Abstract
Background
The neurotrophic receptor tyrosine kinase B (TrkB) has diverse signaling roles in neurons and tumor cells. Accordingly, its suppressive targeting is of interest in neuroblastoma and other tumors, whereas its role in improving survival is focused in neurons. Here we describe targeting of TrkB-binding peptide-conjugated liposomes (PCL) to the TrkB-expressing mouse macrophage-like cell line RAW264, and to all-trans-retinoic acid-treated neuron-like TrkB+ SH-SY5Y human neuroblastoma cells.
Methods
Binding and internalization of PCL was monitored by flow cytometry and confocal fluorescence microscopy.
Results
Internalization of TrkB-targeted PCL by RAW264 cells was enhanced and faster when compared with PCL having the corresponding scrambled peptide. Likewise, binding and augmented uptake were confirmed for TrkB+ SH-SY5Y cells, with targeted PCL appearing in the cytoplasm after 20 minutes of incubation.
Conclusion
We demonstrate here the feasibility of targeting liposomes to TrkB-expressing cells by 18-mer peptides, promoting cellular uptake (at least partly into endosomes) via receptor-mediated pathways.
Keywords: liposomes, targeting, tyrosine kinase B, peptide
Background
Neurotrophic tyrosine kinase receptors are widely expressed in neural as well as other cell types, and are selectively activated by their specific neurotrophin ligands. In most cases, this leads to phosphorylation of downstream kinases and promotion of cell survival, growth, and/or differentiation. Upon inadequate signaling by brain-derived neurotrophic factor, the natural ligand of tyrosine kinase B (TrkB), a loss of cochlear sensory neurons may occur.1 This may also be associated with hereditary conditions involving impairment of either brain-derived neurotrophic factor or TrkB function.1 The search for a cure for TrkB-dependent hearing loss has led to attempts to administer recombinant brain-derived neurotrophic factor.2 However, this approach is challenging due to the limited stability and biological activity of recombinant brain-derived neurotrophic factor2 and parallel stimulation of the proapoptotic p75 receptor by neurotrophins.3,4 Accordingly, the action of neurotrophins can be seen as a “double-edged sword,” with the outcome depending unpredictably on a balance of expression of prosurvival tyrosine kinase receptors and proapoptotic p75 receptors. As with most prosurvival signals, excessive tyrosine kinase stimulation may lead to tumorigenesis.5
The development of nanoparticles for the delivery of therapeutic agents is providing new opportunities to develop more efficient therapies. Encapsulation of small molecule components into nanocarriers such as liposomes allows for, eg, improved delivery of anticancer drugs to tumor tissue.6,7 The current focus on development of cancer therapies is on targeted drug delivery, and several approaches have been described for obtaining therapeutic concentrations of anticancer agents on specific target cells while avoiding healthy tissues.8–10 For this purpose, nanoparticles have been conjugated with targeting moieties, such as antibodies and peptides, recognizing specific structures on tumor cells or tumor vasculature.11,12 Compared with antibodies, peptide ligands are smaller, less immunogenic, and easier to produce, modify, and use. Peptide-conjugated liposomes (PCL) have been developed to bind, eg, the folate receptor, which is overexpressed by tumor cells,6,12,13 and to enter the central nervous system by exploiting receptor-mediated transcytosis using ligands, which aid the passage of liposomes through the blood–brain barrier.14,15 Peptide ligands may additionally improve tumor penetration and decrease clearance of nanoparticles by mononuclear phagocytes.16,17
Studies aiming at specific stimulation of tyrosine kinase receptors without the activation of p75 have led to the discovery of short high-affinity tyrosine kinase-binding peptides.18 Here we describe the design and preparation of liposomes with covalently coupled peptides targeting TrkB-bearing cells.
Materials and methods
Materials
DSPE-polyethylene glycol (PEG)-2000, DSPE-PEG (2000) maleimide, egg phosphatidylcholine, and sphingosine were obtained from Avanti Polar Lipids ( Alabaster, AL). High- performance liquid chromatography grade trifluoroacetic acid was from Fluka (Buchs, Switzerland) and acetonitrile was from Rathburn (Walker Burn, Scotland, UK). HEPES and ethylenediamine tetra-acetic acid were from Sigma-Aldrich (St Louis, MO) and DPPRho was from Molecular Probes (Eugene, OR). The purity of lipids was checked by thin-layer chromatography on silicic acid-coated plates (Merck, Darmstadt, Germany) developed with a chloroform/methanol/water mixture (65:25:4, v/v/v). Examination of the plates after iodine staining,19 and when appropriate, upon ultraviolet illumination, revealed no impurities. Lipid concentrations were determined gravimetrically using a high precision microbalance (SuperG; Kibron Inc, Espoo, Finland). The concentration of DPPRho was determined spectrophotometrically using a molar absorptivity (ɛ465) = 19,000 M−1 cm−1 (in C2H5OH). Other chemicals were of analytical grade and from standard sources.
Peptide design and synthesis
Peptides were designed based on published TrkB-binding sequences (Table 1). In brief, peptides A366, A367, A368, A369, and A370 were as described by Ma et al,20 except for covalently coupled fluorescein. The above peptides were synthesized from the C-terminus while coupled via a hydrazine bridge to an activated resin (Storkbio, Tallinn, Estonia). After cleavage and before deprotection of the N-hydroxysuccinimide ester, the peptides were reacted with fluorescein isothiocyanate (see Figure 1 for a detailed chemical structure).
Table 1.
Sequences of peptides used in the present study
| Peptide code | Peptide sequence |
|---|---|
| A366 | CSMAHPYFAR-hb-fluorescein |
| A367 | CRALIVFTPT-hb-fluorescein |
| A368 | CRANIGGTHA-hb-fluorescein |
| A369 | CRTMLLALLF-hb-fluorescein |
| A370 | CSPGSIHTLV-hb-fluorescein |
| A371 (TrkB targeting) | CTFVKALTMDGKQAAWR |
| Scr-A371 (scrambled control for A371) | CWVLTGFTADRAKQMKA |
| A415 (TrkB targeting) | CENLYFQSGSMAHPYFARa |
| Scr-A415 (scrambled control for A415) | CENLYFQSGAYHMSAPFRa |
| A417 (retroinverse sequence of A415) | CENLYFQSGRAFYPHAMSa |
Note:
Peptides have identical amino acid compositions, as well as identical N-terminal sequence CENLYFQSG, which contains the TeV protease cleavage site (between Q and S).
Abbreviations: hb, hydrazine bridge; TrkB, tyrosine kinase B.
Figure 1.
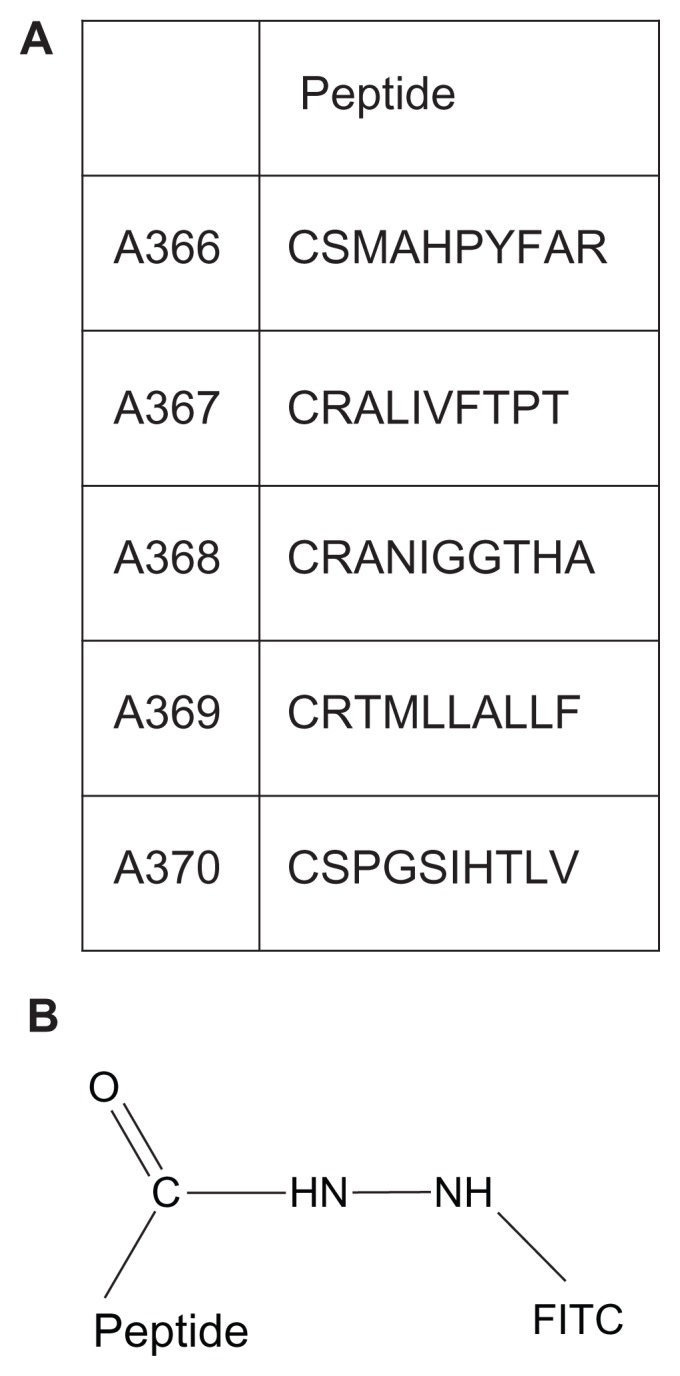
(A) Amino acid sequences of peptides used and conjugated with FITC. (B) FITC is conjugated at the C-termini of the peptides via a hydrazine bridge.
