around histone octamers as nucleosomes and the resulting nucleosome fiber folds in space through a series of protein–protein interaction, which then defines DNA loops. Locally, the chromatin loops are constrained within higher-order structures that typically contain 250–1000 kb of DNA. As in bacteria, this level of chromatin organization provides structure without imposing a rigid chromatin environment. These chromatin domains or DNA foci provide the fundamental subunits of higher-order chromatin folding within CTs and have been shown to reflect stable units of higher-order chromatin folding in fixed (Jackson and Pombo 1998) and live (Bornfleth et al. 1999) mammalian cells. Based on patterns of labeling with replication precursor analogs, these ∼1 Mbp chromatin domains clearly correlate with the independent replicon clusters described above.
The ∼1 Mbp replicon clusters were recognized as major determinants of the S phase structure, even though the molecular mechanisms underlying these domains were unknown. Importantly, recent developments in chromosome conformation capture (3C) techniques have confirmed the basic features of chromosome architecture by demonstrating the existence of steady-state, higher-order chromatin entities in cell populations. Two recent studies used unbiased, genome-wide 3C—called Hi-C—to elaborate chromosome architecture (Lieberman-Aiden et al. 2009; Kalhor et al. 2012). This analysis makes three key points. First, steady-state structures seen at the Mbp scale have a “fractal” architecture allowing these fractal globules to fold and unfold without knotting. Second, globules within the active and inactive chromatin compartments have distinct structural and predicted dynamic properties with a preference to interact with structures in the same chromatin class. And third, DNA within the active chromatin class in particular interacts promiscuously with many other part of the genome in the same class. These conclusions emphasize how chromatin compartments can represent pseudo-stable higher-order domains that are sufficiently dynamic to engage in a variety of interactions across cell populations. The underlying mechanisms that define these structures are likely to be very complex, so that the structure of chromatin domains correlates with numerous features of chromatin function. This is especially evident during cell differentiation, when subtle changes in higher-order chromatin architecture during differentiation provide an excellent correlation with changes in replication timing (Ryba et al. 2010).
S-PHASE PROGRESSION
Replication must be performed with 100% precision to preserve genome integrity. As we have seen, many studies infer that chromosome structure and nuclear architecture contribute to this process. However, to fully understand the replication process, it is necessary to recognize the very complex nature of the regulatory mechanisms and explore how the random nature of the initiation process can be directed by structural features. In doing this, it is also important to realize that in eukaryotic cells as in bacteria the process of DNA replication must occur in such a way that the replication machinery displaces proteins that define the structure of local DNA loops while allowing these to reform once synthesis is complete. This imperative to reform chromatin structure following replication explains why local chromosome structure must be a complex composite of many thousands of dynamic interactions.
In the unicellular model eukaryote budding yeast, origins are genetically defined and are activated according to a temporal program (Raghuraman et al. 2001), which in part at least is controlled by origin activation by different cyclins (McCune et al. 2008). But even in this system, analysis of DNA from single cells shows that initiation can be intrinsically disordered in time (Czajkowsky et al. 2008), implying that origin selection has a significant stochastic component (Patel et al. 2006). In mammalian cells, origin selection is also known to be highly stochastic (see Maya-Mendoza et al. 2009 for review). However, this innate feature of the replication process arises as potential origins outnumber origins that are used in each cell cycle by ∼5:1. Despite this flexibility, there is excellent evidence from many sources that replication in mammalian cells is programmed so that different parts of the genome are replicated at preferred times (Zink 2006; Aladjem 2007; Sclafani and Holzen 2007).
As in yeast, in higher eukaryotes euchromatin is replicated early in the S phase and late in heterochromatin. But how ordered is the timing program and what are the underlying mechanisms? As noted above, studies of nascent sites within nuclei show that a specific subset of potential initiation targets are select with good efficiency at the onset of the S phase (Jackson and Pombo 1998; Ma et al. 1998). Numerous genome-wide studies have shown that active genes are preferentially replicated at the beginning of the S phase and the process of selection correlates with patterns of gene expression (Cadoret et al. 2008; Farkash-Amar et al. 2008), active chromatin marks (Hiratani et al. 2008), and the mostly highly interactive gene regions defined by Hi-C (Ryba et al. 2010). These observations imply that the local chromatin context is a key determinant of initiation at the onset of the S phase, with arguably the most mobile chromatin having the highest probability of engaging synthesis.
Exactly how these concepts feed into the selection of potential origins is unknown. However, the key step in the initiation of synthesis must involve enzyme-mediate activation of potential origins to generate preinitiation complexes (pre-ICs), which are then able to bind the synthetic machinery and initiate replication. The early steps in this process must occur within chromatin domains and subsequent reorganization of these domains is thought to be required for pre-ICs to interact within replication factories. The synchronous activation of contiguous replicons within clusters emphasizes the importance of coordination during synthesis, and although the regulatory mechanisms are unknown, these observations are at least consistent with a role for some form of a factory level control process. As the S phase proceeds, changes in expression of different cyclin–CDK complexes serve to increase the potency of potential origins in heterochromatin at late times of the S phase (Katsuno et al. 2009). Hence, the combination of accessibility of potential origins in open chromatin during early S phase, subsequent increase in activity of remaining potential origins in late S phase and robust checkpoints for DNA completion ensures that synthesis is performed in a way that defines the preservation of both genetic and epigenetic information.
Genomic approaches to the analysis of replication timing support the view that synthesis spreads along chromosomes from dominant initiations sites that operate at the onset of the S phase (reviewed in Cadoret and Prioleau 2010; Farkash-Amar and Simon 2010), with subtle differences in timing occurring during cell differentiation (Hiratani et al. 2008; Ryba et al. 2010). Importantly, the demonstration of replication timing zones with sizes typically in the range ∼0.5–1.0 Mb of DNA across cell populations shows that the replication domains correlate with structural chromatin foci, which also correspond with replicon clusters (Maya-Mendoza et al. 2010). Together, these observations show that cells with a specific differentiation status will follow unique temporal programs of S phase progression, which then become an innate feature of DNA synthesis in the corresponding different cell types. In conclusion, in both eubacteria and eukaryotes the process of DNA replication is principally controlled by chromosome organization. In the different bacterial models discussed above, a key feature of the replication process is to ensure that the sister nucleoids are properly positioned to ensure efficient segregation. Eukaryotes have evolved precise mechanisms to ensure the preservation of genetic integrity during cell division. However, eukaryotes face additional challenges in maintaining their genetic and epigenetic integrity during DNA synthesis, in large part because of the huge size of their genomes. As in eubacteria, fundamental aspects of the synthetic process are defined by chromosome structure and function, with many features of organization contributing to the temporal program through which genetic and epigenetic integrity is maintained. Accordingly, one of the key challenges for the future is to define the molecular basis of higher-order chromosome organization that dictates the spatial and temporal organization of DNA replication.
Footnotes
Editors: Stephen D. Bell, Marcel Mechali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Aladjem MI 2007. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet 8: 588–600 [DOI] [PubMed] [Google Scholar]
- Baddeley D, Chagin VO, Schermelleh L, Martin S, Pombo A, Carlton PM, Gahl A, Domaing P, Birk U, Leonhardt H, et al. 2010. Measurement of replication structures at the nanometer scale using super-resolution light microscopy. Nucleic Acids Res 38: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Kleckner N 2005. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121: 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeson J, Sueoka N 1979. Membrane enrichment of genetic markers close to the origin and terminus during the deoxyribonucleic acid replication cycle in Bacillus subtilis. J Bacteriol 139: 911–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Rudner DZ, Losick R 2003. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536 [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS 1974. Identification of a nuclear protein matrix. Biochem Biophys Res Commun 60: 1410–1417 [DOI] [PubMed] [Google Scholar]
- Berezney R, Coffey DS 1975. Nuclear protein matrix: Association with newly synthesized DNA. Science 189: 291–293 [DOI] [PubMed] [Google Scholar]
- Berkmen MB, Grossman AD 2006. Spatial and temporal organization of the Bacillus subtilis replication cycle. Mol Microbiol 62: 57–71 [DOI] [PubMed] [Google Scholar]
- Berlatzky IA, Rouvinski A, Ben-Yehuda S 2008. Spatial organization of a replicating bacterial chromosome. Proc Natl Acad Sci 105: 14136–14140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A 2005. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C 1999. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys J 77: 2871–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye E, Lobner-Olesen A, Skarstad K 2000. Limiting DNA replication to once and only once. EMBO Rep 1: 479–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadoret JC, Prioleau MN 2010. Genome-wide approaches to determining origin distribution. Chromosome Res 18: 79–89 [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M 2010. Chromosome territories. Cold Spring Harb Perspect Biol 2: a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cseresnyes Z, Schwarz U, Green CM 2009. Analysis of replication factories in human cells by super-resolution light microscopy. BMC Cell Biol 10: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Petrushenko ZM, Rybenkov VV 2008. MukB acts as a macromolecular clamp in DNA condensation. Nat Struct Mol Biol 15: 411–418 [DOI] [PubMed] [Google Scholar]