Abbreviation: FITC, fluorescein isothiocyanate.
The other peptides were: A371, corresponding to the neurotrophin sequence 84–100 TFVKALTMDGKQAAWR, which has been shown to bind to TrkB;21 Scr-A371, with the same amino acid composition as A371, but in a random order; A415, containing the TrkB-binding sequence of A366, with a TeV protease cleavage site (CENLYFQSG, cleavage between Q and S) added to the N-terminus; Scr-A415, a scrambled version of A415 with the TrkB-binding motif in a random order; and A417, the retroinverse version of A415 with the binding motif composed of D-amino acids in a reverse order.
Except for Scr-A371, which was made by t-Boc chemistry, the peptides were synthesized using fluorenylmethyloxycarbonyl chloride chemistry. Their purities (>90%) were verified by high-performance liquid chromatography and the sequences were confirmed by mass spectrometry (Storkbio).
Cell cultures
RAW264 and K562 cells were obtained from a local cryo-depository and descend from original cells purchased from the American Tissue Culture Collection (clones TIB-71™ and CCL-243™, respectively; ATCC, Manassas, VA), each with fewer than five passages. K562 cells are TrkB-negative but positive for the closely related TrkA receptor.22 The cells were maintained in Iscove’s medium supplemented with 5% fetal calf serum (Sigma-Aldrich) without other nutritional supplements or antibiotics. RAW264 cells were maintained at a confluency lower than 50% and K562 cells at a density up to 5 × 105 cells per mL, both in a humidified atmosphere with 5% CO2.
SH-SY5Y cells are the third successive subclone of the SKN-SH human neuroblastoma cell line and were purchased from the Health Protection Agency Culture Collection (Salisbury, UK). When treated with all-trans-retinoic acid (ATRA), the SH-SY5Y cells develop a distinct phenotype with morphological changes and expression of biochemical and functional markers resembling those of neurons.23 These cells are frequently used as a model for studying the molecular mechanisms involved in neuronal differentiation. Treatment of SH-SY5Y with ATRA also induces the expression of TrkB (NTRK2), but not of TrkA (NTRK1), and thus becomes responsive to brain-derived neurotrophic factor and NT-4/5 neurotrophins.23 SH-SY5Y cells were grown in Dulbecco’s modified Eagle’s medium/F12 1:1 medium (PAA, Linz, Austria) supplemented with 2 mM L-glutamine, penicillin 20 U/mL, streptomycin 20 mg/mL, and 15% (by volume) heat-inactivated fetal calf serum (PAA). The cells were maintained at 37°C in a high humidity atmosphere of 95% air and 5% CO2 and were seeded at an initial density of 104 cells/cm2 in culture dishes (Unilab, Innsbruck, Austria). On the day of plating, ATRA (Sigma, Vienna, Austria) was added at a final concentration of 10 μM in Dulbecco’s modified Eagle’s medium/F12 with 15% fetal calf serum, and after that, daily for 5 days in this medium. This resulted in a considerable proportion of cells with a neuronal phenotype and extending neuritic processes, and were utilized for studies of peptide-mediated binding of liposomes.
Analysis of TrkB gene expression in SH-SY5Y cells
Total RNA was isolated from untreated and ATRA-treated SH-SY5Y cells (n = 3), as described.21 Reverse transcription was completed using the iScript cDNA Synthesis Kit (Bio- Rad, Munich, Germany). Real-time quantitative polymerase chain reaction was performed using iQ Sybr Green Supermix (Bio-Rad) in a MyiQ cycler (Bio-Rad). Human TrkB primers were designed using the program Primer Blast (National Library of Medicine, National Institutes of Health, Bethesda, MD) and were: CAA TTG TGG TTT GCC ATC TG (forward) and ACA GTG AAT GGA ATG CAC CA (reverse).
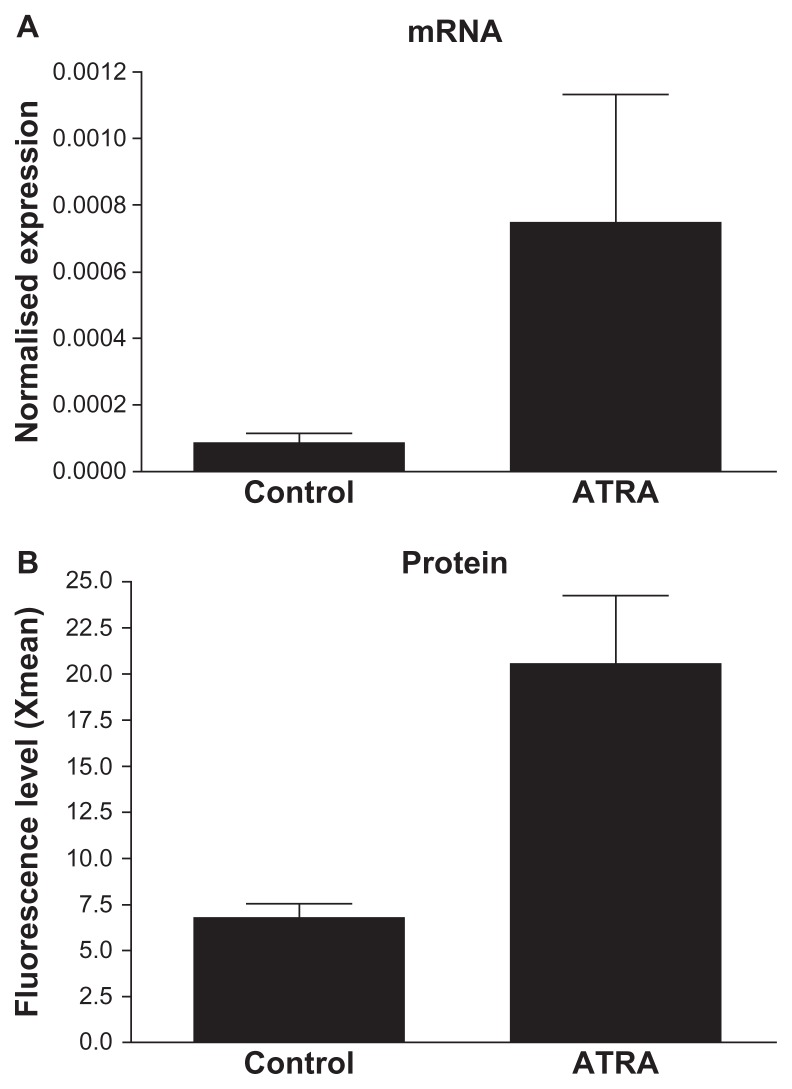
The product length was 345 base pairs and the primers bound to transcript variants a–e did not amplify from genomic DNA but only from cDNA (the amplified region jumped over intron). 18S rRNA was used as an endogenous control.21 The relative gene expression levels were calculated by the threshold (quantification) cycle as described21 (Figure 2).