- Czajkowsky DM, Liu J, Hamlin JL, Shao Z 2008. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J Mol Biol 375: 12–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C 2007. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol 65: 1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng S, Stein RA, Higgins NP 2005. Organization of supercoil domains and their reorganization by transcription. Mol Microbiol 57: 1511–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Dorman CJ 2010. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat Rev Microbiol 8: 185–195 [DOI] [PubMed] [Google Scholar]
- Duigou S, Yamaichi Y, Waldor MK 2008. ATP negatively regulates the initiator protein of Vibrio cholerae chromosome II replication. Proc Natl Acad Sci 105: 10577–10582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Huberman JA 1975. Eukaryotic chromosome replication. Annu Rev Genet 9: 245–284 [DOI] [PubMed] [Google Scholar]
- Espeli O, Mercier R, Boccard F 2008. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68: 1418–1427 [DOI] [PubMed] [Google Scholar]
- Farkash-Amar S, Simon I 2010. Genome-wide analysis of the replication program in mammals. Chromosome Res 18: 115–125 [DOI] [PubMed] [Google Scholar]
- Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I 2008. Global organization of replication time zones of the mouse genome. Genome Res 18: 1562–1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CD, Cozzarelli NR 2005. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol 57: 1636–1652 [DOI] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM 2008. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol 6: e245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes VF, Cozzarelli NR 2000. Closing the ring: Links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci 97: 1322–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmquist GP, Ashley T 2006. Chromosome organization and chromatin modification: Influence on genome function and evolution. Cytogenet Genome Res 114: 96–125 [DOI] [PubMed] [Google Scholar]
- Hoshino T, McKenzie T, Schmidt S, Tanaka T, Sueoka N 1987. Nucleotide sequence of Bacillus subtilis dnaB: A gene essential for DNA replication initiation and membrane attachment. Proc Natl Acad Sci 84: 653–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozak P, Hassan AB, Jackson DA, Cook PR 1993. Visualization of replication factories attached to nucleoskeleton. Cell 73: 361–373 [DOI] [PubMed] [Google Scholar]
- Huberman JA, Riggs AD 1966. Autoradiography of chromosomal DNA fibers from Chinese hamster cells. Proc Natl Acad Sci 55: 599–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Cook PR 1986. Replication occurs at a nucleoskeleton. EMBO J 5: 1403–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A 1998. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Wang SC, Shapiro L 2001. A moving DNA replication factory in Caulobacter crescentus. EMBO J 20: 4952–4963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L 2012. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol 30: 90–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Ozaki S, Keyamura K, Fujimitsu K 2010. Regulation of the replication cycle: Conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol 8: 163–170 [DOI] [PubMed] [Google Scholar]
- Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M 2009. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci 106: 3184–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU 2006. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell 125: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ 2003. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol 49: 731–743 [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD 1998. Localization of bacterial DNA polymerase: Evidence for a factory model of replication. Science 282: 1516–1519 [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD 2000. Movement of replicating DNA through a stationary replisome. Mol Cell 6: 1321–1330 [DOI] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE 2011. Regulation of DnaA assembly and activity: Taking directions from the genome. Annu Rev Microbiol 65: 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Austin S 2002. The P1 plasmid is segregated to daughter cells by a “capture and ejection” mechanism coordinated with Escherichia coli cell division. Mol Microbiol 46: 63–74 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DC, Levin PA, Grossman AD 1997. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci 94: 4721–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R 1998. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol 143: 1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA 1992. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci 103: 857–862 [DOI] [PubMed] [Google Scholar]