Figure 2.
Induction of TrkB expression in SH-SY5Y cells by ATRA-treatment. (A) In SH-SY5Y cells, TrkB was induced by 5 days ATRA-treatment at mRNA level shown by real-time PCR. (B) Its induction and presence on the cell surface was detected at protein level by flow cytometry. Xmean (y-axis) refers to the average detected FITC-fluorescence intensity, based on immunocytochemical reaction and done by using anti-TrKB polyclonal antibody and anti-rabbit Alexa 488-labeled secondary antibody.
Abbreviations: ATRA, all-trans-retiuoic acid; mRNA, messenger RNA; TrKB, tyrosine kinase B; PCR, polymerase chain reaction.
Detection of TrkB in SH-SY5Y cells
Control cells (106) and SH-SY5Y cells treated with ATRA for 5 days23 were prepared for flow cytometry as described previously. 24 Rabbit polyclonal anti-TrkB raised against an epitope corresponding to the amino acids 160–340 mapping within the extracellular domain of human TrkB (SC-8316; Santa Cruz Biotechnology, Santa Cruz, CA) and Alexa 488-labeled antirabbit IgG (Invitrogen, Darmstadt, Germany) were used as the primary and secondary antibody, respectively. Controls were either without antibodies for autofluorescence, with secondary antibody only, or with control rabbit isotype IgG (SC-2027, Santa Cruz). Fluorescence intensities (Coulter Epics XL-MCL, Beckman Coulter, Vienna, Austria) measured for autofluorescence and for the controls were of similar magnitude, while the fluorescence for the TrkB binding antibody was significantly higher.
Binding and uptake of liposomes by SH-SY5Y cells
SH-SY5Y cells were plated in Ibidi dishes (Gerasdorf, Austria, 1 mL cell suspension per dish; 105 cells/mL) and treated for 5 days with ATRA. Subsequently, DPPRho-labeled liposomes (2 μM total final lipid concentration) were added and incubated for up to 24 hours. Cells were then fixed for 20 minutes in freshly prepared 2% formalin solution, followed by DAPI staining, and were observed by confocal microscopy.21
After paraformaldehyde fixation and one phosphate-buffered solution wash, the Ibidi dishes were incubated at room temperature with 50 mM NH4Cl followed by incubation in 0.1% bovine serum albumin in phosphate-buffered solution. After washing in phosphate-buffered solution, the culture dishes were incubated with a mouse anti-early endosomal autoantigene-1 antibody (Catalog number 610457; BD Transduction Laboratories, Austria, Vienna) at a 1:200 dilution in 0.05% saponin-phosphate-buffered solution for at least 1 hour at room temperature. After five 5-minute washes with phosphate-buffered solution, Alexa 488-labeled secondary antimouse IgG antibody (Invitrogen, Karlsruhe, Germany) was added under the same conditions as the primary antibody.25 After five further 5-minute washes with phosphate-buffered solution, the cells were stained with DAPI and covered with Mowiol® (Hoeschst, Frankfurt, Germany).25
Peptide binding and specificity
Testing of the specificities and affinities of the targeting peptides was performed using RAW264 (expressing the TrkB receptor) and K562 cells (TrkB-negative but positive for the closely related TrkA receptor22). Unless otherwise indicated, all peptide-binding experiments were conducted in 5 mM HEPES, 135 mM NaCl, and at pH 7.4. Fluorescein- labeled A366, A367, A368, A369, and A370 peptides were added to a suspension of RAW264 or K562 cells at a concentration of 0.1 μg per 106 cells, incubated for 20 minutes at room temperature in the above buffer, washed four times with phosphate- buffered solution, fixed with 4% paraformaldehyde in phosphate-buffered solution, and then measured using a flow cytometer (FACSAria; Becton Dickinson, Franklin Lakes, NJ). For specificity tests, some of the above-stained cells were incubated with mouse monoclonal antibodies against NTRK2 (clone 4D3 F10, raised against a full length recombinant NTRK2 with a glutathione S-transferase tag; Abnova, Taiwan) for 1 hour at 4°C and pH 7.4 in 5 mM HEPES, 135 mM NaCl, and 5% bovine serum albumin. After three washes with cold phosphate-buffered solution, the cells were stained with Alexa Fluor 633-labeled goat antimouse IgG (Invitrogen) for fluorescence microscopy and with phycoerythrin (Sigma-Aldrich) after a 30-minute incubation at room temperature using the above buffer with bovine serum albumin prior to FACS analysis. Cells were triple-washed with cold phosphate-buffered solution, attached to glass coverslips with a Cytospin (Shandon; Thermo Scientific, Waltham, MA) centrifuge and observed using a confocal microscope (LSM-510; Zeiss, Thornwood, NY) with illumination at 488 nm (R-phycoerythrin) and 633 nm (Alexa Fluor 633) lasers and detection at 520 nm with a bandpass filter and at 650 nm with a longpass filter, respectively.
Kinetics of binding was assessed in serum free Iscove’s medium with cell suspension (107 cell per mL), a constant rotation of 300 rpm, and a constant temperature of 37°C. Aliquots of liposomes were injected into the reaction tube, measurements were started immediately by cytofluorimetry (FACSAria), and the results were calculated by the FCSExpress3 software (DeNovo Software, Los Angeles, CA).
Coupling of peptides to DSPE-PEG (2000) maleimide
DSPE–PEG (2000) maleimide and the indicated peptides were incubated at a 1:1.3 molar ratio in a reaction mixture of 100 mM HEPES at pH 7.0 and methanol (1:1.11 volume ratio), for 3 hours at room temperature with constant stirring. The lipid–peptide conjugates were purified by high-performance liquid chromatography on a reverse phase column (Jupiter 5 μM C4 300A ST 4.6/150; Amersham Biosciences, Uppsala, Sweden) eluted with a linear gradient from 0% to 100% acetonitrile in 0.1% trifluoroacetic acid at a flow rate of 1 mL per minute. Fractions corresponding to the peptide-lipid conjugate were collected and lyophilized. The quantities of lipid–peptide conjugates were calculated by measuring the area under the peaks (observed in high-performance liquid chromatography) with the computer software provided by the instrument manufacturer. The lipid–peptide conjugates were dissolved in methanol for use in subsequent experiments.
Incorporation of peptide-PEG-lipid conjugates into liposomes
Appropriate amounts of the lipid stock solutions and the indicated lipid–peptide conjugate were mixed to obtain the desired compositions. In brief, lipid films composed of sphingosine, egg phosphatidylcholine, DSPE–PEG-2000, peptide–PEG–lipid conjugate, and DPPRho (0.5:0.44:0.02: 0.01:0.03 molar ratio) were prepared. The solvent was removed under a stream of nitrogen and the lipid residues were subsequently maintained under reduced pressure for at least 2 hours. The dry lipid film was then hydrated at 60°C for 1 hour in 5 mM HEPES and 0.1 mM ethylenediaminetetraacetic acid, at pH 7.4. The lipid dispersion at a final concentration of 0.1 mM was subjected for 5 minutes to adaptive focused ultrasound (Covaris S2; KBiosciences, Herts, UK) using instrument settings of duty cycle 20, intensity 10, and 500 cycles per burst.26
Dynamic light scattering
The apparent hydrodynamic particle diameter (Zav) and poly-dispersity index of the liposomes (Table 2) were determined by dynamic light scattering at 25°C (Zetasizer Nano ZS; Malvern Instruments Ltd, Malvern, UK). The instrument uses photon correlation spectroscopy at a scattering angle of 173 degrees to evaluate Zav from the diffusion coefficient (D) using Stokes–Einstein equation dZ = kT/3πηD, where k is the Boltzmann constant, T is the absolute temperature, and η is the viscosity of the solvent. The Zav analysis yields a mean value for size and a width parameter known as the polydispersity index. Zav is also known as the cumulants mean. The cumulants analysis is the fit of a polynomial to the log of the G1 correlation function
Table 2.