- Manders EM, Stap J, Strackee J, van Driel R, Aten JA 1996. Dynamic behavior of DNA replication domains. Exp Cell Res 226: 328–335 [DOI] [PubMed] [Google Scholar]
- Maya-Mendoza A, Tang CW, Pombo A, Jackson DA 2009. Mechanisms regulating S phase progression in mammalian cells. Front Biosci 14: 4199–4213 [DOI] [PubMed] [Google Scholar]
- Maya-Mendoza A, Olivares-Chauvet P, Shaw A, Jackson DA 2010. S phase progression in human cells is dictated by the genetic continuity of DNA foci. PLoS Genet 6: e1000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCready SJ, Godwin J, Mason DW, Brazell IA, Cook PR 1980. DNA is replicated at the nuclear cage. J Cell Sci 46: 365–386 [DOI] [PubMed] [Google Scholar]
- McCune HJ, Danielson LS, Alvino GM, Collingwood D, Delrow JJ, Fangman WL, Brewer BJ, Raghuraman MK 2008. The temporal program of chromosome replication: Genomewide replication in clb5Δ Saccharomyces cerevisiae. Genetics 180: 1833–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister P, Taddei A, Gasser SM 2006. In and out of the replication factory. Cell 125: 1233–1235 [DOI] [PubMed] [Google Scholar]
- Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, Espeli O 2008. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135: 475–485 [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K 1997. Cohesins: Chromosomal proteins that prevent premature separation of sister chromatids. Cell 91: 35–45 [DOI] [PubMed] [Google Scholar]
- Nakamura H, Morita T, Sato C 1986. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res 165: 291–297 [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG 2006. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol 62: 331–338 [DOI] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S 2000. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev 14: 212–223 [PMC free article] [PubMed] [Google Scholar]
- Pardoll DM, Vogelstein B, Coffey DS 1980. A fixed site of DNA replication in eucaryotic cells. Cell 19: 527–536 [DOI] [PubMed] [Google Scholar]
- Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N 2006. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell 17: 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Hardy CD, Arsuaga J, Cozzarelli NR 2004. Topological domain structure of the Escherichia coli chromosome. Genes Dev 18: 1766–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL 2001. Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Rasmussen T, Jensen RB, Skovgaard O 2007. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J 26: 3124–3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ 2008. Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133: 90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol 135: 1685–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokop ME, Auchtung JM, Grossman AD 2004. Control of DNA replication initiation by recruitment of an essential initiation protein to the membrane of Bacillus subtilis. Mol Microbiol 52: 1757–1767 [DOI] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM 2007. Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad K, Boye E, Steen HB 1986. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J 5: 1711–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporbert A, Gahl A, Ankerhold R, Leonhardt H, Cardoso MC 2002. DNA polymerase clamp shows little turnover at established replication sites but sequential de novo assembly at adjacent origin clusters. Mol Cell 10: 1355–1365 [DOI] [PubMed] [Google Scholar]
- Stein RA, Deng S, Higgins NP 2005. Measuring chromosome dynamics on different time scales using resolvases with varying half-lives. Mol Microbiol 56: 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW 1996. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol 6: 1599–1608 [DOI] [PubMed] [Google Scholar]
- Teleman AA, Graumann PL, Lin DC, Grossman AD, Losick R 1998. Chromosome arrangement within a bacterium. Curr Biol 8: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Umbarger MA, Toro E, Wright MA, Porreca GJ, Bau D, Hong SH, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, et al. 2011. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell 44: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, Cornet F, Boccard F 2004. Macrodomain organization of the Escherichia coli chromosome. EMBO J 23: 4330–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Thanbichler M, McGrath PT, West L, Meewan M, McAdams HH, Shapiro L 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci 101: 9257–9262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vologodskii AV, Cozzarelli NR 1994. Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct 23: 609–643 [DOI] [PubMed] [Google Scholar]
- Wang JD, Levin PA 2009. Metabolism, cell growth and the bacterial cell cycle. Nat Rev Microbiol 7: 822–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu X, Possoz C, Sherratt DJ 2006. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev 20: 1727–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CD, Teleman A, Gordon S, Straight A, Belmont A, Lin DC, Grossman AD, Wright A, Losick R 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88: 667–674 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J 1998. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol 27: 777–786 [DOI] [PubMed] [Google Scholar]
- Zink D 2006. The temporal program of DNA replication: New insights into old questions. Chromosoma 115: 273–287 [DOI] [PubMed] [Google Scholar]