Z-average diameters and polydispersity of control liposomes and peptide-conjugated liposomes
| Preparation | Size (nm) | PDI |
|---|---|---|
| Control liposomes (no peptide) | 90 ± 5 | 0.220 ± 0.010 |
| A371-PCL | 110 ± 10 | 0.170 ± 0.015 |
| Scr-A371-PCL | 200 ± 100 | 0.270 ± 0.020 |
| A415-PCL | 91 ± 10 | 0.420 ± 0.050 |
| Scr-A415-PCL | 98 ± 20 | 0.264 ± 0.020 |
| A417-PCL | 82 ± 20 | 0.373 ± 0.20 |
Note: Mean particle diameters (Zav) both with and without the targeting peptide were determined by dynamic light scattering and reveal the liposomes to be monodispersed, with narrow particle size distributions.
Abbreviation: PDI, polydispersity index.
The coefficient of the squared term c, when scaled as 2c/b2 represents the polydispersity index.
Results
Binding of peptides to TrkB-positive cells
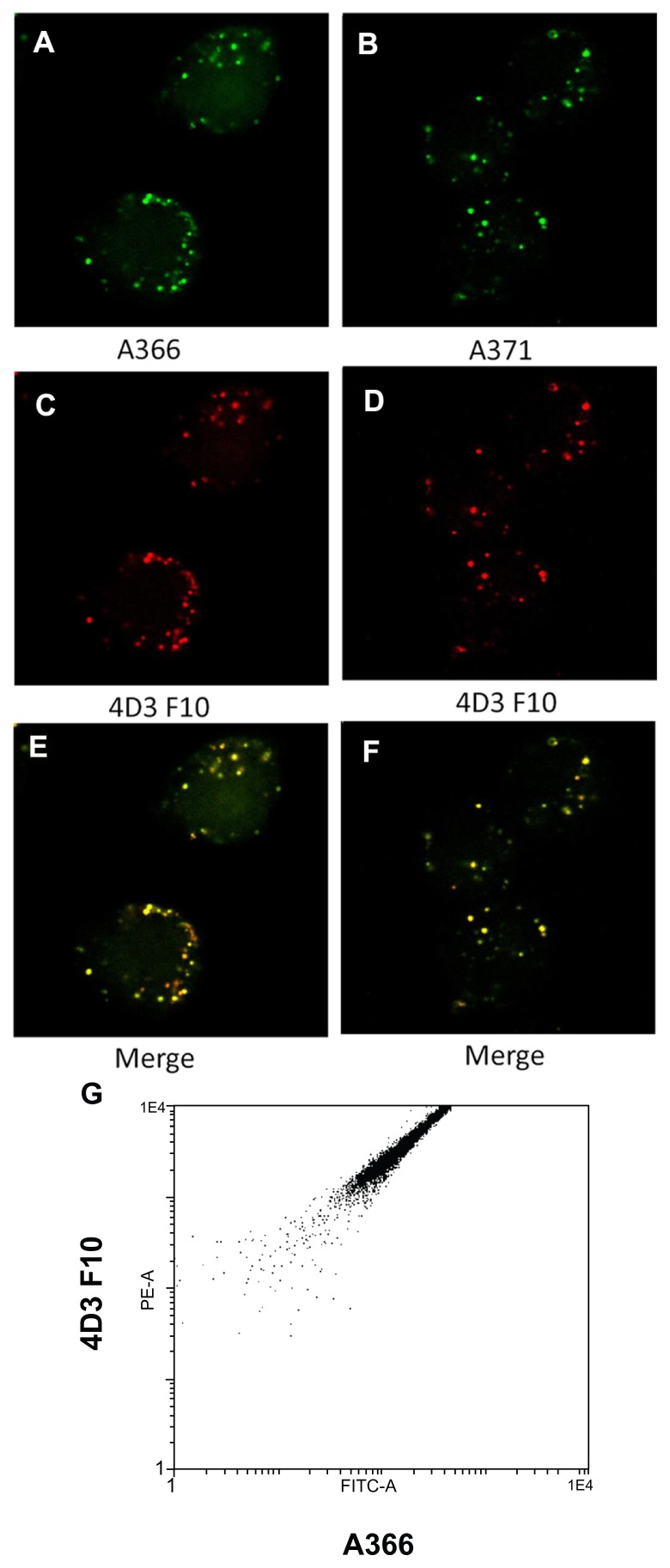
Peptides A367 and A369 were water-insoluble and after initial trials were excluded from further analysis. All the peptides tested showed more binding to RAW264 cells than to K562 cells (Table 3). When the RAW264/K562 ratio for the binding of the peptides was used to indicate specificity for the TrkB versus TrkA receptor, A366 had the highest score (Table 3). The peptides are expected to have a lower selectivity compared with the antibodies. Accordingly, it was important to compare binding of the peptides with binding of TrkB-specific antibodies. Both A366 and A371 showed good colocalization with the commercially available fluorescently-labeled monoclonal antibody, 4D3F10 (Figure 3A–F). Accordingly, these peptides were selected for further analysis. The above results were confirmed quantitatively by flow cytometry, revealing a good correlation between the signals from labeled antibodies and A366 (Figure 3G).
Table 3.
Binding of designed peptides to TrkB+ and TrkA+ cells, with relative specificities given as the TrkB+/TrkA+ ratio
| RAW264 | K562 | RAW264/K562 ratio | |
|---|---|---|---|
| Autofluorescence | 11.28 ± 64.37 | 18.67 ± 66.71 | |
| A366 | 1296.03 ± 45.88 | 403.02 ± 48.44 | 3.22 |
| A368 | 1217.71 ± 43.06 | 626.78 ± 46.78 | 1.94 |
| A370 | 616.14 ± 80.27 | 81.78 ± 45.55 | 1.06 |
| A371 | 486.27 ± 130.25 | 265.88 ± 96.1 | 1.83 |
Note: The numbers relate to mean value for fluorescence intensity ± coefficient of variation and are given in the customary arbitrary fluorescence units (au).
Abbreviation: Trk, tyrosine kinase.
Figure 3.
(A–F) Colocalization of fluorescein-labeled TrkB binding peptides (A366 and A371, green) and Alexa 633-labeled monoclonal anti-TrkB antibodies (red) to RAW264 cells. (G) Two-dimensional FACS plot made with similarly stained RAW264 cells.
Abbreviations: FITC-A, fluorescein isothiocyanate-labeled A366 peptide; PE-A, phycoerythrin-conjugated AD3F10 antibody; TrkB, tyrosine kinase B.
Binding and internalization of PCL by cultured cells
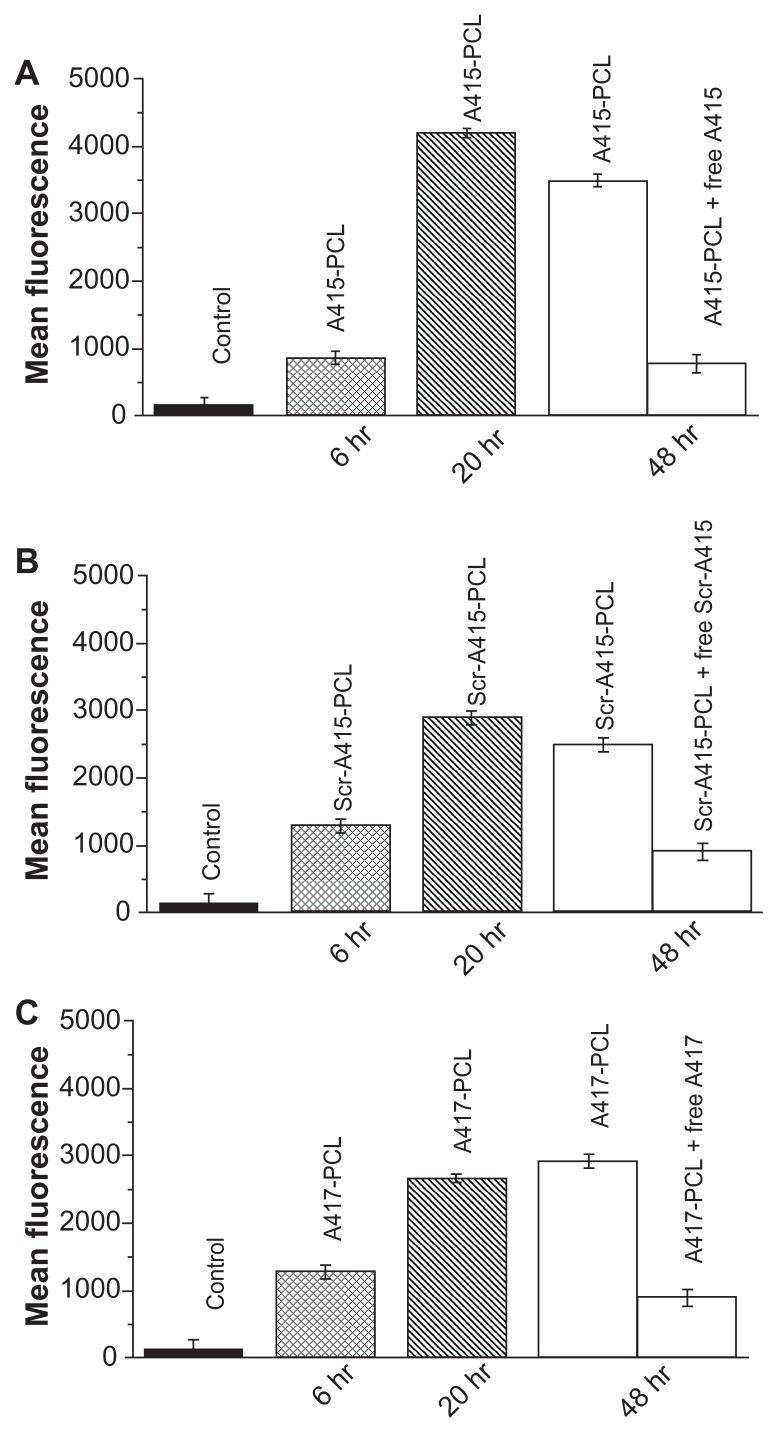
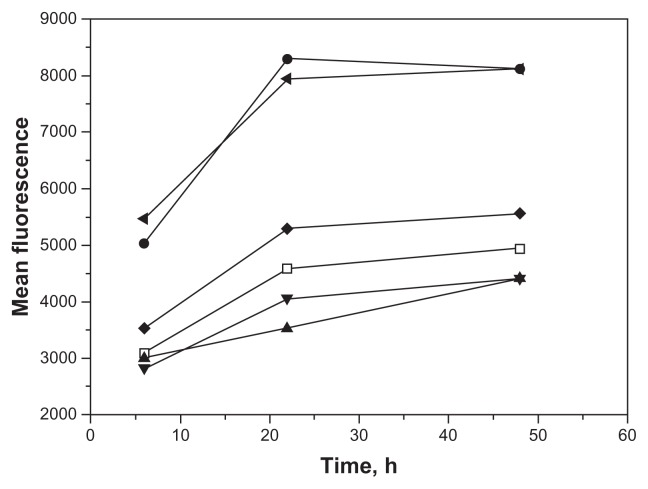
Peptides A415, Scr-A415, and A417 were developed from peptide C1 (CSMAHPYFARC), which was obtained from phage display of a custom library against NIH 3T3 cells expressing the full-length rat TrkB receptor.20 A415 (CENLYFQSGSMAHPYFAR) contained the cysteine-truncated binding sequence of C1, with a cleavage site for TeV protease added to the N-terminus (CENLYFQSG, cleavage between Q and S). The addition of the TeV cleavage site and conjugation to liposomes may perturb the receptor binding properties of this peptide. To check for possible interference, liposomes with peptides A415, Scr-A415, and A417 covalently coupled to the PEG-lipid were studied for their binding to RAW264 and K562 cells. Liposomes with A415 and A417 showed specificity similar to that of the original peptide A366 (Figure 4). To distinguish between binding and internalization, 100E per mL of TeV protease (Invitrogen) was added to the cells during incubation with PCL at different time points (0 minutes, TeV protease added at the same time as liposomes). PCL bound to the cell surface via an interaction between the peptide and TrkB receptors could be released by TeV protease, cleaving the peptide off from the liposome. However, once PCL were internalized, treatment with TeV no longer influenced the fluorescence signal from the cells. Our data suggest a critical time for the internalization of the A415-bearing liposomes into the cells to be between 1 and 6 hours (Figure 5). PCL were tested for toxicity using standard toxicological assays, and were found to be nontoxic, even at concentrations higher than those used in the present study (unpublished data).
Figure 4.
(A) Binding of A415-PCL to TrkB positive cells (6 or 20 hours). Binding of A415-PCL to TrkB-positive cells in the presence (1 mM) or absence of competing free peptides (48 hours). (B) Binding of Scr-A415-PCL to TrkB-positive cells (6 or 20 hours). Binding of Scr-A415 PCL to TrkB-positive cells in the presence (1 mM) or absence of competing free peptides (48 hours). (C) Binding of A417-PCL to TrkB-positive cells (6 or 20 hours). Binding of A417-PCL to TrkB-positive cells in the presence (1 mM) or absence of competing free peptides (48 hours).
Notes: Control depicts the level of background fluorescence of the cells.
Abbreviations: PCL, peptide-conjugated liposomes; TrkB, tyrosine kinase B.
Figure 5.
Early cleavage of the targeting peptide reduces internalization of PCL.
Notes: Fluorescent A415 PCL were treated with TeV protease at 0, 20 minutes, and one and 6 hours after their addition to the cell cultures and fluorescence intensities were measured at 6, 22, and 48 hours after the addition of PCL. Control liposomes without peptide (□), A415-PCL (●), A415-PCL+TeV (0 minutes, ▲), A415-PCL+TeV (20 minutes, ▼), A415-PCL+TeV (one hour, ◆), and A415-PCL+TeV (6 hours ◀).
Abbreviation: PCL, peptide-conjugated liposomes.
Kinetics of PCL binding to cells
To understand better the relationship between binding and internalization of PCL, we analyzed PCL binding in real time by FACS. A415 and Scr-A415 harboring PCL were added to a suspension of RAW264 cells and measurements were taken continuously over a 20-minute period. The results reveal significantly faster kinetics of binding for the liposomes with the covalently coupled targeting A415 peptide compared with the scrambled Scr-A415 peptide (Figure 6).
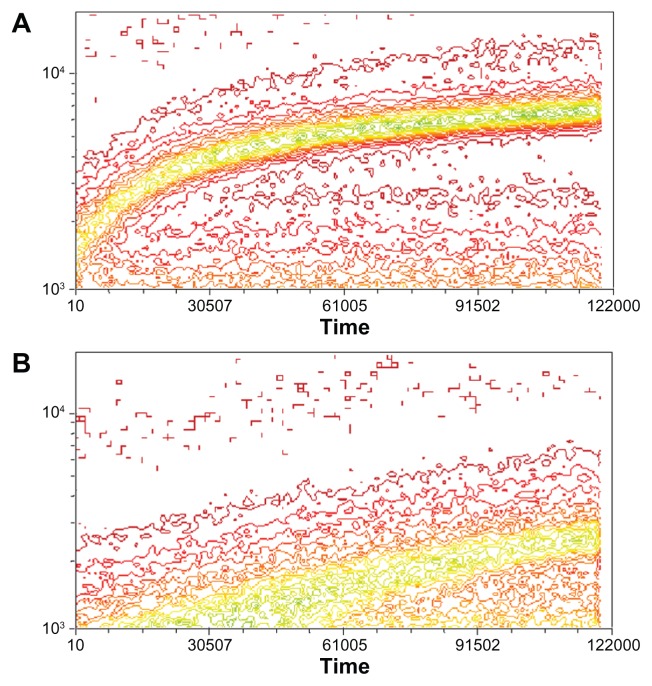
Figure 6.
Kinetics of binding of PCL with fluorescein-labeled A415 (A) or the scrambled peptide Scr-A415 (B) added into a suspension of RAW264 cells with constant rotation.
Notes: Fluorescence of individual cells was monitored for 20 minutes by FACS and is illustrated as a contour plot, with time in seconds (×100) on horizontal axis and number of binding events on the vertical axis, with colors representing relative density of cells in each location within the plot showing increasing frequency from red to green. The data show significantly faster binding kinetics for the liposomes with the covalently coupled targeting peptide A415 compared with the scrambled peptide.
Abbreviation: PCL, peptide-conjugated liposomes.
TrkB expression induced in SH-SH5Y cells
The SH-SY5Y neuroblastoma cell line was used as a rapid and simple procedure to obtain nearly pure populations of human neuron-like cells.23 Using a combination of immunoprecipitation and Western blotting, Encinas et al23 reported that significant levels of functional TrkB became detectable on the cell surface after 3 days of treatment with ATRA. The levels of TrkB were maximal at day 5 of treatment, and decreased slightly during the subsequent days. We evaluated the quantitative characteristics of cells derived from the protocol developed by Encinas et al23 and assessed TrkB mRNA and protein expression after 5 days. Both mRNA and cell surface expression of TrkB protein (analyzed in unfixed cells by FACS) was induced by ATRA (Figure 2). Expression of the TrkB+ gene was significantly induced (P < 0.001) by treatment with ATRA. Our quantitative results thus confirm the findings of Encinas et al,23 validating SH-SY5Y cells as a neuronal cell culture model for TrkB+ cells. In our hands, TrkB expression was comparable with that described by Encinas et al23 and Nishida et al.27
Uptake of liposomes by TrkB+ SH-SY5Y cells
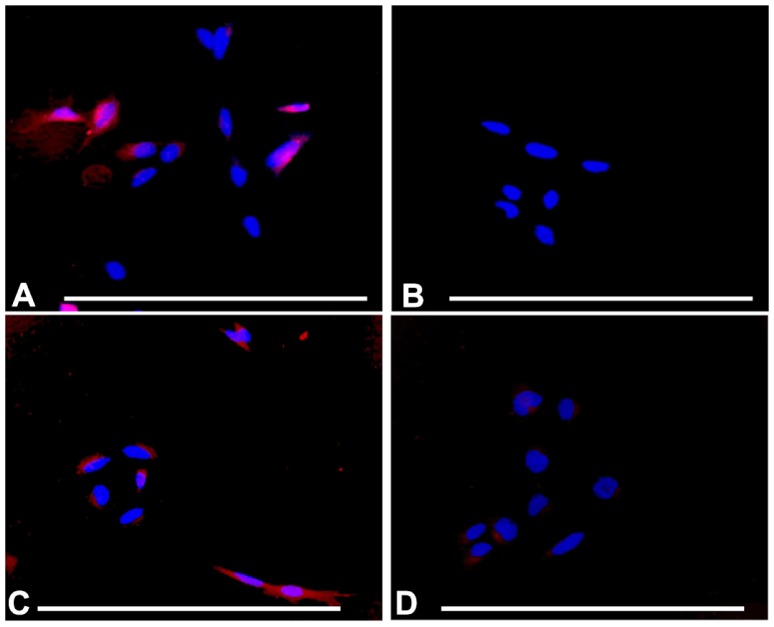
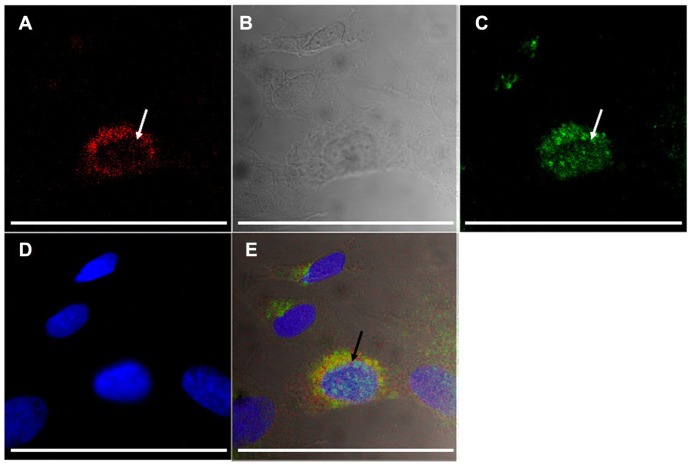
In order to assess further the specificity of PCL, DPPRho-labeled PCL were added to ATRA-SH-SY5Y cells and incubated for a period from 20 minutes to 4 hours. Selective uptake of A415-PCL was evident by ATRA-treated SH- SY5Y cells, showing cytoplasmic localization upon a short incubation (20 minutes) at 2 μM total lipid concentration. Scr-A415-PCL were used as a control for the A415-PCL. At later time points (4 hours), Scr-A415-PCL were also detected in cells (Figure 7). We further investigated the intracellular localization of A415-conjugated liposomes in TrkB+ SH-SY5Y cells using antibodies for early endosomal antigen 1 protein that binds phospholipid vesicles containing phosphatidylinositol 3-phosphate (anti-early endosomal autoantigene-1 IgG), and could detect partial colocalization of early endosomes and A415-conjugated PCL (Figure 8).
Figure 7.
Selective uptake of A415-PCL by ATRA-treated SH-SY5Y cells, the selectivity being dependent on incubation time. (A) ATRA-treated SH-SY5Y cells incubated with A415-PCL for 20 minutes. (B) ATRA-treated SH-SY5Y cells incubated with Scr-A415-PCL for 20 minutes. (C) ATRA-treated SH-SY5Y cells incubated with A415-PCL for 4 hours. (D) ATRA-treated SH-SY5Y cells incubated with Scr-A415-PCL for 4 hours.
Note: Bar corresponds to100 μm.
Abbreviations: ATRA, all-trans-retinoic acid; PCL, peptide-conjugated liposomes.
Figure 8.
Uptake of A415-PCL by ATRA-treated SH-SY5Y TrkB+ cells after 8 hours of incubation, showing partial colocalization with the anti-early endosomal 1 antibody (arrows). Fluorescence of A415-PCL-labeled with TRITC (A), differential contrast image (B), immunocytochemistry of early endosomal autoantigene-1 (fluorescein isothiocyanate-labeled, (C), nuclear counterstaining (D), and merge of panels A–D (E).
Note: Bar illustrates 100 μm.
Abbreviations: ATRA, all-trans-retinoic acid; PCL, peptide-conjugated liposomes.
Discussion
Nanoparticles offer several advantages in the delivery of therapeutic agents, including an unprecedented freedom to modify some of the most fundamental properties of therapeutic carriers, such as solubility, diffusivity, biodistribution, release characteristics, and immunogenicity. Over recent decades, different types of nanoparticles have been studied for their use in therapeutic applications. At present, perhaps the most interesting nanoparticles for clinical use are liposomes,28 which are spherical vesicles surrounded by a bilayer membrane shell composed of natural or synthetic lipids.29–31 The possibility of preparing both biocompatible and biodegradable compositions, together with encapsulation of either hydrophilic or hydrophobic therapeutic agents, makes liposomes excellent carriers. Liposomes can be coated with biocompatible and antibiofouling polymers, such as PEG, to prolong their half-life in the circulation. Precise molecular engineering has already resulted in long circulation times, good bioavailability, and low toxicity.32,33 To improve the efficacy of therapeutic nanoparticles further requires functionalizing them with targeting ligands. The latter allow delivery of drug-encapsulating nanoparticles to defined tissue sites, while having minimal undesired effects elsewhere.
Delivery of a nanoparticle cargo via TrkB targeting should affect TrkB-harboring cells in specific ways, depending on the active ingredient, eg, stem cells to boost their migration and differentiation, postmitotic neuronal cells to promote their function and survival, and neoplastic cells to devour. Given that coexistence of at least two of three groups of these cells is expected in every organism, the cargo must provide some level of specificity in its action.
In this study, we investigated if a small targeting peptide might convey selectivity to PCL for binding and cellular internalization. TrkB-targeting and scrambled control peptides, as well as comparable TrkB-expressing and TrkB-nonexpressing cells were used. We demonstrate that targeting by small 18-mer peptides coupled to liposomes ensured their relatively selective intracellular uptake. The PCL used receptor-mediated pathways and were at least partly found in endosomes (Figure 8). Uptake of PCL with scrambled nontargeting peptides also occurred; however, the kinetics were slower compared with PCL having targeting peptides. Accordingly, the peptide-based targeting of liposomes described here is not completely selective, and may require additional rounds of tuning to avoid unwanted uptake. Nevertheless, our data demonstrate augmented cellular uptake of PCL and targeting towards the TrkB receptor. Our results further suggest that nerve cell differentiation and survival could be improved by a combination of therapeutic agents (eg, neurotrophic factors) and PCL through the TrkB receptor, which could provide an efficient strategy for treatment of auditory nerve abnormality-induced hearing loss.
Targeting of TrkB+ cells, such as SH-SY5Y neuroblastoma-derived cells, using liposomes might make it possible to design more complex nanoparticles delivering, eg, siRNA and kinase inhibitors to modify chemoresistant PI3K/Akt pathway signaling in targeted TrkB+ cells, resulting in reduced side effects when compared with use of inhibitors lacking targeting. Neuroblastoma is a neural crest precursor cell-derived solid tumor in children. Patients with a poor prognosis are often resistant to chemotherapy and have tumors that express TrkB. Moreover, the growth/survival factor receptor conveys signals which stimulate mechanisms leading to chemoresistance in neuroblastoma.34
Interestingly, TrkB plays a crucial role in tumor progression, not only in neuroblastoma, but also in nonneuronal tumors.35 Neuroblastoma tumors express high levels of brain-derived neurotrophic factor, as well as neurotrophin receptors (TrkA, TrkB, and TrkC), which have been implicated as important prognostic markers, linking the biology of the tumor to patient outcome. More specifically, high expression levels of TrkA and TrkC receptors have been linked to favorable biological features and a good prognosis. Instead TrkB is expressed in aggressive malignant tumors, and several studies suggest that high levels of TrkB and activation by its ligand, brain-derived neurotrophic factor, promote tumor cell survival, proliferation, and chemoresistance. Accordingly, TrkB-targeted cytotoxic agents could provide possibilities for treating neuroblastoma and other tumors. Similar to other receptor tyrosine kinases, TrkB receptors homodimerize in response to ligand binding, and overexpression of TrkB in neuroblastoma should promote selective binding of PCL compared with cells having low expression of TrkB. Our data indicate improved liposome uptake in TrkB+ cells if targeting peptides are used, which might contribute to selection of highly TrkB-positive cells over cells expressing low levels of TrkB. TrkB receptor activation in normal cells further suggests improved survival of neurons.
While there is an interest in understanding the signaling pathways involved in receptor-mediated and nonreceptor-mediated PCL uptake and its trafficking within the neuronal cells of the inner ear, we have not studied intracellular signaling pathways in this context. In a recent study, Roy et al36 showed that a peptide mimetic of NGFβ conjugated to the surface of polymersome-embedded nanoparticles was capable of activating TrkB receptors, such that TrkB phosphorylation occurs, thus providing evidence that nanoparticles carrying TrkB-targeting peptides are capable of inducting TrkB-related signal pathways. These data also suggest that nanoparticles may offer a novel treatment strategy for neurodegenerative disorders.
Kupferman et al35 recently reported the use of an in vivo mouse model of head and neck squamous cell carcinoma, in which the downregulation of TrkB suppresses tumor growth. They found that the TrkB receptor may be a critical component in multistep tumor progression in head and neck squamous cell carcinoma, and may be an attractive target for much needed new therapies for this disease. While PCL may have potential for this purpose, the peptides used here are not selective enough, and binding of scrambled peptides to TrkB+ cells was evident at longer incubation times. Likewise, binding of targeting peptides to TrkB− cells was not negligible.
The above issues make PCL targeting for selective suppression of tumor cells by the peptide described inefficient, because unwanted cells will be targeted as well. At the same time, the same targeting conditions might be promising to support neuron survival, where unwanted target cells may be better tolerated, because support not suppression is the aim.
In neurons, TrkB has been described to convey protective cell survival signaling, depending on the ligand.37 However, administration of TrkB ligands to neurons may not result in neuroprotection, or neuronal survival, and apoptotic signaling by TrkB ligands such as brain-derived neurotrophic factor, has also been demonstrated via activation of an alternative receptor, p75NTR.38 In this context, TrkB-targeting liposomes without p75NTR-targeting should be capable of intracellular delivery of survival genes, without adverse induction of apoptosis, thus representing a promising option for neuroprotection. It is this latter scenario in which A415- PCL are improved compared with other available targeting peptide-bearing nanoparticles. The previously described A371-conjugated liposomes were unable to distinguish between p75NTR and TrkB.21 There have been several successful attempts to make peptides which are agonistic towards TrkB.39,40 Roy et al recently showed the A371 peptide to be identical to the hNgf_EE peptide, which specifically targets TrkB and p75NTR receptors.21 However, specificity for TrkB cannot be claimed. The A415 peptide is based on the A366 peptide, which was developed by Ma et al20 using TrkB expressed in NIH 3T3 cells, and a phage-displayed random peptide library. A366 antagonizes the activity of brain-derived neurotrophic factor in a dose-dependent manner, and is more TrkB-specific than A371 (which might also bind to p75). Taken together, A371 preferably binds to TrkB and also p75 receptors, while A415 is able to antagonize brain-derived neurotrophic factor, and is thus more TrkB-specific. This is why we selected A415 and not A371 for the present study. We deliberately avoided peptides having physiological activity and utilized peptides which demonstrated affinity toward this receptor.20 Moreover, we changed the sequences of the peptides to ensure easy and unambiguous conjugation, as well as controlled release of the peptides from the surface of the PCL. Internalization and potential targetability of A371-functionalized liposome nanoparticles were observed in primary cochlear cell cultures, cochlear explants, and adult rat cochlea, as reported in our previous publication.41 We are continuing studies of the binding of other peptide-coated liposomes to auditory nerve cells (data to be published).
Our data demonstrate that structural modifications did not hamper the TrkB-binding activity of the peptides, allowing PCL to deliver cargo to different TrkB+ cell types. Depending on their cargo, these PCL could provide an efficient means either for survival treatment of neuronal tissues or for proapoptotic treatment of neoplasms, conveying either antiapoptotic or proapoptotic agents, respectively.
Acknowledgments
This work was supported by the integrated NANOEAR European Union FP6 project (NMP4-CT-2006-026556). SR was also supported by the Magnus Ehrnrooth Foundation, and RS was supported by the Instrumentarium Foundation. The authors thank past and current members of the NANOEAR consortium who contributed to our discussions on targeting peptides and the assay conditions used in the present study, and the skilled laboratory personnel of the Pyykkö Laboratory for assistance with the cell experiments.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Schimmang T, Tan J, Muller M, et al. Lack of Bdnf and TrkB signalling in the postnatal cochlea leads to a spatial reshaping of innervation along the tonotopic axis and hearing loss. Development. 2003;130:4741–4750. doi: 10.1242/dev.00676. [DOI] [PubMed] [Google Scholar]
- 2.Chikar JA, Colesa DJ, Swiderski DL, et al. Over-expression of BDNF by adenovirus with concurrent electrical stimulation improves cochlear implant thresholds and survival of auditory neurons. Hear Res. 2008;245:24–34. doi: 10.1016/j.heares.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabizadeh S, Oh J, Zhong LT, et al. Induction of apoptosis by the low- affinity NGF receptor. Science. 1993;261:345–348. doi: 10.1126/science.8332899. [DOI] [PubMed] [Google Scholar]
- 4.Friedman WJ. Neurotrophins induce death of hippocampal neurons via the p75 receptor. J Neurosci. 2000;20:6340–6346. doi: 10.1523/JNEUROSCI.20-17-06340.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geiger TR, Peeper DS. Critical role for TrkB kinase function in anoikis suppression, tumorigenesis, and metastasis. Cancer Res. 2007;67:6221–6229. doi: 10.1158/0008-5472.CAN-07-0121. [DOI] [PubMed] [Google Scholar]
- 6.Mozafari MR, Pardakhty A, Azarmi S, et al. Role of nanocarrier systems in cancer nanotherapy. J Liposome Res. 2009;19:310–321. doi: 10.3109/08982100902913204. [DOI] [PubMed] [Google Scholar]
- 7.Sajja HK, East MP, Mao H, et al. Development of multifunctional nanoparticles for targeted drug delivery and noninvasive imaging of therapeutic effect. Curr Drug Discov Technol. 2009;6:43–51. doi: 10.2174/157016309787581066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allen TM, Cullis PR. Drug delivery systems: entering the mainstream. Science. 2004;303:1818–1822. doi: 10.1126/science.1095833. [DOI] [PubMed] [Google Scholar]
- 9.Cho K, Wang X, Nie S, et al. Therapeutic nanoparticles for drug delivery in cancer. Clin Cancer Res. 2008;14:1310–1316. doi: 10.1158/1078-0432.CCR-07-1441. [DOI] [PubMed] [Google Scholar]
- 10.Hood JD, Bednarski M, Frausto R, et al. Tumor regression by targeted gene delivery to the neovasculature. Science. 2002;296:2404–2407. doi: 10.1126/science.1070200. [DOI] [PubMed] [Google Scholar]
- 11.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002;2:750–763. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 12.Lee TY, Lin CT, Kuo SY, et al. Peptide-mediated targeting to tumor blood vessels of lung cancer for drug delivery. Cancer Res. 2007;67:10958–10965. doi: 10.1158/0008-5472.CAN-07-2233. [DOI] [PubMed] [Google Scholar]
- 13.Carlsson J, Kullberg EB, Capala J, et al. Ligand liposomes and boron neutron capture therapy. J Neurooncol. 2003;62:47–59. doi: 10.1007/BF02699933. [DOI] [PubMed] [Google Scholar]
- 14.Newton HB. Advances in strategies to improve drug delivery to brain tumors. Expert Rev Neurother. 2006;6:1495–1509. doi: 10.1586/14737175.6.10.1495. [DOI] [PubMed] [Google Scholar]
- 15.Gutman RL, Peacock G, Lu DR. Targeted drug delivery for brain cancer treatment. J Control Release. 2000;65:31–41. doi: 10.1016/s0168-3659(99)00229-1. [DOI] [PubMed] [Google Scholar]
- 16.Medina OP, Kairemo K, Valtanen H, et al. Radionuclide imaging of tumor xenografts in mice using a gelatinase-targeting peptide. Anticancer Res. 2005;25:33–42. [PubMed] [Google Scholar]
- 17.Mori T. Cancer-specific ligands identified from screening of peptide- display libraries. Curr Pharm Des. 2004;10:2335–2343. doi: 10.2174/1381612043383944. [DOI] [PubMed] [Google Scholar]
- 18.Longo FM, Yang T, Knowles JK, et al. Small molecule neurotrophin receptor ligands: novel strategies for targeting Alzheimer’s disease mechanisms. Curr Alzheimer Res. 2007;4:503–506. doi: 10.2174/156720507783018316. [DOI] [PubMed] [Google Scholar]
- 19.Palumbo G, Zullo F. The use of iodine staining for the quantitative analysis of lipids separated by thin layer chromatography. Lipids. 1987;22:201–205. doi: 10.1007/BF02537303. [DOI] [PubMed] [Google Scholar]
- 20.Ma Z, Wu X, Cao M, et al. Selection of trkB-binding peptides from a phage-displayed random peptide library. Sci China C Life Sci. 2003;46:77–86. doi: 10.1007/BF03182687. [DOI] [PubMed] [Google Scholar]
- 21.Roy S, Johnston AH, Newman TA, et al. Cell-specific targeting in the mouse inner ear using nanoparticles conjugated with a neurotrophin-derived peptide ligand: potential tool for drug delivery. Int J Pharm. 2010;390:214–224. doi: 10.1016/j.ijpharm.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Suarez O, Hannestad J, Esteban I, et al. Expression of the TrkB neurotrophin receptor by thymic macrophages. Immunology. 1998;94:235–241. doi: 10.1046/j.1365-2567.1998.00505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Encinas M, Iglesias M, Liu Y, et al. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem. 2000;75:991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 24.Dudas J, Mansuroglu T, Batusic D, et al. Thy-1 is an in vivo and in vitro marker of liver myofibroblasts. Cell Tissue Res. 2007;329:503–514. doi: 10.1007/s00441-007-0437-z. [DOI] [PubMed] [Google Scholar]
- 25.Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev Cell. 2002;3:803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- 26.Tejera-Garcia R, Ranjan S, Zamotin V, et al. Making unilamellar liposomes using focused ultrasound. Langmuir. 2011;27:10088–10097. doi: 10.1021/la201708x. [DOI] [PubMed] [Google Scholar]
- 27.Nishida Y, Adati N, Ozawa R, et al. Identification and classification of genes regulated by phosphatidylinositol 3-kinase- and TRKB-mediated signalling pathways during neuronal differentiation in two subtypes of the human neuroblastoma cell line SH-SY5Y. BMC Res Notes. 2008;1:95. doi: 10.1186/1756-0500-1-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang AZ, Gu F, Zhang L, et al. Biofunctionalized targeted nanoparticles for therapeutic applications. Expert Opin Biol Ther. 2008;8:1063–1070. doi: 10.1517/14712598.8.8.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Granick S. How to stabilize phospholipid liposomes (using nanoparticles) Nano Lett. 2006;6:694–698. doi: 10.1021/nl052455y. [DOI] [PubMed] [Google Scholar]
- 30.Bangham AD, Horne RW. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. J Mol Biol. 1964;8:660–668. doi: 10.1016/s0022-2836(64)80115-7. [DOI] [PubMed] [Google Scholar]
- 31.Lasic DD, Needham D. The “stealth” liposome: a prototypical biomaterial. Chem Rev. 1995;95:2601–2628. [Google Scholar]
- 32.Emerich DF, Thanos CG. Targeted nanoparticle-based drug delivery and diagnosis. J Drug Target. 2007;15:163–183. doi: 10.1080/10611860701231810. [DOI] [PubMed] [Google Scholar]
- 33.Groneberg DA, Giersig M, Welte T, et al. Nanoparticle-based diagnosis and therapy. Curr Drug Targets. 2006;7:643–648. doi: 10.2174/138945006777435245. [DOI] [PubMed] [Google Scholar]
- 34.Li Z, Thiele CJ. Targeting Akt to increase the sensitivity of neuroblastoma to chemotherapy: lessons learned from the brain-derived neurotrophic factor/TrkB signal transduction pathway. Expert Opin Ther Targets. 2007;11:1611–1621. doi: 10.1517/14728222.11.12.1611. [DOI] [PubMed] [Google Scholar]
- 35.Kupferman ME, Jiffar T, El Naggar A, et al. TrkB induces EMT and has a key role in invasion of head and neck squamous cell carcinoma. Oncogene. 2010;29:2047–2059. doi: 10.1038/onc.2009.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roy S, Johnston AH, Syed TM, et al. Activation of TrkB receptors by NGFβ mimetic peptide conjugated polymersome nanoparticles. Nanomedicine. 2012;8:271–274. doi: 10.1016/j.nano.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 37.Royo NC, LeBold D, Magge SN, et al. Neurotrophin-mediated neuroprotection of hippocampal neurons following traumatic brain injury is not associated with acute recovery of hippocampal function. Neuroscience. 2007;148:359–370. doi: 10.1016/j.neuroscience.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng HK, Teng KK, Lee R, et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J Neurosci. 2005;25:5455–5463. doi: 10.1523/JNEUROSCI.5123-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Leary PD, Hughes RA. Design of potent peptide mimetics of brain-derived neurotrophic factor. J Biol Chem. 2003;278:25738–25744. doi: 10.1074/jbc.M303209200. [DOI] [PubMed] [Google Scholar]
- 40.Fletcher JM, Hughes RA. Novel monocyclic and bicyclic loop mimetics of brain-derived neurotrophic factor. J Pept Sci. 2006;12:515–524. doi: 10.1002/psc.760. [DOI] [PubMed] [Google Scholar]
- 41.Zou J, Zhang Y, Zhang W, et al. Internalization of liposome nanoparticles functionalized with TrkB ligand in rat cochlear cell populations. Eur J Nanomed. 2009;2:7–13. [Google Scholar]